Abstract
Many isolates of the plant-parasitic nematode Meloidogyne hapla reproduce by facultative meiotic parthenogenesis. Sexual crosses can occur, but, in the absence of males, the diploid state appears to be restored by reuniting sister chromosomes of a single meiosis. We have crossed inbred strains of M. hapla that differ in DNA markers and produced hybrids and F2 lines. Here we show that heterozygous M. hapla females, upon parthenogenetic reproduction, produce progeny that segregate 1:1 for the presence or absence of dominant DNA markers, as would be expected if sister chromosomes are rejoined, rather than the 3:1 ratio typical of a Mendelian cross. Codominant markers also segregate 1:1 and heterozygotes are present at low frequency (<3%). Segregation patterns and recombinant analysis indicate that a homozygous condition is prevalent for markers flanking recombination events, suggesting that recombination occurs preferentially as four-strand exchanges at similar locations between both pairs of non-sister chromatids. With this mechanism, meiotic parthenogenesis would be expected to result in rapid genomic homozygosity. This type of high negative crossover interference coupled with positive chromatid interference has not been observed in fungal or other animal systems in which it is possible to examine the sister products of a single meiosis and may indicate that meiotic recombination in this nematode has novel features.
ROOT-knot nematodes (Meloidogyne spp.) are obligate parasites capable of feeding inside the roots of >2000 plant species and causing extensive crop losses worldwide (Sasser and Freckman 1987; Roberts 1995). Control of the damage caused by root-knot nematodes in agricultural settings often requires the use of toxic pesticides (Barker and Koenning 1998). There is considerable interest in identifying genes involved in parasitism and in determining host range to develop more environmentally friendly control strategies (Williamson and Gleason 2003). Many of the species of greatest agricultural importance reproduce solely by mitotic parthenogenesis and have various degrees of polyploidy and aneuploidy (Triantaphyllou 1985; Trudgill and Blok 2001). This mode of reproduction has frustrated attempts to characterize pathogenicity traits as classical genetic analysis is not possible. However, other root-knot nematodes, including most isolates of the widely distributed species Meloidogyne hapla, reproduce by facultative meiotic parthenogenesis (Triantaphyllou 1966). In this mode of reproduction, sexual crosses occur, but parthenogenetic progeny are also produced.
The obligately parasitic life cycle of the root-knot nematode has also been a limitation for its study (Williamson and Gleason 2003). The first of four molts occurs in the egg and the nematodes hatch as second-stage juveniles (J2s), which are the infective form. These J2 penetrate plants near the root tip and move to a feeding site in the vascular tissue where they initiate the formation of feeding cells called giant cells in the host. These cells serve as the nutrient source for the nematode, now completely embedded in the root. The nematode becomes sedentary and undergoes three more molts as it develops into a bulbous female. Its posterior eventually breaks through the root surface. Egg production begins ∼25 days after infection and continues for a few weeks during which the eggs are deposited as a gelatinous mass. One advantage of this organism for genetic analysis is that these gelatinous egg masses, each containing up to several hundred eggs from a single female, can be easily collected from roots. Males are environmentally determined after the J2 reach their feeding sites and develop only under stressful conditions such as crowding and poor nutrition (Triantaphyllou 1973). Approximately 3 weeks after infection, males become vermiform and motile and leave the root. These males are attracted to and can fertilize females that remain in the root.
The temporal order of meiosis products along the distal–proximal axis of adult gonads in M. hapla, as in Caenorhabditis elegans, has enabled cytological characterization of the progress through meiosis (Triantaphyllou 1985; Albertson et al. 1997; McCarter et al. 1999). Previous researchers have observed synaptonemal complexes and recombination nodules during the pachytene stage of prophase I of M. hapla (Goldstein and Triantaphyllou 1978). Bivalents are seen at metaphase I, and homologs appear to separate at the first meiotic division as in a typical meiosis (Triantaphyllou 1966; van der Beek et al. 1998). Cytological studies report that, if sperm are present when the oocyte passes through the spermatheca, oocyte maturation occurs to form a pronucleus and two polar bodies, and the sperm nucleus fuses with the haploid egg pronucleus to form a sexual product. In the absence of fertilization, the diploid state was reported to be restored by reuniting sister chromosomes of a single meiosis (Triantaphyllou 1966). This process differs fundamentally from the hermaphrodite selfing of the free-living nematode C. elegans in which a single organism produces both egg and sperm.
An organism for which both outcrossing and inbreeding can be utilized is desirable for genetic analysis, and M. hapla has potential to be such a system. Previous work has shown that genetic crosses are possible between strains that differed in pathogenicity on a particular host and that pathogenicity segregates in the progeny (Chen and Roberts 2003), but molecular markers were not used to monitor the crosses. Genetic crosses have been carried out with other species of plant parasitic nematodes, primarily cyst nematodes; however, the obligate outcrossing of those species was a limitation for analysis of traits in these tiny organisms (Janssen et al. 1991; Dong and Opperman 1997; Atibalentja et al. 2005). We have produced inbred strains of M. hapla by sequential transfer of single egg masses (Liu and Williamson 2006). Cytological examination showed that these strains are meiotic and have a chromosome complement of n = 16. Comparison of genomic DNA using AFLP, a DNA fingerprinting technique that reveals polymorphisms by selective PCR amplification of restriction fragments from a total digest of genomic DNA (Vos et al. 1995), showed an average of 4% of fragments to be polymorphic. For the work presented here, we selected two strains, VW8 and VW9, that differ in DNA markers and in ability to reproduce on specific plant hosts to initiate a genetic analysis.
To determine the feasibility of developing a genetic map for this important parasite, we developed a strategy to carry out a genetic cross and to monitor the cross and segregation pattern using molecular markers. Here we demonstrate that a genetic cross is possible and that both outcrossing and selfing can occur. In addition, we report a novel marker segregation pattern and present a model to explain this pattern.
MATERIALS AND METHODS
Nematode strains:
M. hapla strains VW8 and VW9 were produced by sequentially transferring single egg masses of isolates from diverse geographic locations on tomato plants (Liu and Williamson 2006). All nematode cultures were maintained on tomato cultivar VFNT cherry.
DNA markers:
AFLP markers that distinguished DNA from VW8 and VW9 were identified using the protocol of Vos et al. (1995) with minor modifications (Liu and Williamson 2006). Polymorphic fragments were isolated and sequenced as previously described (Liu and Williamson 2006). Primers were designed on the basis of these sequences and then tested for strain specificity to develop PCR-based markers. Two such markers, H1 and H2, specific for VW9 and VW8, respectively, were developed from the sequence of two AFLP markers. H1 (primers AGCGTTCAAAAAACCGTCCAT and AGGGCTATAAATATGCTGACC) and H2 (primers CAATTCACCACCTTTCA and TAAATTCCCTCGTTTTAC) were amplified using standard procedures. Amplification of 100 single J2 worms from each of strains VW8 and VW9 showed the strains to be uniform for the presence or absence of the primers.
Nematode crosses:
The strategy for producing F2 lines is diagrammed in Figure 1. VW9 males were produced after inoculating tomato plants in 1-liter cups with 20,000 J2s/plant. Four weeks post-inoculation, the tomato roots were washed and then soaked in 10% commercial bleach for 5 min and rinsed thoroughly in water. The roots were placed on Baermann funnels in a mist chamber to collect males (Barker 1985). Males were collected every other day over a period of 10 days. One week after the male-producing culture was initiated, ∼200 infective VW8 J2s were inoculated onto tomato plants to produce females. Two to three thousand males were hand picked under a dissecting scope and placed onto the tomato plant infected with VW8 over a period of 10 days. Two weeks after the final application of VW9 males, tomato roots were stained with Erioglaucine (Sigma-Aldrich, St. Louis) to allow easy visualization of the egg masses (Omwega and Roberts 1992). Egg masses were hand picked and collected separately into 1.5-ml microfuge tubes containing 0.3 ml sterile water; eggs were allowed to hatch for 24 hr. Approximately 20 J2s/egg mass were handpicked, crushed with a barbed broach (Maillefer), and together digested with proteinase K (100 ng/ml) in DNA extraction buffer at 50° for 1 hr (Williamson et al. 1997). PCR amplification was carried out using the VW9 specific marker H1.
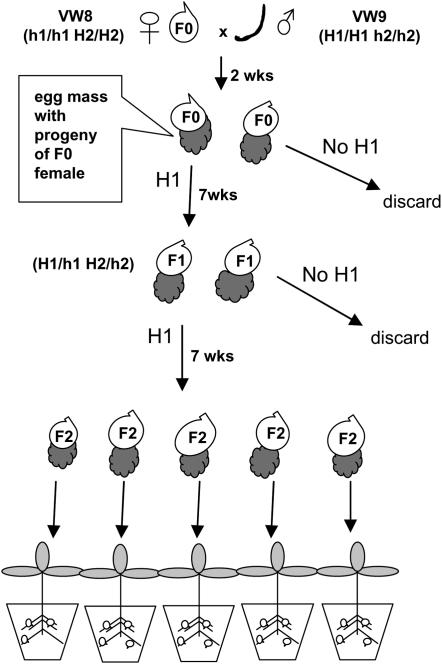
Figure 1.—
Strategy for producing F2 lines of M. hapla. A culture with young females (F0) of strain VW8 (which lacks PCR marker H1) is inoculated with males (curved line) of strain VW9 (which carries marker H1). After 2 weeks, egg masses are collected from VW8 F0 females and tested for the presence of marker H1. Juveniles from egg masses with marker H1 are inoculated onto plants and allowed to develop parthenogenetically into F1 females. Egg masses from F1 females are tested for the presence of marker H1. Eggs from F2 egg masses are inoculated onto individual plants. This figure is adapted from Williamson and Liu (2006).
The remaining J2s of each egg mass shown to be positive with male-derived marker H1 were inoculated onto tomato plants and cultured under conditions for parthenogenetic reproduction. About 7 weeks post-inoculation, egg masses that were produced by the females that developed from the J2s were picked into individual microfuge tubes and allowed to hatch for 24 hr. DNA was extracted from a fraction of J2s of each egg mass and tested for markers H1 and H2. Egg masses of females carrying both H1 and H2 were inoculated onto separate tomato plants. Egg masses that developed on the roots of these plants were picked and individually inoculated onto separate tomato plants for propagation of F2 lines. Seven weeks post-innoculation, bulk eggs were collected from each tomato plant as previously described (Branch et al. 2004). An aliquot of eggs of each F2 line was used to reinfect tomato plants and the remainder was used for marker analysis.
Marker segregation and linkage analysis in F2 lines:
The PCR amplifications to detect H1 and H2 were repeated three times for each DNA sample, and control amplifications were carried out with nonpolymorphic markers to verify that the ratios were not due to PCR failure. For each AFLP marker lane, we compared the intensity of several nonpolymorphic fragments to that of a nearby polymorphic band to assess whether lack of a band was due to amplification failure (see Figure 2C, for example). All templates were amplified with selective primers and run on sequencing gels at least two times. A homogeneity test (Weir 1990) on F2 lines indicated that it was appropriate to pool the marker data from the progeny of the three hybrid females for linkage analysis. Linkage groups were identified using JoinMap 3.0 (Van Ooijen and Voorrips 2001) with a minimum LOD value of 4.0 and verified by examination of allele segregation in individual F2 lines.
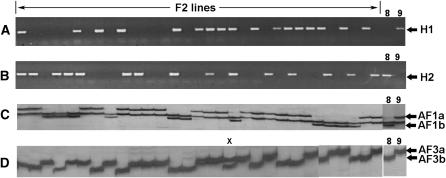
Figure 2.—
DNA marker segregation in F2 lines from hybrid females of M. hapla. For each marker, phenotypes of parental lines VW8 and VW9 are shown at the right. Lanes marked “F2 lines” show DNA from individual lines amplified with the PCR primers H1 (A) or H2 (B). Segregation pattern of allelic AFLP markers AF1a and AF1b in F2 lines is shown in C. The monomorphic band between AF1a and AF1b is a useful control for PCR amplification. (D) Segregation pattern of allelic AFLP markers AF3a and AF3b in F2 lines. The one heterozygous line is designated with an “x.”
Cytology:
Dissection of gonads and staining with Hoechst 33258 were carried out as previously described (Liu and Williamson 2006) except that slides were acid washed and coated with 1 mg/ml poly-l-lysine solution (Sigma-Aldrich) to improve adhesion of dissected material to the glass surface. Slides were stored in the dark until examination using a Nikon Microphot-SA fluorescent microscope.
RESULTS AND DISCUSSION
Marker-assisted production of F2 lines:
PCR marker H1, which is specific to the M. hapla strain VW9, was detected in 18 of 48 egg masses derived from a culture of VW8 females after applying males of VW9. The presence of marker H1 indicated that fertilization by VW9 males had occurred with the females that produced those egg masses. J2s hatching from each of the 18 egg masses were used to inoculate tomato plants under conditions supporting development into females. Each egg mass could contain a mix of sexual and parthenogenetic progeny. The exclusion of parthenogenetic progeny was accomplished in the next generation.
Egg masses were harvested from a total of 100 potential F1 females that developed from the J2s hatched from the 18 egg masses, and a subset of eggs from each egg mass was analyzed for markers H1 and H2. Marker H1 was detected in DNA from 17 of 100 egg masses examined, and all egg masses were positive for marker H2. The egg masses from three hybrid females (i.e., females producing eggs with both H1 and H2 markers) were inoculated onto separate tomato plants. After 7 weeks, we collected 183 egg masses from progeny of these females, representing >50% of the total number of eggs inoculated onto the roots. Individual egg masses were inoculated onto separate tomato plants and 183 F2 lines were successfully established.
DNA extracted from bulk eggs produced by each F2 line was assessed with markers H1 and H2 (cf. Figure 2, A and B). Lines segregated independently for these two markers; however, the data fit better to a 1:1 segregation pattern than to the 3:1 ratio typical for a Mendelian cross (Table 1). To further investigate marker segregation, we tested for the presence in the F2 progeny of 15 AFLP markers that we had determined were polymorphic between the parental strains (Table 1, markers AF1a–AF12). Again, a 1:1 segregation pattern was favored in all cases (Table 1). The majority of the AFLP markers that we tested were dominant; that is, a product was amplified from one parent but not the other. However, segregation patterns suggested that three pairs of markers were codominant with amplified products that differed in size in the inbred strains (Figure 2, C and D). DNA sequences of the putative pairs of codominant markers were determined to be identical except for a single indel of 15, 6, or 73 nt for markers AF1, AF3, and AF5, respectively (GenBank accession nos. EF506917–EF506922). For these codominant markers, heterozygotes were greatly underrepresented at 2.0, 1.6, and 2.0%, respectively, compared to the 50% expected in a standard Mendelian cross. To be certain that the apparent heterozygotes were not due to runover from adjacent lanes, the samples showing heterozygous patterns for markers AF1, AF3, and AF5 were amplified and assessed on a separate gel.
TABLE 1.
Marker segregation in F2 lines
| F2 linesc
|
|||||
|---|---|---|---|---|---|
| Markera | Primersb | Fragment size (bp) | % positive | P (1:1) | P (3:1) |
| H1 | See text. | 359 | 55.5 | 0.272 | 6.65E-06 |
| H2 | See text. | 250 | 48.6 | 0.782 | 1.11E-09 |
| AF1a | E+AT/M+GA | 258 | 49.2 | 0.867 | 2.42E-09 |
| AF1b | E+AT/M+GA | 243 | 50.8 | 0.867 | 2.41E-08 |
| AF2 | E+AT/M+GG | 231 | 42.5 | 0.136 | 6.58E-14 |
| AF3a | E+TA/M+GG | 233 | 54.2 | 0.402 | 1.54E-06 |
| AF3b | E+TA/M+GG | 227 | 45.8 | 0.402 | 1.57E-11 |
| AF4 | E+TA/M+GG | 256 | 50.8 | 0.867 | 2.41E-08 |
| AF5a | E+CG/M+AG | 257 | 40.0 | 0.046 | 6.32E-16 |
| AF5b | E+CG/M+AG | 184 | 60.0 | 0.046 | 5.32E-04 |
| AF6 | E+CG/M+AG | 218 | 57.7 | 0.124 | 6.41E-05 |
| AF7 | E+AT/M+GA | 190 | 53.0 | 0.548 | 3.79E-07 |
| AF8 | E+AT/M+GG | 100 | 48.6 | 0.782 | 1.11E-09 |
| AF9 | E+AT/M+GG | 180 | 43.1 | 0.167 | 1.73E-13 |
| AF10 | E+CG/M+AG | 90 | 47.3 | 0.583 | 1.47E-10 |
| AF11 | E+ACC/M+G | 280 | 47.2 | 0.579 | 1.41E-10 |
| AF12 | E+ACC/M+G | 230 | 49.4 | 0.912 | 3.59E-09 |
| AF22a | E+CAA/M+TA | 205 | 47.2 | 0.579 | 1.41E-10 |
| AF22b | E+CAA/M+TA | 220 | 52.8 | 0.579 | 2.87E-07 |
| AF23a | E+ACG/M+CTT | 93 | 47.8 | 0.657 | 3.24E-10 |
| AF23b | E+ACG/M+CTT | 79 | 52.2 | 0.657 | 1.44E-07 |
| AF24a | E+CG/M+CTT | 93 | 50.3 | 0.955 | 1.14E-08 |
| AF24b | E+CG/M+CTT | 79 | 49.7 | 0.955 | 5.28E-09 |
| AF25a | E+CA/M+GA | 154 | 47.0 | 0.543 | 9.47E-11 |
| AF25b | E+CA/M+GA | 155 | 53.0 | 0.543 | 3.94E-07 |
Markers labeled “a” and “b” after the same numerical designation are allelic.
E and M stand for universal AFLP primers (E: GACTGCGTACCAATTC; M: GATGAGTCCTGAGTAA). Sequences of H1 and H2 primers are listed in materials and methods.
P represents the P-value of the chi-squared goodness-of-fit test for 1:1 or 3:1 segregation.
Cytology of meiosis in M. hapla:
Previous cytological studies of M. hapla meioses concluded that, in parthenogenetic reproduction, restoration of ploidy occurs by fusion of sister nuclei after meiosis II (Triantaphyllou 1966; van Der Beek et al. 1998). Such a mechanism, in the absence of meiotic recombination, is predicted to yield a 1:1 segregation pattern and progeny homozygous for parental markers. An alternative explanation for the 1:1 segregation ratio is diploidization of haploid gametes, as has been proposed for parthenogenesis in some Drosophila species (Matsuda and Tobari 2004). However, this explanation is not consistent with the published cytological observations in M. hapla that telophase II sister chromosomes merge into a single nucleus.
To confirm and extend previous observations, we examined gonads of M. hapla strain VW9 stained with Hoescht 33258 from females derived from plant cultures infected with high or low nematode numbers. In several cases, we were able to isolate the gonad arm intact or as large sections, allowing us to follow the meiotic progression. Examination of multiple gonads produced the following summary interpretations. Before entering the spermatheca, oocytes in both the high and low population cultures progress through stages of prophase I much as is observed for C. elegans. In gonads from high-nematode population cultures, large numbers of sperm were seen in the spermatheca (not shown). Oocytes that passed through the spermatheca in these gonads completed meiosis in the first two eggs postspermatheca and began embryogenesis, suggesting that oocyte maturation is triggered by a signal from the sperm as it is in C. elegans (McCarter et al. 1999; Miller et al. 2001). In contrast, for gonads from M. hapla females cultured at low population levels, <5% of spermatheca examined carried sperm, as expected due to the environmental sex determination.
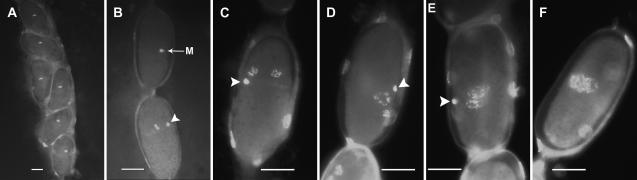
Meiotic maturation is dramatically extended in gonads lacking sperm compared to those in which sperm are present. The first few oocytes postspermatheca appeared to be in late prophase I and had not progressed in meiosis from those entering the spermatheca. These were followed by many (>10) sequential oocytes in metaphase I (Figure 3A). Completion of meiosis I and initiation of meiosis II appear to occur rapidly on the basis of the presence of only one or two oocytes at these stages (Figure 3B). Several sequential oocytes containing three strongly staining bodies are seen as the oocytes progress toward the vagina (Figure 3, C–F). In two of these bodies, condensed chromosomes are visible and can sometimes be counted with each containing the haploid chromosome number of 16 (Figure 3C; Triantaphyllou 1985; Liu and Williamson 2006). The third body is condensed and likely corresponds to the polar body or polar nucleus I. As eggs near the vagina and eggshells became more distinct, the two telophase chromosome complements become less condensed, appear closer together, and in some examples merge together (Figure 3, D–F). Polar body I, which is present in a variable location in the cell and generally in a different plane of focus from the pronuclei, appears to be expelled as the two nuclei merge (Figure 3, E and F). Our observations support previous studies that meiosis execution is complete and that the meiosis II products from sister chromosomes reunite into a single pronucleus during parthenogenetic reproduction. In C. elegans and in some other nematode species, the meiotic process can be examined in real time due to the transparency of the worms (McCarter et al. 1999; Lahl et al. 2006). Our efforts to examine the process in vivo in M. hapla were not successful due to the parasitic lifestyle and opaque bodies of the females.
Figure 3.—
Cytology of meiotic maturation in M. hapla. (A–F) Successive images from the same gonad stained with Hoescht 33258 that represent progression from oocytes just posterior to the spermatheca. The gonad was from a culture of strain VW9 infected with a low nematode innoculum. No sperm were seen in the spermatheca of this gonad. (A) Multiple oocytes arrested in metaphase I. (B) The last metaphase I oocyte (labeled M) is followed by an oocyte that appears to be in anaphase II. The following oocyte (C) appears to be arrested in telophase II with two pronuclei and a condensed polar body or polar nucleus. As the oocytes move toward the vagina, the two pronuclei are seen progressively closer and merge together (D–F). The polar nucleus or polar body from meiosis I is indicated by an arrowhead. In E, the polar body appears to be extruded from the cell and no polar body is apparent in F. Bar in A–F, 0.02 mm.
Recombination patterns in F2 progeny:
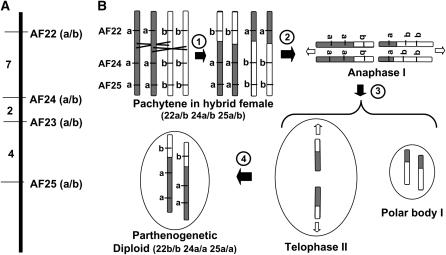
To investigate marker segregation, we identified additional codominant markers by screening our parental strains and a subset of the F2 lines with 320 AFLP primer sets. Twenty-four sets of codominant markers were identified and used to screen the F2 lines. For all codominant markers, heterozygotes were severely underrepresented. We identified one linkage group with four codominant markers, AF22a–AF25b (Table 1; Figure 4A) and two groups with two linked codominant makers each. Both alleles of these codominant markers were cloned and sequenced; again, marker pairs were found to differ in sequence by only indels or internal substitutions (GenBank accession nos. EF506923–EF506930). By examining the marker alleles for the four linked codominant markers in each F2 line, we identified 15 progeny classes (Table 2). The largest classes with 79 and 78 lines were the parental types, supporting the integrity of the linkage group. Surprisingly, the most common type of recombinant is homozygous for each of the closest markers flanking the recombination (classes 3–7 in Table 2). The less common recombinant types appear to represent a crossover between a single pair of non-sister chromatids (classes 8–10) or nonreciprocal exchanges (classes 11–14). We cannot rule out the possibility that some of the individuals in our F2 lines are the products of outcrossing the hybrid females with rare males in the population or due to errors in picking egg masses that have stuck together. In fact, one F2 line was heterozygous for all four markers as might be expected in these cases. However, our data generally are not consistent with such outcrossings. To determine whether recombinants with apparent four-strand exchanges are typical for other loci in the genome, we examined progeny classes in our F2 lines for two other pairs of linked, codominant markers. Apparent four-strand crossovers were similarly overrepresented in these loci (Tables 3 and 4).
Figure 4.—
Model for recombination and segregation in M. hapla. (A) Linkage group with four codominant AFLP markers. Numbers to the left represent the percentage of recombinant chromosomes between markers based on the data in Table 2. (B) Model to explain double recombinants between markers AF22 and AF24. (1) A four-strand double exchange occurs between markers AF22 and AF24. (2) Open arrows show the direction of movement of chromosomes during anaphase I and telophase II. (3) One set of sister chromosomes forms polar body I and the other set undergoes a second meiotic division arresting in telophase II. (4) In the absence of sperm nucleus fusion, the sister chromosomes are rejoined in a single pronucleus homozygous for all markers.
TABLE 2.
Marker allele classes for linkage group 1 in M. hapla F2 lines
| Marker allelea
|
No. of lines | |||||||
|---|---|---|---|---|---|---|---|---|
| AF22 | AF24 | AF23 | AF25 | |||||
| Class 1 | a/a | a/a | a/a | a/a | 79 | |||
| Class 2 | b/b | b/b | b/b | b/b | 78 | |||
| Class 3 | a/a | ×× | b/b | b/b | b/b | 9 | ||
| Class 4 | a/a | a/a | a/a | ×× | b/b | 3 | ||
| Class 5 | b/b | b/b | b/b | ×× | a/a | 3 | ||
| Class 6 | b/b | ×× | a/a | a/a | a/a | 1 | ||
| Class 7 | a/a | a/a | ×× | b/b | b/b | 1 | ||
| Class 8 | a/b | × | b/b | b/b | b/b | 1 | ||
| Class 9 | a/a | × | a/b | a/b | a/b | 1 | ||
| Class 10 | b/b | × | a/b | a/b | a/b | 1 | ||
| Class 11 | b/b | × | a/b | × | b/b | b/b | 1 | |
| Class 12 | b/b | × | a/b | a/b | × | b/b | 1 | |
| Class 13 | b/b | ×× | a/a | ×× | b/b | b/b | 1 | |
| Class 14 | a/a | a/a | ×× | b/b | ×× | a/a | 1 | |
| Class 15 | a/b | a/b | a/b | a/b | 1 | |||
| Total | 182b | |||||||
Crosses between columns indicate proposed recombination interval to generate each class. “×” represents a two-strand and “××” a four-strand exchange.
DNA from 1 of the 183 F2 lines did not amplify in this test with two primer sets for the above markers and so was omitted from the analysis.
TABLE 3.
Marker allele classes for linkage group 2 in M. hapla F2 lines
| Marker allelea
|
||||
|---|---|---|---|---|
| AF5 | AF21 | No. of lines | ||
| Class 1 | a/a | a/a | 102 | |
| Class 2 | b/b | b/b | 58 | |
| Class 3 | b/b | ×× | a/a | 14 |
| Class 4 | a/a | × | a/b | 4 |
| Class 5 | a/a | ×× | b/b | 2 |
| Class 6 | a/b | × | a/a | 2 |
| Class 7 | a/b | b/b | 1 | |
| Total | 183 | |||
Crosses between columns indicate proposed recombination interval to generate each class. “×” represents a two-strand and “××” a four-strand exchange.
TABLE 4.
Marker allele classes for linkage group 3 in M. hapla F2 lines
| Marker allelea
|
||||
|---|---|---|---|---|
| AF26 | AF30 | No. of lines | ||
| Class 1 | a/a | a/a | 99 | |
| Class 2 | b/b | b/b | 59 | |
| Class 3 | b/b | ×× | a/a | 8 |
| Class 4 | a/a | ×× | b/b | 5 |
| Class 5 | a/b | × | a/a | 2 |
| Class 6 | a/b | × | b/b | 1 |
| Class 7 | b/b | × | a/b | 1 |
| Class 8 | a/b | a/b | 1 | |
| Total | 176b | |||
Crosses between columns indicate proposed recombination interval to generate each class. “×” represents a two-strand and “××” a four-strand exchange.
DNA from 7 of the 183 F2 lines did not amplify in this test with one or more primer sets of the above markers and so were omitted from the analysis.
Model to explain recombination data:
Recombination is an integral part of meiosis in most organisms, including C. elegans (Hillers and Villeneuve 2003; Page and Hawley 2003). Our data indicate that recombination and segregation do occur in meiosis in M. hapla. It has been predicted, and demonstrated in some organisms in which there is sister chromosome fusion within the tetrad, that markers centromere proximal to the first chiasma would be homozygous, but that heterozygosity would be maintained for distal markers (Asher 1970; White 1973; Johnson et al. 1995; Hood and Antonvics 2004).
While C. elegans chromosomes are holocentric in mitosis, in meiosis I, one end of each pair of sister chromosomes, usually the one farthest from the crossover, leads the way to the pole (Albertson et al. 1997). If this also occurs in M. hapla meiosis, which previous work suggests it does (Triantaphyllou 1966), one would expect recombination events between non-sister chromatids to result in a heterozygous condition distal to the recombination point. In C. elegans, a single crossover occurs for each chromosome pair (Hillers and Villeneuve 2003) and this crossover usually occurs at a significantly off-center position. If this is the case for M. hapla, it may contribute to the homozygosity. However, even considering this, the amount of heterozygosity that we observe is unexpectedly low.
In our study, the largest classes of recombinants are homozygous for both flanking markers. One explanation for this is that recombination of each sister chromatid with its non-sister homolog generally occurs in similar, but not necessarily identical, regions of the chromosome (high negative crossover interference coupled with positive chromatid interference) as diagrammed in Figure 4B. Such a crossover pattern would explain the low frequency of heterozygotes that we observe. However, four-strand crossovers have been predicted to cause loss of sister-chromatid cohesion during meiotic pairing and missegregation (Nilsson and Sall 1995), and this pattern of crossovers has not been observed in other organisms. Other possible explanations for the segregation pattern include postmeiotic endoduplication, but, as mentioned above, this is not consistent with the cytological observations from our work and others (Triantaphyllou 1966; van der Beek et al. 1998). Crossovers that occurred before meiosis I would also produce the observed segregation patterns for linked markers. However, these would be expected to be rare. It may be that a novel mechanism of recombination or chromosome resolution that has not been previously described occurs in M. hapla, perhaps related to the prolonged meiosis in parthogenetic reproduction. Two features of the M. hapla genome that may facilitate an unusual segregation pattern are the holocentric nature of the chromosomes and their unusually small size. The genome size of M. hapla has been estimated to be ∼50 Mb (Mitreva et al. 2005) and the haploid chromosome number is 16. On the basis of this, the average size of a chromosome is ∼3 Mb, smaller than that of the fungus Schizosaccharomyces pombe (Fan et al. 1989). So far, genetic and genomic studies on this parthogenetic nematode have been limited; additional genetic and cytological studies, together with the increasing availability of DNA sequence information, is likely to increase our understanding of the meiotic process in this organism.
Consequences of facultative meiotic parthenogenesis:
While fusion of haploid pronuclei from a single meiosis is found in diverse groups of organisms, the tendency toward homozygosity is often reduced by specific mechanisms that rejoin products separated at meiosis I (Asher 1970; Hood and Antonvics 2004). The paucity of heterozygosity of DNA markers in our M. hapla F2 lines suggests that meiotic parthenogenesis would result in rapid genomic homozygosity. However, M. hapla males are environmentally determined, appear under conditions of stress and crowding and, as we have demonstrated, can cross with females to produce heterozygous progeny. Field isolates of M. hapla are diverse in pathogenicity and molecular markers (Janssen et al. 1997; Mitkowski and Abawi 2003; Liu and Williamson 2006). It is likely that asexual reproduction at low population levels and on rapidly growing hosts alternates with periods of male-favoring conditions of high population and poor nutrition on unhealthy or senescent hosts. Cyclical parthenogenesis occurs in other nematodes as well as in aphids and other insects as a strategy with advantages for dispersal and reproduction (White 1973; Viney 2006).
The data presented here indicate that it is feasible to produce a genetic map for M. hapla and, because of the unusual genetic system, to maintain F2 lines. Strains VW8 and VW9 differ in host range and attraction to particular plant hosts (Liu and Williamson 2006). Analysis of segregation of these traits, coupled with information from the ongoing efforts to sequence the M. hapla genome (Mitreva et al. 2005), should allow us to identify genes involved in parasitism and host specificity of this widespread and destructive group of plant parasites.
Acknowledgments
We thank Sean Burgess, Chuck Langley, Neil Hunter, JoAnne Engebrecht, George Bruening, Mel Green, and Frank McNally for helpful comments and discussions on the manuscript and Andrew Walker and Ed Caswell-Chen for advice. We also thank Carlos Gutierrez for assistance with sequence analysis of markers. The research presented here was funded by National Science Foundation awards 011080 and IOB-05-20824 to V.M.W.
References
- Albertson, D. G., A. M. Rose and A. M. Villeneuve, 1997. Chromosome organization, mitosis, and meiosis, pp. 47–78 in C. elegans II, edited by D. L. Riddle, T. Blumenthal, B. J. Meyer and J. R. Priess. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- Asher, J. H., 1970. Pathenogenesis and genetic variability. II. One-locus models for various diploid populations. Genetics 66: 369–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atibalentja, N., S. Bekal, L. L. Domier, T. L. Niblack, G. R. Noel et al., 2005. A genetic linkage map of the soybean cyst nematode Heterodera glycines. Mol. Gen. Genomics 273: 273–281. [DOI] [PubMed] [Google Scholar]
- Barker, K. R., 1985. Nematode extraction and bioassays, pp. 19–35 in An Advanced Treatise on Meloidogyne, Vol II, edited by K. R. Barker, C. C. Carter and J. N. Sasser. North Carolina University Graphics, Raleigh, NC.
- Barker, K. R., and S. R. Koenning, 1998. Developing sustainable systems for nematode management. Annu. Rev. Phytopathol. 36: 165–205. [DOI] [PubMed] [Google Scholar]
- Branch, C., C. F. Hwang, D. A. Navarre and V. M. Williamson, 2004. Salicylic acid is part of the Mi-1-mediated defense response to root-knot nematode in tomato. Mol. Plant Microbe Int. 17: 351–356. [DOI] [PubMed] [Google Scholar]
- Chen, P., and P. A. Roberts, 2003. Genetic analysis of (a)virulence in Meloidogyne hapla to resistance in bean (Phaseolus vulgaris). Nematology 5: 687–697. [Google Scholar]
- Dong, K., and C. H. Opperman, 1997. Genetic analysis of parasitism in the soybean cyst nematode Heterodera glycines. Genetics 146: 1311–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, J. B., Y. Chikashige, C. L. Smith, O. Niwa, M. Yanagida et al., 1989. Construction of a Not I restriction map of the fission yeast Schizosaccharomyces pombe genome. Nucleic Acids Res. 17: 2801–2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein, P., and A. C. Triantaphyllou, 1978. Occurrence of synaptonemal complexes and recombination nodules in a meiotic race of Meloidogyne hapla and their absence in a mitotic race. Chromosoma 68: 91–100. [Google Scholar]
- Hillers, K. J., and A. M. Villeneuve, 2003. Chromosome-wide control of meiotic crossing over in C.elegans. Curr. Biol. 13: 1641–1647. [DOI] [PubMed] [Google Scholar]
- Hood, M. E., and J. Antonvics, 2004. Mating within the meiotic tetrad and the maintenance of genomic heterozygosity Genetics 166: 1751–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen, R., J. Bakker and F. J. Gommers, 1991. Mendelian proof for a gene-for-gene relationship between virulence of Globodera rostochiensis and the H1 resistance gene in Solanum tuberosum ssp. andigena CPC 1673. Rev. Nematol. 14: 213–219. [Google Scholar]
- Janssen, G., A. van Norel, B. Verkerk-Bakker and R. Janssen, 1997. Intra- and interspecific variation of root-knot nematodes, Meloidogyne spp., with regard to resistance in wild tuber-bearing Solanum species. Fundam. Appl. Nematol. 20: 449–457. [Google Scholar]
- Johnson, S. L., D. Africa, S. Horne and J. H. Postlethwait, 1995. Half-tetrad analysis in zebrafish: mapping the ros mutation and the centromere of linkage group I. Genetics 139: 1727–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahl, V., B. Sadler and E. Schierenberg, 2006. Egg development in parthenogenetic nematodes: variations in meiosis and axis formation. Int. J. Dev. Biol. 50: 393–398. [DOI] [PubMed] [Google Scholar]
- Liu, Q. L., and V. M. Williamson, 2006. Host-specific pathogenicity and genome differences between inbred strains of Meloidogyne hapla. J. Nematol. 38: 158–164. [PMC free article] [PubMed] [Google Scholar]
- Matsuda, M., and Y. N. Tobari, 2004. Genetic analyses of several Drosophila ananassae-complex species show a low-frequency major gene for parthenogenesis that maps to chromosome 2. Genet. Res. 83: 83–89. [DOI] [PubMed] [Google Scholar]
- McCarter, J., B. Bartlett, T. Dang and T. Schedl, 1999. On the control of oocyte meiotic maturation and ovulation in Caenorhabditis elegans. Dev. Biol. 205: 111–128. [DOI] [PubMed] [Google Scholar]
- Miller, M. A., V. Q. Nguyen, M. H. Lee, M. Kosinski, T. Schedl et al., 2001. A sperm cytoskeletal protein that signals oocyte meiotic maturation and ovulation. Science 291: 2144–2147. [DOI] [PubMed] [Google Scholar]
- Mitkowski, N. A., and G. S. Abawi, 2003. Reproductive fitness on lettuce of populations of Meloidogyne hapla from New York State vegetable fields. Nematology 5: 77–83. [Google Scholar]
- Mitreva, M., M. L. Blaxter, D. M. Bird and J. P. McCarter, 2005. Comparative genomics of nematodes. Trends Genet. 21: 573–581. [DOI] [PubMed] [Google Scholar]
- Nilsson, N.-O., and T. Sall, 1995. A model of chiasma reduction of closely formed crossovers. J. Theor. Biol. 173: 93–98. [DOI] [PubMed] [Google Scholar]
- Omwega, C. O., and P. A. Roberts, 1992. Inheritance of resistance to Meloidogyne in common bean and the genetic basis of its sensitivity to temperature. Theor. Appl. Genet. 83: 720–726. [DOI] [PubMed] [Google Scholar]
- Page, S. L., and R. S. Hawley, 2003. Chromosome choreography: the meiotic ballet. Science 301: 785–789. [DOI] [PubMed] [Google Scholar]
- Roberts, P. A., 1995. Conceptual and practical aspects of variability in root-knot nematodes related to host plant resistance. Annu. Rev. Phytopathol. 33: 199–221. [DOI] [PubMed] [Google Scholar]
- Sasser, J. N., and D. W. Freckman, 1987. A world prospective on nematology: the role of the society, pp. 7–14 in Vistas on Nematology, edited by J. A. Veech and D. W. Dickson. Society of Nematologists, Hyattsville, MD.
- Triantaphyllou, A. C., 1966. Polyploidy and reproductive patterns in the root-knot nematode Meloidogyne hapla. J. Morphol. 118: 403–414. [DOI] [PubMed] [Google Scholar]
- Triantaphyllou, A. C., 1973. Environmental sex differentiation of nematodes in relation to pest management. Annu. Rev. Phytopathol. 11: 441–462. [Google Scholar]
- Triantaphyllou, A. C., 1985. Cytogenetics, cytotaxonomy and phylogeny of root-knot nematodes, pp. 113–126 in An Advanced Treatise on Meloidogyne, Vol. 1, edited by J. N. Sasser and C. C. Carter. North Carolina State University Graphics, Raleigh, NC.
- Trudgill, D. L., and V. C. Blok, 2001. Apomictic, polyphagous root-knot nematodes: exceptionally successful and damaging biotrophic root pathogens. Annu. Rev. Phytopathol. 39: 53–77. [DOI] [PubMed] [Google Scholar]
- Van der Beek, J., J. Los and L. Pijanacker, 1998. Cytology of parthenogenesis of five Meloidogyne species. Fundam. Appl. Nematol. 21: 393–399. [Google Scholar]
- Van Ooijen, J. W., and R. E. Voorrips, 2001. JoinMap® 3.0: Software for the Calculation of Genetic Linkage Maps. Plant Research International, Wageningen, The Netherlands.
- Viney, M. E., 2006. The biology and genomics of Strongyloides. Med. Microbiol. Immunol. 195: 49–54. [DOI] [PubMed] [Google Scholar]
- Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. van der Lee et al., 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23: 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir, B. S., 1990. pp. 71–114 in Genetic Data Analysis: Methods for Discrete Population Genetic Data. Sinauer Associates, Sunderland, MA.
- White, M. J. D., 1973. Animal Cytology and Evolution, Ed. 3. Cambridge University Press, Cambridge, UK.
- Williamson, V. M., and C. A. Gleason, 2003. Plant-nematode interactions. Curr. Opin. Plant Biol. 6: 327–333. [DOI] [PubMed] [Google Scholar]
- Williamson, V. M., and Q. Liu, 2006. Genetics and Pathogenicity of root-knot nematodes, pp. 659–665 in Biology of Plant-Microbe Interactions, Vol. 5, edited by F. Sanchez, C. Qinto, I. M. Lopez-Lara, and O. Geiger. International Society for Molecular Plant-Microbe Interactions, St. Paul.
- Williamson, V. M., E. P. Caswell-Chen, B. B. Westerdahl, F. F. Wu and G. Caryl, 1997. A PCR assay to identify and distinguish single juveniles of Meloidoygne hapla and M. chitwoodi. J. Nematol. 29: 9–15. [PMC free article] [PubMed] [Google Scholar]