Abstract
In budding yeast, B-type cyclin (Clb)-dependent kinase activity is essential for S phase and mitosis. In newborn G1 cells, Clb kinase accumulation is blocked, in part because of the Sic1 stoichiometric inhibitor. Previous results strongly suggested that G1 cyclin-dependent Sic1 phosphorylation, and its consequent degradation, is essential for S phase. However, cells containing a precise endogenous gene replacement of SIC1 with SIC1-0P (all nine phosphorylation sites mutated) were fully viable. Unphosphorylatable Sic1 was abundant and nuclear throughout the cell cycle and effectively inhibited Clb kinase in vitro. SIC1-0P cells had a lengthened G1 and increased G1 cyclin transcriptional activation and variable delays in the budded part of the cell cycle. SIC1-0P was lethal when combined with deletion of CLB2, CLB3, or CLB5, the major B-type cyclins. Sic1 phosphorylation provides a sharp link between G1 cyclin activation and Clb kinase activation, but failure of Sic1 phosphorylation and proteolysis imposes a variable cell cycle delay and extreme sensitivity to B-type cyclin dosage, rather than a lethal cell cycle block.
B-TYPE cyclin regulation is central to cell cycle control. In budding yeast, DNA replication and entry into mitosis are driven by B-type cyclins (Clb's) activating the cyclin-dependent kinase Cdc28, and mitotic Clb-Cdc28 is antagonistic to mitotic exit (Nasmyth 1996). The Cdk activity cycle is interdigitated with regulation of the anaphase-promoting complex (APC) (Zachariae and Nasmyth 1999), and multiple oscillatory mechanisms collaborate to provide alternate periods of low and high Clb-Cdk activity (Morgan and Roberts 2002; Cross 2003). The lowest period of Clb-Cdk activity is in newborn G1 cells. In G1, Cdh1 promotes APC-dependent ubiquitination and proteolysis of mitotic Clb's such as Clb2 (Schwab et al. 1997; Visintin et al. 1997). Independently, activation upon mitotic exit of SIC1 transcription leads to accumulation of Sic1 protein, a potent stoichiometric inhibitor of Clb-Cdk (Schwob et al. 1994; Knapp et al. 1996). An added mechanism is low transcription of all CLB genes in early G1 (Wittenberg and Reed 2005). Thus Clb kinase activation in early G1 is stringently regulated.
In late G1, a burst of gene expression dependent on the factors SBF/MBF is triggered by Cln3-Cdk (Wittenberg and Reed 2005). Among the targets of SBF/MBF are the G1 cyclins Cln1 and Cln2. These cyclins form a Cdk complex that is insensitive to Sic1 and Cdh1 and that initiate Sic1 and Cdh1 phosphorylation on multiple sites (there are 9 Cdk sites in Sic1 and 11 in Cdh1). Sufficient phosphorylation of these proteins results in their inactivation. Phosphorylated Cdh1 loses the ability to interact with the APC (Zachariae et al. 1998). Phosphorylated Sic1 is specifically recognized by the ubiquitination E3 complex SCF-Cdc4, leading to ubiquitination and proteolysis of phosphorylated Sic1 (Verma et al. 1997a; Nash et al. 2001). While there is no unique combination of sites in Sic1 required for Cdc4 binding, at least 6 of the 9 sites may need to be phosphorylated for efficient binding (Verma et al. 1997a; Nash et al. 2001; Orlicky et al. 2003).
Upon Clb-Cdk activation, Sic1 and Cdh1 phosphorylation can be carried out by Clb-Cdk instead of Cln-Cdk; thus Cln-Cdk can be viewed as flipping a switch that allows a transition between two otherwise stable states, one of low and one of high Clb-Cdk activity (Morgan and Roberts 2002; Cross 2003).
Sic1 binds tightly to the Clb-Cdc28 complexes required for DNA replication (Mendenhall 1993; Schwob et al. 1994), so the essentiality of removal of Sic1 is probably dependent on the relative stoichiometry of Sic1 and B-type cyclins if Sic1 were not degraded. Strains with temperature-sensitive mutations in SCF-Cdc4 components arrest in G1 at high temperature with high levels of stable Sic1 and low Clb kinase activity. Deletion of the SIC1 gene in these mutants allowed DNA replication, leading to the conclusion that the stable Sic1 was directly responsible for blocking DNA replication in the absence of SCF-Cdc4 (Schwob et al. 1994; Knapp et al. 1996). In another approach, moderate (∼1.5-fold) overexpression of a Sic1 mutant stabilized by four phosphorylation-site mutations blocked DNA replication in a G1 block-release protocol; similar overexpression of wild-type Sic1 had no effect because the protein was ubiquitinated and degraded (Verma et al. 1997a). These experiments led to the conclusion that at endogenous levels of Sic1 and B-type cyclins, Sic1 degradation is essential for DNA replication.
Sic1 stabilized by SCF inactivation is not in great excess for blocking S phase in cdc4 mutants, since heterozygous sic1/SIC1 cdc4/cdc4 diploids carried out DNA replication, unlike homozygous SIC1/SIC1 cdc4/cdc4 diploids (Knapp et al. 1996). Measurement of Sic1 and all B-type cyclins on a common scale indicated that, in a normal cell cycle, Sic1 did not accumulate in excess to peak B-type cyclin levels (Cross et al. 2002). However, CLB5 overexpression was reported not to accelerate DNA replication in G1 cells due to the presence of Sic1, suggesting at least some excess of Sic1 over Clb5 in these conditions (Schwob et al. 1994).
cln1 cln2 cln3 cells, lacking all G1 cyclins and normally blocked permanently in G1, are partially rescued by deletion of SIC1 (Tyers 1996), suggesting that, in the cln1 cln2 cln3 background, Sic1 is present at a high-enough level to titrate out any available Clb5 or other B-type cyclin; however, the absence of Cln3 and consequent absence of efficient CLB5 transcription (Wittenberg and Reed 2005) makes this situation difficult to compare accurately to wild type.
The G1 cyclins Cln1 and Cln2 probably carry out the bulk of physiological Sic1 phosphorylation; strains lacking CLN1 and CLN2 are highly sensitive to SIC1 gene dosage (Tyers 1996). Strains lacking CLN1 and CLN2 probably degrade Sic1 slowly, and DNA replication is delayed until these cells reach a larger cell size; this delay is Sic1 dependent (Dirick et al. 1995). This suggests that undegraded Sic1 delays DNA replication, but does not prove that undegraded Sic1 cannot block DNA replication indefinitely, since these cells do ultimately replicate DNA; however, the cells also likely ultimately degrade Sic1, probably due to Sic1 phosphorylation mediated by other cyclins (Nishizawa et al. 1998; Moffat and Andrews 2004).
Sic1 mutants with phosphorylation sites removed are as effective at Clb5-Cdc28 kinase inhibition as wild type (Nash et al. 2001). In contrast, phosphorylation is absolutely required for binding Cdc4 (Verma et al. 1997a; Nash et al. 2001). The N-terminal region of Sic1 containing multiple phosphorylation sites is necessary and sufficient for ubiquitination, whereas the C-terminal region is necessary and sufficient for Clb5 binding (Verma et al. 1997b). Indeed, only the C-terminal 70 amino acids of Sic1 are required for in vivo cell cycle inhibition upon overexpression (Hodge and Mendenhall 1999). Thus, the sole function of the phosphorylation sites may be to promote cell-cycle-regulated Sic1 degradation.
In an interesting parallel to Sic1, the mammalian Cdk inhibitor p27, which accumulates to high levels in G1, is also degraded following SCF-dependent ubiquitination, which is dependent on Cdk phosphorylation of threonine 187. While this degradation could contribute to the G1/S transition, the T187A mutation in p27 does not cause a significant cell cycle block; however, this mutant revealed a previously unappreciated G1 mode of p27 degradation (Malek et al. 2001). Thus, the consequence of complete stabilization of p27 remains unknown.
Here, we characterize a precise gene replacement of SIC1 with a mutant allele lacking all nine Cdk sites, SIC1-0P, to rigorously test the proposed essentiality of Sic1 phosphorylation and the ensuing proteolysis (Schwob et al. 1994; Verma et al. 1997a) at endogenously controlled levels and in an otherwise wild-type background.
MATERIALS AND METHODS
Plasmids:
Standard methods were used throughout. Starting materials for plasmid constructions were the following: MT2728, GAL-SIC1-0P (Nash et al. 2001); MT907, EcoRI–BglII fragment containing the wild-type SIC1 gene in RS316 (from M. Tyers); MDM168, GAL-SIC1 in YIplac204 (from A. Amon); RD609, GAL-SIC1-del3P (Verma et al. 1997a) from R. Verma; and plasmids containing GAL-SIC1-GFP with and without Lys-Arg mutations in N-terminal Sic1 ubiquitination sites (Petroski and Deshaies 2003a,b) (from R. Deshaies). The EcoRI–HpaI fragment from MT907 was subcloned into MDM168, replacing the GAL1 promoter with the wild-type SIC1 promoter. SIC1-0P was introduced in place of SIC1-wt by amplifying the endogenous promoter sequence from MT907 and the SIC1-0P sequence from MT2728 by polymerase chain reaction (PCR), combining the two PCR products by splice-overlap PCR and subcloning an EcoRI–HpaI fragment into MDM168. All cloned PCR products were sequenced. SIC1-wt and SIC1-0P inserts were subcloned into RS406, yielding FC667 and FC663. The SpeI–HpaI fragment from RD609 containing T33A, S76A, was subcloned into FC667 to reconstruct the four-site phosphorylation-site mutant (SIC1-del3P) under control of the endogenous SIC1 promoter in FC672. (One difference was that GAL-SIC1-del3P in RD609 was T2A T5GP, while FC672 was T2A T5A.) FC675 (SIC1-2P) was made by recombining FC663 and FC667 at a SpeI site, resulting in restoring T2 and T5 phosphorylation sites to SIC1-0P in FC663. Fusions of SIC1 and mutant derivatives to GFP were carried out by subcloning a SIC1-GFP fragment derived from GAL-SIC1-GFP plasmids (Petroski and Deshaies 2003a) into SIC1 plasmids. SIC1 lacking six N-terminal ubiquitin-acceptor lysines (SIC1-K0N: R32K, R36K, R50K, R53K, R84K, R88K) was constructed by subcloning a SpeI–HpaI fragment from the GAL-SIC1-K0N plasmid (Petroski and Deshaies 2003a) into FC667.
Strain constructions:
A swi5∷kanMX sic1∷HIS3 strain (W303 background) was constructed and transformed with PflFI-cut FC663 (targeting integration to the SIC1 promoter 5′ to sic1∷HIS3, creating SIC1-0P∷URA3∷sic1∷HIS3). Transformants were colony purified and Ura− popouts selected on 5-FOA medium. His− popouts were candidates for being SIC1-0P exact integrants. These popouts were confirmed by Southern blotting and by PCR amplification followed by sequencing across the entire locus, including the entire 5′ and 3′ noncoding regions. No mutations were found other than the expected phosphorylation-site mutations (and a deletion relative to the standard genomic sequence of one E residue from a poly(E) stretch in Sic1; this deletion is found in all our clones of SIC1). Similar methods were used for genomic introduction of SIC1-5P, SIC1-2P, and SIC1-K0N; correct integration was confirmed by restriction fragment length polymorphism or sequence analysis of diagnostic PCR products. Protein A-tagged versions (SIC1-0P-PrA, SIC1-5P-PrA, SIC1-K0N-PrA) were constructed by similar methods in a swi5∷kanMX SIC1-wt-PrA∷HIS3MX background, containing the SIC1 gene endogenously tagged with protein A; this is a functional fusion, described previously (Cross et al. 2002). In this case, FOA-R His+ popouts were tested by restriction digestion and sequencing of diagnostic PCR products.
Other strain constructions employed standard yeast mating and tetrad analysis. CLN2pr-GFP (the endogenous CLN2 promoter driving destabilized GFP, with a functional copy of CLN2 present in tandem) was described previously (Mateus and Avery 2000; Bean et al. 2006). GALS-CLB5 was from J. Bloom (unpublished results) and GALS-CLB2 was from C. Lookingbill (unpublished results); the GALS promoter is a weakened galactose-regulatable promoter that provides tight regulation without strong overexpression (Mumberg et al. 1995). Viability analysis of various genotypes in tetrad analysis was carried out by assuming a 2:2 segregation of markers to assign genotypes to inviable segregants. Only tetrads for which genotypes could be assigned to all spores, viable and inviable, were used for the quantitation. In this analysis, very tiny colonies (too small to genotype by replica plating) were scored as “inviable.” In some cases, SIC1 genotypes were confirmed by PCR and restriction digestion to check for phosphorylation-site mutations.
Time-lapse fluorescence microscopy:
We used a Leica DMIRE2 inverted motorized fluorescence microscope, with HCX Plan Apo, ×100, numerical aperture 1.40, oil immersion objective, in a heated 30° incubation chamber, imaging cells on agar slabs with a Hamamatsu ORCA ER 1394 digital CCD camera with the gain set to 50 (scale 0–250) for GFP images. We used Image Pro Plus 4.5 IPP to adjust brightness and contrast of phase and fluorescence images, to resize the phase image to the same size as the binned fluorescent images, to overlay phase and fluorescence images to create false-colored composites (phase, white; GFP, green), and to readjust brightness and contrast of the phase and fluorescence channels of the composite image (no nonlinear adjustments made during processing). For Sic1-GFP images, an additional adjustment in brightness was uniformly applied to all images in Photoshop, to better document the Sic1-wt-GFP signal. CLN2pr-GFP contained yeast-enhanced GFP3; SIC1-GFP contained GFP(S65T,Q80R). Data collection and analysis using CLN2pr-GFP were done as described (Bean et al. 2006), except that signal intensity and signal:noise ratio were enhanced by 2 × 2 binning of CCD camera pixels before data collection (our unpublished data).
For analysis of CLN2pr-GFP movies, assignment of budding times and genealogy (assignment of bud to a mother cell) was done manually using a custom-designed graphical user interface. All subsequent analysis of CLN2pr-GFP expression (standardized peak intensity and duration, time from budding to peak expression), cell cycle times (intervals between successive mother-bud emergences), and cell sizes at budding (pixel areas) were carried out completely automatically using the image and data analysis software described previously (Bean et al. 2006), with the addition of a new software routine to determine the histograms of GFP peak widths (time between half-maxima rising and falling) for all defined peaks. Data points >3 SD from the mean were removed before calculating statistics in Table 1, to avoid outlier bias (this correction typically removed zero to two events and had no effect on the conclusions drawn). CLN2pr-GFP peak amplitudes refer to average pixel intensities over the segmented cell border, and all values are standardized to a reference wild type run in parallel in every experiment. The peaks are the maxima of smoothing spline fits, after trough-to-trough background subtraction, as described (Bean et al. 2006). CLN2pr-GFP peak width is defined as the time from the rising to the falling attainment of 50% of the peak level in the background-subtracted spline fit. Cell size at budding is defined as the number of pixels in the segmented mother-cell boundary at the time that bud emergence was scored. Mother-cell cycle time is defined as the interval between successive budding of a mother cell. The former statistics were all determined automatically by the automated data analysis software from the complete data set (Bean et al. 2006). Bud long axis/short axis ratio was determined using images from the same movies and length analysis was determined using ImageProPlus, measuring bud dimensions 60 min after bud emergence.
TABLE 1.
CLN2pr-GFP expression and other statistics from quantitative time-lapse fluorescence microscopy
| Measure | SIC1 genotype | Mean ± SD (n) | Wild type vs. SIC1-0P (t-test) |
|---|---|---|---|
| CLN2pr-GFP peak amplitude (AU) | Wild type | 1.04 ± 0.23 (57) | P < 0.001 |
| SIC1-0P | 1.46 ± 0.40 (65) | ||
| CLN2pr-GFP peak width (in minutes) | Wild type | 47 ± 5.6 (57) | P < 0.001 |
| SIC1-0P | 54 ± 5.6 (70) | ||
| Budding to CLN2pr-GFP peak (in minutes) | Wild type | 21 ± 8 (88) | P < 0.001 |
| SIC1-0P | 27 ± 6 (89) | ||
| Cell size at budding (pixels) | Wild type | 1983 ± 311 (104) | P < 0.001 |
| SIC1-0P | 2914 ± 587 (127) | ||
| Mother-cell cycle time (in minutes) | Wild type | 95 ± 19 (59) | P < 0.001 |
| SIC1-0P | 121 ± 25 (85) | ||
| Bud long axis/short axis length ratio (60 min after budding) | Wild type | 1.1 ± 0.06 (50) | P < 0.001 |
| SIC1-0P | 1.7 ± 0.389 (81) |
Data were obtained as described in Bean et al. (2006). Nine-hour recordings (images every 3 min), starting with seven mutant and six wild-type founder cells containing CLN2pr-GFP were made, images were automatically segmented and assigned background-subtracted GFP signal values for each cell body, and bud emergence timing and mother–daughter relationships were assigned using the graphical user interface (Bean et al. 2006). Computations are described in materials and methods. Bud dimensions were determined using ImageProPlus on randomly selected cells 60 min after bud emergence. AU, arbitrary units.
Nuclear residence of Sic1-GFP and bud emergence were scored manually from composite-phase/fluorescence movies, as described previously for Whi5-GFP (Bean et al. 2006). Sic1-wt-GFP signal was low and somewhat variable; occasional cells in which a Sic1-GFP signal could not be reliably scored (usually mother cells) were omitted from the analysis. The low signal also made scoring of the timing of Sic1-GFP nuclear residence ambiguous in some cases, potentially adding a few frames' error to this estimate. These scoring problems do not affect the qualitative conclusions reported. Sic1-0P-GFP and Sic1-5P-GFP gave a very bright signal that led to no scoring ambiguity.
Other methods:
Flow-cytometry analysis (Epstein and Cross 1992) and Western blotting (Cross et al. 2002) were done as described. Purification of GST-Sic1 was carried out by lysozyme-sonication lysis of Escherichia coli expressing GST fusion proteins (GST-Sic1 or GST-Sic1-0P; plasmids from M. Tyers) followed by purification on glutathione–sepharose and elution with glutathione for soluble preparations. Estimated concentrations of full-length soluble GST-Sic1 and GST-Sic1-0P (∼0.6 mg/ml) were determined on the basis of amido-black-stained gel-transfer membranes compared to threefold serial dilutions of BSA standard (our unpublished data). For pull-down purification of Clb5-PrA with IgG–sepharose or with glutathione–sepharose carrying GST-Sic1 fusion proteins, yeast extracts were prepared from strain VAY79 (cdc20 GALL-CDC20 CLB5-PrA∷HIS3MX) after block of the strain in glucose for 3.5 hr by breakage of cell pellets derived from 100 ml of culture in 400 μl LSHNN buffer (10 mm HEPES, pH 7.5, 50 mm NaCl, 10% glycerol, 0.1% NP-40) with 400 μl of glass beads by shaking in a FastPrep bead beater for two periods of 20 sec at a setting of 5, separated by 1 min on ice. Extracts were clarified by a 1-min microfuge spin and precipitated with bead-bound affinity reagent (GST-Sic1 or IgG) (1 hr on ice, followed by three washes in LSHNN, one wash in the same buffer with 250 mm NaCl, and, for kinase assays, one wash and resuspension in kinase buffer (10 mm HEPES, pH 7.5, 10 mm MgCl2,1 mm DTT). For kinase assays, 15 μl of IgG-bound Clb5-PrA (purified from ∼20 ml of culture) were incubated on ice for 15 min with 600, 60, or 6 ng of GST-Sic1. Reaction mix (5 μl) containing 2 μg of histone H1, 5 μm ATP, and tracer [32P]ATP was added. Reactions were incubated for 10 min at 30°. The final concentration of Sic1 in the assays was estimated to be ∼500, 50, and 5 nm.
RESULTS
Expression of unphosphorylatable Sic1 from the endogenous promoter is not lethal, but results in a lengthened G1:
The expected lethality of endogenous expression of unphosphorylatable Sic1 (see Introduction) should be alleviated in the absence of the Swi5 transcription factor that activates SIC1 expression, since Sic1-dependent failure to replicate in a cdc4 background was rescued by swi5 deletion (Knapp et al. 1996). Therefore, we carried out a two-step gene replacement of sic1∷HIS3 with SIC1-0P [containing none of the nine consensus Cdk sites in Sic1 (Nash et al. 2001) (Figure 1)] in a swi5∷kanMX background. Gene replacements were confirmed by Southern blotting and by sequencing of PCR products spanning the entire locus (Figure 1; our unpublished data). This strategy results in exact replacement of the coding sequence of SIC1 with SIC1-0P, under control of the endogenous promoter, with no associated vector or marker sequences. We crossed SIC1-0P swi5∷kanMX to sic1∷HIS3 SWI5, so that we could assess viability of SIC1-0P SWI5 segregants (G418-S His−) among the progeny by tetrad analysis. In contrast to expectation, such progeny were not inviable; they were recovered at expected Mendelian proportions, exhibiting a moderately reduced growth rate, and an increased proportion of cells in G1 as determined by flow-cytometry analysis (Figure 2).
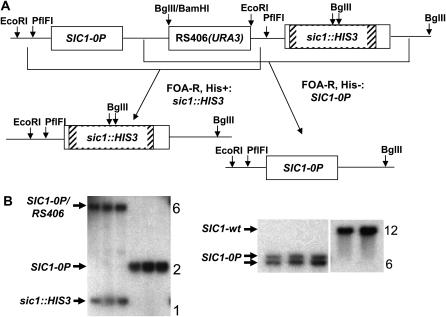
Figure 1.—
Scheme for replacement of SIC1 with SIC1-0P. (A) SIC1-0P, including a 5′ promoter sequence, in the cloning vector RS406 (URA3) (plasmid FC663) was digested with PflFI, cutting in the SIC1 promoter, and used to transform a sic1∷HIS3 strain. The expected resulting structure is shown on top (not to scale). Homologous recombination events resulting in Ura3− popout derivatives were selected on the basis of 5-FOA resistance and were found to be a mix of His+ and His−. These were interpreted as being due to recombination in the regions indicated by brackets. (B) Selected Southern blot analysis: (Left) EcoRI+BglII digestion of DNA from three integrants of RS406-SIC1-0P at the sic1∷HIS3 locus (first three lanes) and FOA-resistant His− popouts derived from these integrants (second three lanes), probed with SIC1 DNA from EcoRI to KpnI (at border of HIS3 insertion in sic1∷HIS3), confirming replacement of sic1∷HIS3 with SIC1-0P in the His− popouts. (Right) DNA from the same three FOA-resistant His− popouts and two SIC1-wt controls digested with ApaI and probed with a SIC1 coding sequence probe, confirming an ApaI site introduced at the S80A phosphorylation-site mutation (near the center of an ∼12-kb genomic ApaI fragment containing SIC1). Deduced identities of the bands are indicated; band sizes are approximately as expected from known genomic sequence.
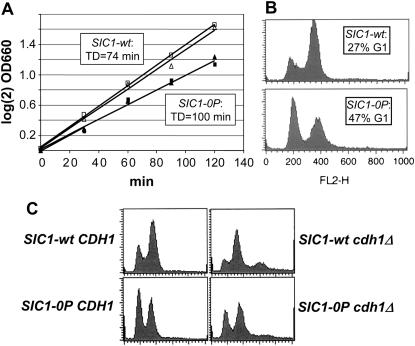
Figure 2.—
SIC1-0P cells are viable with an elongated G1. (A) SIC1-0P SWI5 and SIC1-wt SWI5 strains were constructed and grown in YEPD at 30°. At intervals, the OD660 was determined to measure the doubling times, indicated in the insets. (B) In the middle of the growth curve shown in A, samples were taken for flow-cytometry analysis. An approximate quantitation of the proportion of 1C DNA content (G1) cells was made (insets). (C) A SIC1-0P strain was crossed to a cdh1∷LEU2 sic1∷HIS3 pURA3-SIC1 strain, and tetrads were dissected. Flow-cytometry data from representative segregants (all lacking the pURA3-SIC1 plasmid) are presented.
SIC1 is essential in the absence of CDH1, an activator of the anaphase-promoting complex (Schwab et al. 1997; Visintin et al. 1997). To establish that SIC1-0P was functional, we crossed SIC1-0P strains to cdh1∷LEU2 strains. Viable SIC1-0P cdh1∷LEU2 segregants were readily obtained, while sic1∷HIS3 cdh1∷LEU2 segregants constructed in parallel were inviable, as expected. Asynchronous cdh1∷LEU2 strains had a reduced G1 population compared to wild type; SIC1-0P significantly increased G1 in the cdh1∷LEU2 background (Figure 2), indicating that Sic1-0P restrains S-phase entry even in the absence of Cdh1.
Viability of cells expressing Sic1 lacking only some phosphorylation sites:
Removal of as few as four of the nine Cdk sites from Sic1 can eliminate its interaction with SCF-Cdc4 (Verma et al. 1997a; Nash et al. 2001). Therefore, we carried out gene replacements of endogenous SIC1 with the four-site mutant of Verma et al. (1997a), called by them SIC1-del3P, which we have reconstructed under control of the endogenous promoter; we name this mutant here SIC1-5P for consistency, indicating the retention of five sites. This mutant was shown to possess full Cdk inhibitory activity and to completely escape ubiquitination by SCF-Cdc4 (Verma et al. 1997a). We recovered viable SWI5 strains with the SIC1-5P mutation as well as with numerous other combinations of phosphorylation-site mutations, including a mutant containing only the T2 and T5 sites (SIC1-2P) (our unpublished data).
Sic1-0P binds Clb5 and inhibits Clb5-associated kinase activity:
We carried out bacterial expression and purification of recombinant GST fusion proteins containing Sic1-wt or Sic1-0P. We assessed the ability of the GST fusion proteins to stably bind to Clb5 (tagged with protein A); GST-Sic1-0P was equivalent to GST-Sic1-wt in these assays (Figure 3A), confirming previous findings that Clb5 binding is independent of phosphorylation, and indeed independent of the N-terminal regions containing the phosphorylation sites (Verma et al. 1997a). We also observed essentially comparable inhibition of Clb5-associated kinase activity by soluble GST-Sic1-0P and GST-Sic1-wt (Figure 3B). Clb5 kinase inhibition assays of a number of phosphorylation-site mutants, including Sic1-0P, carried out using physiological concentrations of recombinant inhibitor and kinase, showed comparable inhibition by all Sic1 mutants to wild type (Nash et al. 2001); our results confirm this finding.
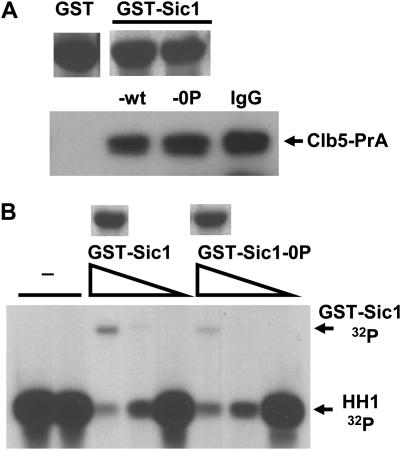
Figure 3.—
Sic1-0P binds and inhibits Clb5-Cdc28. (A) GST, GST-Sic1, and GST-Sic1-0P were purified from E. coli on glutathione–sepharose beads. VAY79 (cdc20 GAL-CDC20 CLB5-PrA) was blocked in glucose medium to deplete Cdc20 and allow accumulation of high levels of Clb5-PrA. Extracts made from this culture were incubated for 1 hr on ice with glutathione–sepharose beads containing equivalent amounts of GST, GST-Sic1, or GST-Sic1-0P (parallel amido black stains for GST fusions at top of figure) or with IgG–sepharose beads to directly purify Clb5 via the PrA tag, and beads were washed by repeated centrifugation. Western blot analysis of bead-bound Clb5-PrA is shown. (B) Clb5-PrA was purified from cell extracts as in A using IgG–sepharose. Aliquots (15 μl) of Clb5-PrA–IgG–agarose bead suspension in kinase buffer were incubated for 15 min on ice with no addition, with 1 μl of glutathione elution buffer, or with glutathione elution buffer containing 10-fold serial dilutions of soluble GST-Sic1 fusions (an estimated addition of ∼600, 60, and 6 ng of GST-Sic1 or GST-Sic1-0P; parallel amido black stains of GST fusions at top). Then 5 μm ATP, 50 μCi 32P-labeled ATP, and 2 μg histone H1 were added (final 21 μl). Reactions were incubated for 10 min at 30°, stopped with SDS sample buffer, and separated by gel electrophoresis before exposure to film to assess histone H1 kinase activity. Phosphorylation of GST-Sic1, and less efficient phosphorylation of GST-Sic1-0P, were also detected, as indicated.
Sic1 phosphorylation-site mutants are stable throughout the cell cycle:
Lack of cell cycle arrest with Sic1-0P or Sic1-5P at endogenous expression levels could be due to proteolysis of these proteins by a phosphorylation-independent mechanism. While no such mechanism was detected previously (Verma et al. 1997a; Nash et al. 2001), these previous experiments depended upon Sic1 overexpression, which could have saturated this hypothetical phosphorylation-independent mechanism.
To test stability of unphosphorylatable Sic1 at endogenous expression levels, we constructed SIC1-0P-PrA and SIC1-5P-PrA at the endogenous locus, by integration excision at a previously characterized functional SIC1-PrA locus (Cross et al. 2002). In these strains, we included a GALS-CLB5 cassette. Wild-type Sic1-PrA exhibited a pattern of accumulation consistent with cell cycle regulation of its abundance (Figure 4): almost no Sic1-PrA accumulation in cultures delayed in S/G2 by Clb5 overexpression (on galactose medium, where GALS-CLB5 was expressed) (Jacobson et al. 2000), a moderate amount in normally cycling cultures (on glucose medium, without GALS-CLB5 overexpression), and a high amount in cells blocked in G1 by the mating pheromone α-factor. Sic1-5P-PrA and Sic1-0P-PrA, in contrast, were present at comparably high levels with all these treatments.
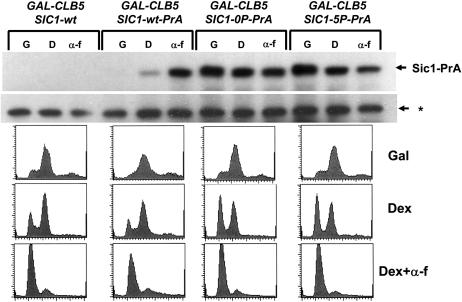
Figure 4.—
Sic1-0P and Sic1-5P fail to exhibit regulated changes in abundance displayed by wild-type Sic1. Strains carrying SIC1-wt, SIC1-0P, or SIC1-5P, all fused to a C-terminal protein A tag and expressed from the endogenous locus and also carrying a GALS-CLB5 cassette, were grown in galactose medium to overexpress Clb5 (G), shifted to glucose medium for 2.5 hr to shut off GALS-CLB5 (D), or to glucose medium plus α-factor (α-f) for 2.5 hr to arrest cells in G1. (Top) Western blots to detect Sic1-PrA or a control cross-reacting band (*) as loading control. Flow-cytometry profiles for the samples are presented below.
Upon release of the α-factor block, Sic1-wt-PrA was lost rapidly, around the time of bud emergence, reappearing faintly around the time of the succeeding mitosis. In contrast, Sic1-0P-PrA was stable throughout the cell cycle (Figure 5). Cell cycle progression in the two cultures was similar on the basis of budding, nuclear division, Clb5-myc accumulation, and DNA replication (Figure 5). Sic1-5P-PrA gave essentially the same results as Sic1-0P-PrA with respect to protein stability and cell cycle progression in this protocol (supplemental Figure 1 at http://www.genetics.org/supplemental/). Little or no G1 delay was detectable in comparing the Sic1 mutants to wild type in this protocol, even though SIC1-0P-PrA and SIC1-5P-PrA strains exhibited a clear increase in G1 population in asynchronous flow-cytometry profiles compared to wild type (Figure 4). We speculate that this may be due to the protracted G1 arrest due to α-factor.
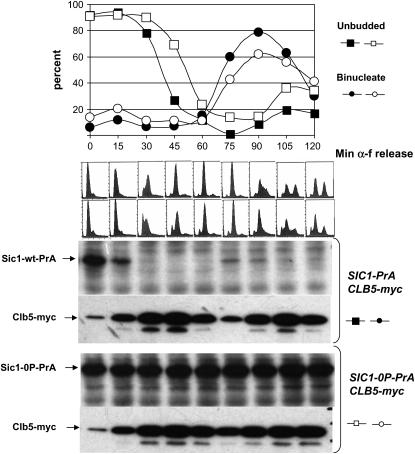
Figure 5.—
Sic1-0P is stable throughout the cell cycle. SIC1-wt-PrA or SIC1-0P-PrA GALS-CLB5 myc-CLB5 strains pregrown in galactose medium were switched to glucose medium plus α-factor (α-f) to arrest cells in G1, as in Figure 4. After a 2.5 hr arrest, cells were centrifuged, washed twice to remove α-factor, and reinoculated into fresh glucose medium. Time points were taken every 15 min. (Bottom) Western blot analysis of Sic1-PrA and myc-Clb5. The proportions of unbudded and binucleate cells were determined microscopically (top), and the approximate period of DNA replication (between 15 and 45 min after release) was determined by FACS (middle).
Constitutive nuclear localization of Sic1 phosphorylation-site mutants:
Sic1 is localized to the nucleus during a brief period between mitosis and G1/S (Nash et al. 2001). This period corresponds roughly to the time when Sic1 is present in the cell, but Sic1 nuclear localization could still be regulated independently of proteolysis. Since the B-type cyclins regulated by Sic1 have predominantly nuclear localization, the lack of cell cycle arrest by stabilized Sic1 could be due to Sic1 leaving the nucleus even if undegraded. To test this, we fused wild-type Sic1, Sic1-0P, and Sic1-5P to GFP, expressed from the endogenous locus, and analyzed these strains by time-lapse fluorescence microscopy.
Wild-type Sic1-GFP appeared at about the same time in mother and daughter nuclei (Figure 6), presumably as mitotic exit initiated, and then was detectable for only ∼11 ± 6 min in mother-cell nuclei. Daughter cells, with a longer G1, exhibited an ∼30 ± 15 min period of Sic1 nuclear residence. In both mothers and daughters, budding followed loss of the nuclear Sic1 signal by ∼10–15 min (Figure 6; our unpublished data).
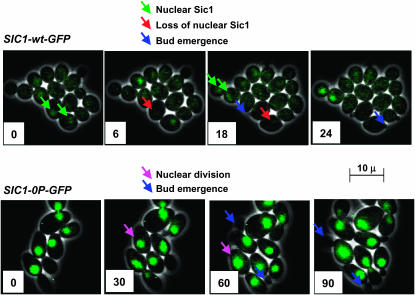
Figure 6.—
Nuclear Sic1-wt-GFP is detected only during M/G1; Sic1-0P-GFP is nuclear throughout the cell cycle. Cells containing SIC1-wt-GFP or SIC1-0P-GFP, expressed from the endogenous locus were analyzed by time-lapse fluorescence microscopy, with illumination every 3 min. Selected frames from movies are shown to illustrate Sic1-wt-GFP appearing as a nuclear signal in large-budded cells (top, green arrows) and then disappearing rapidly from the mother-cell nucleus and more slowly from the daughter-cell nucleus (top, red arrows). Sic1-wt-GFP disappearance occurs shortly before bud emergence (top, blue arrows). Sic1-0P-GFP was nuclear throughout the cell cycle, including before and after bud emergence (bottom, blue arrows) and before and after nuclear division (pink arrows). The disappearance of Sic1-0P-GFP from nuclei was never observed. In both series, for clarity, only some events are labeled with arrows.
In contrast, Sic1-0P-GFP was nuclear throughout the cell cycle, including throughout nuclear division and bud emergence (Figure 6). Sic1-5P-GFP gave essentially the same results as Sic1-0P-GFP (supplemental Figure 2 at http://www.genetics.org/supplemental/).
Single-cell analysis of the consequences of blocking Sic1 phosphorylation:
We characterized the cell cycle of SIC1-0P cells by quantitative time-lapse fluorescence microscopy using CLN2pr-GFP, encoding unstable GFP under control of the endogenous CLN2 promoter (Mateus and Avery 2000; Bean et al. 2006). SIC1-0P cells had CLN2pr-GFP peaks of greater intensity than wild type, and the interval between bud emergence and the CLN2-GFP peak intensity was significantly increased in the mutant (Figure 7; Table 1). This may reflect a less-efficient shutoff of CLN2 expression by mitotic Clb activity (Amon et al. 1993), due to Clb inhibition by stable Sic1.
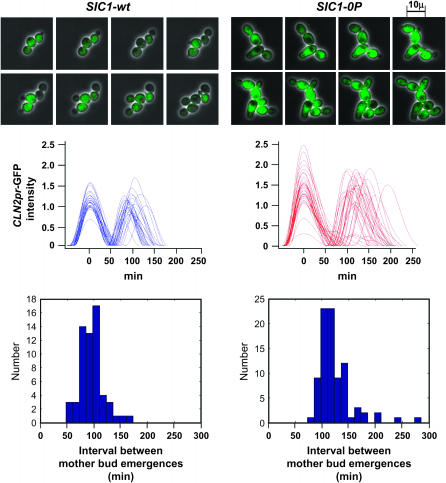
Figure 7.—
Analysis of the cell cycle in SIC1-0P cells by quantitative time-lapse fluorescence microscopy using a CLN2pr-GFP marker. Time-lapse fluorescence microscopy on SIC1-wt or SIC1-0P cells also containing CLN2pr-GFP (unstable GFP under control of the CLN2 promoter) (Mateus and Avery 2000; Bean et al. 2006) was carried out as described (Bean et al. 2006) with 3-min resolution. (Top) Composite phase/fluorescence images from representative movies are presented [images taken every 15 min, starting 60 min (SIC1-wt) or 101 min (SIC1-0P) after plating]. The images were collected in parallel and processed identically. Background fluorescence corresponding to autofluorescence in unlabeled cells was subtracted. Graphs (middle) present data extracted automatically from all data collected (annotated files for microcolonies developed over 9 hr of exponential growth from seven mutant and six wild-type founder cells). The traces (left) represent the smoothed and background-subtracted CLN2pr-GFP intensity profiles for all cases where the peak finder found two successive peaks on a common scale for wild type and SIC1-0P. The histogram (bottom) indicates the distribution of times between successive mother-cell buddings, measuring the mother-cell cycle time.
The ratio of bud length to width was significantly higher in SIC1-0P cells than in wild type (Table 1). This could be due to a longer period of polarized bud growth caused by excessive Cln1/2 and/or reduced mitotic Clb activity (Lew and Reed 1993). Interestingly, this ratio was much more variable in SIC1-0P cells than in wild-type (standardized variance ratio of 17, P < 0.001), suggesting that this effect varied significantly in different cells in the SIC1-0P population. Overall, these data suggest that SIC1-0P preferentially elongates a period of the cell cycle between budding and transcriptional activation of CLN2 and the later shutoff of CLN2 and switch to an isotropic bud growth pattern, both of which are dependent on activity of mitotic B-type cyclins (Amon et al. 1993; Lew and Reed 1993).
We also carried out movies with wild-type and SIC1-0P strains containing MCM2-GFP (data not shown). The Mcm complex enters the nucleus upon mitotic exit and then undergoes regulated nuclear export, dependent on Clb kinase activation (Labib et al. 1999). The period of Mcm2-GFP nuclear residence was significantly elongated in the SIC1-0P cells (this period was scored at 57 ± 21 and 45 ± 20 min for wild-type daughters and mothers; for SIC1-0P, the period was scored at 77 ± 15 and 62 ± 12 min for daughters and mothers). The period between bud emergence and Mcm2-GFP nuclear exit was greatly elongated in the SIC1-0P cells (11 ± 11 and 14 ± 15 min for wild-type daughters and mothers, and 46 ± 13 and 37 ± 13 min for SIC1-0P daughters and mothers).
Since activation of G1 cyclins, including CLN1 and CLN2, drives bud emergence (Moffat and Andrews 2004), while Clb kinase activation is required for Mcm complex nuclear exit (Labib et al. 1999), these data on timing of Mcm2-GFP nuclear exit support an increased delay between SBF-dependent CLN2 transcriptional activation and the later activation of Clb kinases in SIC1-0P cells, consistent with conclusions reached using CLN2pr-GFP time-lapse fluorescence microscopy.
The overall cell division cycle measured by the interval between successive mother-cell bud emergences was longer in the SIC1-0P mutant, and the mutant cells were larger (Figure 7, Table 1), probably because of the longer average cell cycle time. Separate movies using wild-type or SIC1-0P strains expressing Myo1-GFP as a bud neck/cytokinesis marker (Bi et al. 1998) indicate that the longer cell cycle in SIC1-0P strains is due to a preferential elongation of the part of the cell cycle between budding and cytokinesis; the unbudded period is significantly shorter in SIC1-0P cells than in wild-type cells (Table 2). Most likely the unbudded period is shortened because the delay in the budded period results in large progeny (Table 1). Such large cells may escape size control over Start (Hartwell and Unger 1977) and therefore bud soon after division.
TABLE 2.
Preferential elongation of the budded part of the cell cycle in SIC1-0P cells
| Measure | SIC1 genotype | Mean ± SD (n) | Wild type vs. SIC1-0P (t-test) |
|---|---|---|---|
| Unbudded period, daughters | Wild type | 49 ± 27 (13) | P < 0.001 |
| SIC1-0P | 16 ± 6 (29) | ||
| Budded period, daughters | Wild type | 71 ± 17 (10) | P < 0.001 |
| SIC1-0P | 119 ± 40 (14) | ||
| Cell cycle, daughters | Wild type | 119 ± 28 (10) | P < 0.25 |
| SIC1-0P | 134 ± 41 (14) | ||
| Unbudded period, mothers | Wild type | 25 ± 8 (15) | P < 0.001 |
| SIC1-0P | 14 ± 9 (27) | ||
| Budded period, mothers | Wild type | 65 ± 17 (10) | P < 0.001 |
| SIC1-0P | 103 ± 19 (14) | ||
| Cell cycle, mothers | Wild type | 90 ± 16 (10) | P < 0.005 |
| SIC1-0P | 116 ± 18 (10) |
MYO1-GFP cells, either SIC1-wt or SIC1-0P, were analyzed by time-lapse microscopy, and the times of budding and Myo1 ring formation and cytokinesis, as marked by Myo1 ring closure and disappearance (Bi et al. 1998), were scored manually. The unbudded period was defined as the time from cytokinesis to budding, and the budded period as the time from budding to cytokinesis. The total cell cycle was the time from one cytokinesis to the next. Data for mothers and daughters were analyzed separately.
In addition to the significant increase in average division time caused by SIC1-0P (Table 1), there were a number of outliers with highly increased division times in the SIC1-0P strain (Figure 7, bottom right); delays of this magnitude are much less frequent in wild type.
Sic1 stabilization makes major B-type cyclins essential:
The simplest model to account for viability of SIC1-0P cells, given that Sic1-0P is an effective inhibitor (Figure 3), is that Clb cyclins accumulate to a sufficiently high level to titrate out available Sic1-0P. This idea leads to the prediction that reduction of CLB gene dosage should result in SIC1-0P lethality.
The S-phase B-type cyclin Clb5 is a major target of Sic1 (Schwob et al. 1994). CLB5 deletion was nearly lethal in tetrad analysis when combined with SIC1-0P (Figure 8). This lethality was partially suppressed by simultaneous deletion of SWI5. Tetrad analysis with SIC1 mutants containing various combinations of phosphorylation-site mutations (Figure 8) yielded the interesting result that some SIC1 mutants retaining phosphorylation sites gave a more profound block to viability in the absence of clb5 than did SIC1-0P, in some cases resulting in complete inviability of even swi5 segregants. This suggests the hypothesis that stable Sic1 retaining some phosphorylation sites might be a more potent in vivo inhibitor than fully unphosphorylatable Sic1. Despite this, all SIC1 mutants could be recovered in tetrad analysis in a CLB5 SWI5 background with high viability.
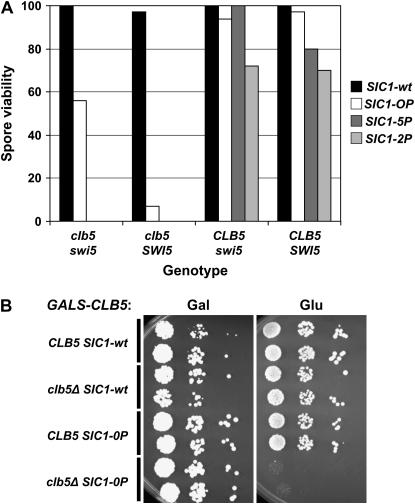
Figure 8.—
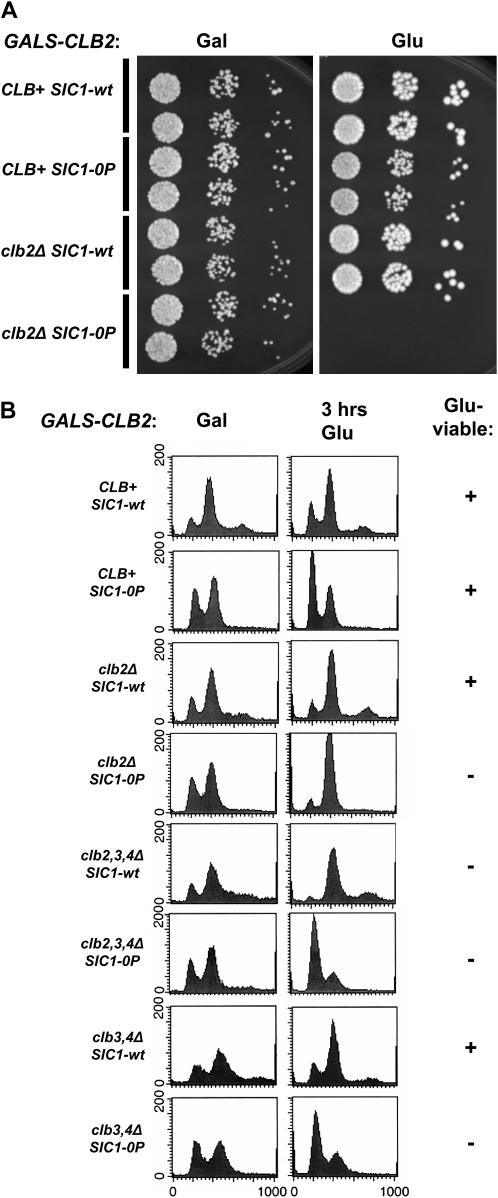
Phosphorylation-site mutations in SIC1 result in lethality in the absence of CLB5. (A) Tetrad analysis was performed on diploids of genotypes swi5∷kanMX/+ clb5∷URA3/+, sic1∷HIS3/SIC1-wt, sic1∷HIS3/SIC1-0P, sic1∷HIS3/SIC1-5P, or sic1∷HIS3/SIC1-2P: SIC1-5P, T2A, T5A, T33A, S76A; SIC1-2P, T33A, S69A, S76A, T80A, S174A, NS T192A. Spore viability for various genotypes was assessed assuming 2:2 segregation for all markers. Rare viable segregants of the genotype clb5 SWI5 SIC1-0P were small, slow-growing colonies. (B) GALS-CLB5 SIC1-0P clb5∷URA3 strains and controls were constructed by tetrad analysis on galactose medium. Serial dilutions on galactose medium (Gal; GALS-CLB5 on) and glucose medium (Glu; GALS-CLB5 off) were incubated for 3 days at 30°.
We could recover viable SWI5 clb5 SIC1-0P segregants that also contained GALS-CLB5 on galactose medium (GALS-CLB5 on). These strains were essentially inviable on glucose medium (Figure 8B). Upon shifting to glucose medium for 3 hr to shut off GALS-CLB5, clb5 SIC1-0P GALS-CLB5 strains exhibited an accumulation of G1 cells much higher than that observed with clb5 SIC1-wt GALS-CLB5 controls, suggesting that initiation of replication was delayed by SIC1-0P in this background (supplemental Figure 3 at http://www.genetics.org/supplemental/). Most of the cells had long buds, reminiscent of cells lacking B-type cyclin activity (Schwob et al. 1994). The clb5 SIC1-0P block was somewhat leaky (as reflected by sporadic recovery of weakly viable clb5 SIC1-0P segregants in tetrad analysis) (Figure 8A); therefore, we did not characterized the GALS-CLB5 shutoff phenotype in great detail.
Four B-type cyclins, CLB1–4, act to promote mitosis. The Clb1/Clb2 and Clb3/Clb4 pairs are close sequence homologs (Fitch et al. 1992). Clb2 and Clb3 are present in higher protein copy number than Clb1 and Clb4 (Cross et al. 2002). We tested combinations of cyclin deletions with SIC1-0P by tetrad analysis, scoring the viability of segregants of various genotypes. clb2 SIC1-0P and clb3 clb4 SIC1-0P segregants were completely inviable in tetrad analysis. clb4 SIC1-0P segregants were moderately slow growing; clb3 SIC1-0P segregants formed extremely slow-growing microcolonies. These results, combined with the lethality of clb5 SIC1-0P segregants, suggest that SIC1-0P imposes a requirement for a critical level of B-type cyclin without requirements as to Clb sequence class.
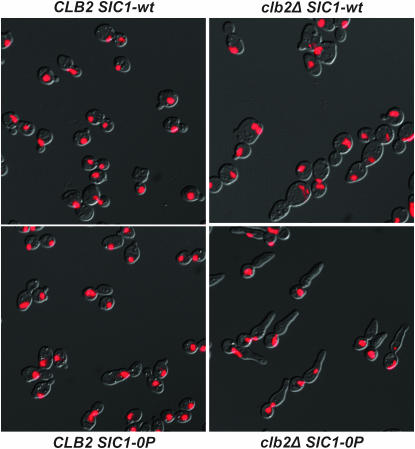
We could recover viable clb2 SIC1-0P GALS-CLB2 strains by tetrad analysis on galactose medium (GALS-CLB2 on). These strains were tightly inviable when transferred to glucose medium (GALS-CLB2 off) (Figure 9). We determined the lethal phenotype of clb2 SIC1-0P by transferring clb2 SIC1-0P GALS-CLB2 strains from galactose to glucose medium for 3 hr. The cells exhibited an arrest with 2C DNA content, indicating completion of DNA replication but failure to complete mitosis (Figure 9); most of the cells had long buds and a single nucleus (Figure 10). This phenotype resembles that of cells lacking the mitotic cyclins CLB1–4 (Fitch et al. 1992).
Figure 9.—
Requirements for mitotic CLB2–4 in a SIC1-0P background. (A) GALS-CLB2 SIC1-0P clb2∷LEU2 strains and controls were constructed by tetrad analysis on galactose medium. Serial dilutions on galactose medium (Gal; GALS-CLB on) and glucose medium (Glu; GALS-CLB off) were incubated for 3 days at 30°. (B) Flow-cytometry analysis of GALS-CLB2 strains, either SIC1-wt or SIC1-0P, containing the indicated additional CLB gene deletions. Strains were grown in galactose medium (Gal) or transferred to glucose medium (Glu) for 3 hr. (Right) Viability of the indicated genotype on glucose medium (GALS-CLB2 off) as determined by a replica-plating patch assay.
Figure 10.—
Lethal phenotype of clb2 SIC1-0P cells. Cells of the indicated genotype, also containing a GALS-CLB2 cassette, were grown in galactose medium to log phase and transferred to glucose medium for 3 hr. Cells were fixed in ethanol and digested with RNase and protease and nuclear DNA was stained with propidium iodide; samples were examined by DIC and fluorescence microscopy, and composite images were generated. All exposure settings were identical for these images.
clb2 clb3 clb4 SIC1-wt GAL-CLB2 strains arrest with 2C DNA upon turnoff of GAL-CLB2 (Fitch et al. 1992) (Figure 9), since they complete replication due to Clb5, -6 activity (Schwob et al. 1994), but then fail to complete mitosis due to deficiency of mitotic cyclins Clb1–4. Strikingly, clb2 clb3 clb4 SIC1-0P GALS-CLB2 strains arrested with 1C DNA (Figure 9), indicating that Clb5 and Clb6 are insufficient for driving replication in the clb2 clb3 clb4 SIC1-0P background. This 1C arrest, with long buds (our unpublished data), resembles that of cells lacking all B-type cyclins (Schwob et al. 1994).
clb3 clb4 SIC1-0P GALS-CLB2 strains showed inviability on glucose medium; clb3 clb4 SIC1-wt strains, in contrast, are viable (Fitch et al. 1992). clb3 clb4 SIC1-0P GALS-CLB2 strains transferred to glucose medium for 3 hr showed a predominantly 1C arrest (Figure 9), suggesting that, in this background, Sic1-0P effectively inhibits the S-phase cyclins Clb5, -6 as well as the remaining mitotic B-type cyclins Clb1, -2, resulting overall in Clb-dependent kinase activity insufficient for driving DNA replication.
Viability of cells expressing Sic1 lacking ubiquitin-accepting lysines:
Removal of six lysines from the N-terminal half of Sic1 significantly inhibits Sic1 ubiquitination and turnover; these lysines may be the primary acceptor sites for SCF-Cdc4-dependent ubiquitination (Petroski and Deshaies 2003a). We replaced SIC1 with SIC1-K0N (with these six lysines replaced by arginines) under control of the endogenous promoter in a swi5 background, as described above for SIC1-0P (see Figure 1). Subsequent tetrad analysis employing essentially the assay described in Figure 8 revealed ∼100% viability of SIC1-K0N SWI5 cells; these cells exhibited a G1 delay by flow cytometry (supplemental Figure 5 at http://www.genetics.org/supplemental/). Deletion of CLB5 sharply reduced viability of SIC1-K0N segregants, essentially as observed with SIC1-0P (∼17% compared to ∼100% for SIC1-wt clb5 strains and ∼7% for SIC1-0P clb5 segregants; see Figure 8), and the rare viable SIC1-K0N clb5 segregants were notably slow growing in tetrad analysis. These results thus recapitulate the effects of removing Sic1 phosphorylation sites by removing instead ubiquitin-accepting lysine residues in Sic1 but leaving the phosphorylation sites intact.
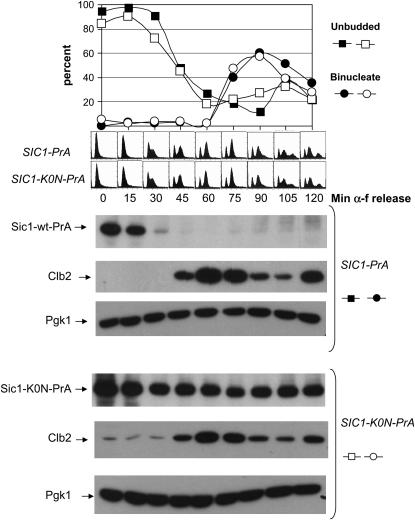
In an α-factor block-release experiment using cells expressing endogenous levels of Sic1-K0N-PrA compared to Sic1-wt-PrA, little difference was observed in the time of DNA replication between the two strains, and replication occurred after Sic1-wt-PrA was degraded. In contrast, most cells had completed DNA replication in the presence of high levels of Sic1-K0N-PrA, although partial reduction in Sic1-K0N-PrA levels were detectable later in the cell cycle (Figure 11). The partial, slow residual ubiquitination and degradation of Sic1-K0N presumably operates through inefficiently used C-terminal lysine–ubiquitin acceptors (Petroski and Deshaies 2003a).
Figure 11.—
Sic1-K0N is relatively stable throughout the cell cycle. SIC1-wt-PrA or SIC1-K0N-PrA strains were grown to log phase in glucose medium and then α-factor (α-f) was added to arrest cells in G1, as in Figure 4. After a 2.5 hr arrest, cells were centrifuged and washed twice to remove α-factor and reinoculated into fresh glucose medium. Time points were taken every 15 min. (Bottom) Western blot analysis of Sic1-PrA, Clb2, and Pgk1 as a loading control. The proportions of unbudded and binucleate cells were determined microscopically, and the approximate period of DNA replication (between 15 and 45 min after release) by flow cytometry (middle panels).
Sic1-K0N-GFP (supplemental Figure 6 at http://www.genetics.org/supplemental/) exhibited intermediate behavior between Sic1-wt and Sic1-0P (Figure 6) or Sic1-5P (supplemental Figure 2 at http://www.genetics.org/supplemental/). Sic1 appeared at the time of mitotic exit in mother and daughter nuclei similarly to wild type, but the Sic1 nuclear signal then decayed slowly compared to wild type. As noted above, the period of Sic1-wt-GFP nuclear residence was ∼11 ± 6 min in mother-cell nuclei and ∼30 ± 15 min in daughter-cell nuclei. In contrast, Sic1-K0N-GFP nuclear residence was ∼35 ± 27 min in mother-cell nuclei and ∼55 ± 26 min in daughter-cell nuclei. The Sic1-K0N-GFP nuclear signal almost always persisted until well after bud emergence (∼ +25 ± 25 min), while the Sic1-wt-GFP signal always disappeared before bud emergence (∼ −12 ± 8 min). Ultimate loss of Sic1-K0N-GFP nuclear signal is likely due at least in part to partial degradation of Sic1-K0N (Figure 11), although a contribution of regulated nuclear export of Sic1-K0N-GFP cannot be ruled out. In any case, this behavior of Sic1-K0N-GFP in single cells strongly suggests persistence of significant levels of Sic1-K0N until after DNA replication, because of the known correlation between bud emergence and DNA replication, as confirmed, for example, in the α-factor block-release experiment in Figure 11.
Thus, overall, results with Sic1-K0N suggest that even phosphorylatable Sic1 that fails to be degraded is unable to block DNA replication.
DISCUSSION
Sic1 phosphorylation is not essential:
It was proposed that Sic1 phosphorylation is essential for the G1/S transition (Schwob et al. 1994; Verma et al. 1997a). Our data present a serious challenge to this hypothesis: SIC1-0P cells are viable despite stability and constitutive nuclear localization of a high level of functional Sic1. Replication is delayed in SIC1-0P cells, probably due to delayed activation of Clb kinases.
How can we reconcile our findings with previous results suggesting the essentiality of Sic1 phosphorylation and degradation? The finding (Schwob et al. 1994) that sic1 cdc4 mutants fail to arrest in G1, unlike SIC1 cdc4 mutants, strongly suggests that undegraded Sic1 at endogenous levels is sufficient for blocking replication, at least in a cdc4 mutant background. One possible explanation of the discrepancy may be that fully phosphorylated Sic1, such as accumulates in a cdc4 block, is a more effective inhibitor in vivo than unphosphorylated Sic1. SIC1 mutants containing some phosphorylation sites intact are more deleterious in tetrad analysis than SIC1-0P (Figure 8), which could be consistent with this explanation. However, our data show that Sic1-0P was as effective as wild type at Clb5-Cdc28 binding and inhibition in vitro (Figure 3), confirming previous results (Verma et al. 1997a; Nash et al. 2001). Sic1 lacking the primary ubiquitin-acceptor lysines is phosphorylated, but nevertheless is significantly stabilized. Therefore, as an additional test of the idea that Sic1-0P might be a partially defective in vivo inhibitor due to lack of phosphorylation sites, we tested whether Sic1 lacking these lysines (Sic1-K0N), expressed from the endogenous SIC1 promoter, would have a more deleterious phenotype than SIC1-0P; however, this allele allowed full viability with a G1 delay phenotype, similar to that observed with SIC1-0P (supplemental Figure 5 at http://www.genetics.org/supplemental/). This result is consistent with the idea that stable Sic1, even if phosphorylatable, delays but does not block DNA replication, when expressed from the endogenous promoter. A complication in this interpretation is the partial degradation of Sic1-K0N (Petroski and Deshaies 2003a) (Figure 11) and its loss from the nucleus late in the cell cycle (supplemental Figure 6 at http://www.genetics.org/supplemental/). Still, the results with Sic1-K0N are supportive of the results with Sic1-0P in suggesting that Sic1 proteolysis is not essential. Sic1-K0N-expressing cells are able to synthesize DNA on schedule in an α-factor block-release protocol while significant levels of Sic1-K0N persist (Figure 11), although some drop in the Sic1-K0N level is notable later in the time course. While SIC1-K0N CLB5 and SIC1-0P CLB5 segregants are fully viable, SIC1-K0N segregants lacking clb5 are strongly reduced in viability (see above), as are SIC1-0P clb5 segregants (Figure 8), suggesting that both these mutations sensitize the cell to lowered levels of B-type cyclins.
Another possibility is that the requirement for Sic1 phosphorylation is somehow restricted to the SCF-Cdc4-deficient background. We found a strong interaction between cdc4-1 and SIC1-0P, such that the double mutants were significantly slowed in growth rate even at permissive temperature (our unpublished data), and this interaction limited our ability to pursue this issue in more depth. This is because we were concerned that the double mutants might accumulate suppressors or modifiers due to cryptic selection from slow growth, rendering results with them difficult to interpret. SIC1 is haplo-insufficient in a cdc4 background for restraining replication (Knapp et al. 1996), suggesting that even if Sic1 is in excess of Clb kinases in this background, this excess is within a factor of 2. The alternative that Sic1 degradation is critical only at higher temperatures is probably not correct since SIC1-0P cells are not temperature sensitive for viability and they show no further increase in G1 accumulation by flow-cytometry analysis upon shift to 37° (data not shown).
Finally, we cannot rule out that the phosphorylation-site mutations (and perhaps the Lys-Arg mutations in the SIC1-K0N mutant) subtly perturb the structure of Sic1 such that it is a less effective inhibitor: these putative alterations in structure might not even be related to phosphorylation, but rather related to changes in function of the unphosphorylated protein by some of the mutational changes. This hypothetical reduction in efficiency of Sic1 would have to result in minimal or undetectable changes in efficiency of in vitro inhibition (Figure 3) (Nash et al. 2001; Petroski and Deshaies 2003a) and would also call into question the generally accepted modular structure of Sic1, with the N-terminal regions dedicated to the phosphodegron and only the C-terminal 70 amino acids being involved in Cdk inhibition (Hodge and Mendenhall 1999; Nash et al. 2001). For these reasons, we do not favor the hypothesis that the phosphorylation-site mutations weaken direct Cdk inhibition, although we cannot rule out this hypothesis at this time.
SIC1-del3P (mutated in four phosphorylation sites, T2, T5, T33, and T76) blocked entry into S phase when overexpressed only very moderately (Verma et al. 1997a). We reconstructed this mutant under control of the endogenous promoter (our SIC1-5P). Like SIC1-0P, SIC1-5P encodes a highly stable, constitutively nuclear protein that elongates G1 but does not result in inviability or in a block to replication in an α-factor block-release experiment (Figure 4; supplemental Figures 1 and 2 at http://www.genetics.org/supplemental/).
Lethality of SIC1-0P when combined with deletion of the normally nonessential B-type cyclins CLB2, CLB3, or CLB5 suggests that unphosphorylatable Sic1 is just below a lethal threshold, so even moderate overexpression of unphosphorylatable Sic1 (Verma et al. 1997a) may block DNA replication. Consistent with this idea, transformation of a sic1∷HIS3 strain with a CEN (low-copy-number) plasmid containing SIC1-0P (endogenous promoter) gives notably slow-growing colonies compared to vector or wild-type SIC1 plasmid (supplemental Figure 4 at http://www.genetics.org/supplemental/).
In summary, our data very strongly suggest that Sic1 phosphorylation is not essential. While it is impossible, using Sic1 phosphorylation-site mutants, to rule out the hypothesis that allowing Sic1 phosphorylation but then preventing its degradation might be essential, the latter situation is essentially an artifact of fully phosphorylated Sic1 encountering an experimentally disrupted Sic1 removal system. Evolutionarily, it appears likely that Sic1 initially evolved as a Cdk inhibitor that later incorporated phosphorylation-dependent degradation, rather than the other way around; thus, SIC1-0P is much more likely to be similar to the precursor of the fully evolved system. Therefore, our data emphasize that a fully functional Cdk inhibitory system is compatible with the complete absence of regulated proteolysis or localization of the inhibitor, and we speculate that such a system probably evolved first, before the development of the regulated proteolysis system.
Sic1 phosphorylation provides a Cln-dependent sluice gate to a Sic1 dam restraining B-type cyclin-dependent kinase:
We explain viability of SIC1-0P cells by proposing that stable Sic1 transiently blocks Clb-Cdk activation, but that ultimately the total level of Clb-Cdk accumulates to above the level of the Sic1 blockade. Since Sic1 is a stably bound stoichiometric inhibitor, early-accumulating Clb-Cdk could effectively titrate Sic1, remaining bound even when later-accumulating Clb-Cdk is fully active. This model accounts for the lethal phenotypes of SIC1-0P when it is combined with various CLB deletions: normally nonessential CLB genes become essential, presumably because there is insufficient residual Clb protein to titrate Sic1-0P (Figure 9). The final phenotype (1C or 2C arrest; Figure 9) presumably reflects some combination of differential replication-promoting vs. mitosis-promoting activity of different Clb proteins and the varying contribution of different Clb proteins to total Clb levels attained at various points in the cell cycle (Miller and Cross 2001; Cross et al. 2002). This model for titration of stable Sic1 is formally similar to the proposal that cyclin D complexes activated early in the mammalian cell cycle titrate the p27 inhibitor and thus indirectly activate later-accumulating cyclin E and cyclin A complexes (Sherr and Roberts 1999).
Nasmyth and Hunt (1993) proposed a “dams and sluices” analogy for Cdk inhibitors. A simple dam blocks water flow until the water level rises above the dam. A sluice gate is a regulatable opening at the base of the dam. A dam with a sluice gate provides all the functionality of a simple dam, blocking downstream water flow until the level becomes high, but can also release all of the upstream accumulated water, independent of its level. Sic1 can be analogized as a dam for accumulated Clb's and Cln-dependent phosphorylation as opening the sluice gate. Our data reveal the nonlethal, but still significant, flaws in the Sic1-0P system (a dam without a sluice gate): this system demands that Clb-Cdk accumulates to high levels, unlike the wild-type system, accounting for the elongated G1 period and the genetic requirement for CLB2, CLB3, and CLB5 observed in the SIC1-0P background.
Rates of production of many different proteins may vary significantly across a cell population (Bar-Even et al. 2006; Newman et al. 2006). The Cln-Cdk sluice-gate control of Sic1 levels may buffer the system to variation in rates of accumulation of Sic1 or Clb proteins. Such variation may account for the sporadic occurrence of highly delayed cell cycles in SIC1-0P cells (Figure 7).
Cln-Cdk removal of Sic1 is catalytic, and Cln-Cdk is probably in stoichiometric excess to the Sic1 catalytic target (Cross et al. 2002). As a consequence, wild-type cells can cope with at least 10 extra copies of the SIC1 gene without significant slowing of growth rate (Thornton and Toczyski 2003; Moriya et al. 2006). This ability is dependent on Cln1 and Cln2, since cln1 cln2 cells are inviable with only a few extra copies of the SIC1 gene (Tyers 1996). What sets the peak level of Sic1 in wild-type cells, given that much more could be dealt with expeditiously? The level (∼1000 copies/cell) is similar to the subsequent peak levels of Clb5 and then Clb2 (Cross et al. 2002). Therefore, these levels may provide another level of robustness: the relatively low levels of Sic1 ensure that the simple dam mechanism for providing a Clb-Cdk-free period in early G1 could operate as a backup. Occasional cell cycles with inefficient expression of Cln1 and Cln2 would provide a selection keeping the peak level of Sic1 reasonably tuned to the peak levels of the target Clb cyclins.
Thus the Sic1 proteolytic removal system may have evolved due to the requirement for a transient block to Clb-Cdk activity in G1, with the additional constraints of robustness to high expression of Sic1 and to low expression of the Clb targets of Sic1 and the Cln antagonists of Sic1 that normally drive its removal.
A recent computational model (Chen et al. 2004) predicts that, in cells expressing unphosphorylatable Sic1 from the endogenous promoter, DNA replication would occur after a very long G1 period, consisting of more than two wild-type doubling times (K. Chen, personal communication). Some revision of the model is required to fit the present data, since the elongated G1 in SIC1-0P is significantly less than a normal wild-type doubling time; however, minor alterations, such as doubling the expression level of CLB5 or halving the expression level of SIC1, will largely rescue the overly long G1 phenotype predicted by the model (our unpublished data). Inclusion of Clb3 in the model, at appropriate expression levels (Cross et al. 2002), might be an effective and realistic revision, since accumulation of Clb3 in mid-cell cycle would help to titrate stable Sic1, accounting for the observed dependence on Clb3 for viability in the SIC1-0P background.
Precise gene replacement for precise answers?
Analogously to the value of null alleles for determining the function of a protein in an otherwise wild-type system, the role of a specific sequence within a protein may be best analyzed with a precise gene replacement of the wild type with a version mutated in the specific sequence, with no other changes in promoter or flanking sequences. Here, this strategy has provided data strongly suggesting that contradictory to prior expectations, Sic1 phosphorylation is not essential for removing a lethal block to the G1/S transition; rather, it performs the nonessential but important role of increasing cell cycle precision and robustness.
Acknowledgments
Thanks go to A. Amon, R. Deshaies, M. Tyers, and R. Verma for providing plasmids. Thanks also go to J. Bloom and C. Lookingbill for strains and to K. Chen for communicating unpublished modeling results. This work was supported by National Institutes of Health grant PHS GM047238 to F.R.C.
References
- Amon, A., M. Tyers, B. Futcher and K. Nasmyth, 1993. Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell 74: 993–1007. [DOI] [PubMed] [Google Scholar]
- Bar-Even, A., J. Paulsson, N. Maheshri, M. Carmi, E. O'Shea et al., 2006. Noise in protein expression scales with natural protein abundance. Nat. Genet. 38: 636–643. [DOI] [PubMed] [Google Scholar]
- Bean, J. M., E. D. Siggia and F. R. Cross, 2006. Coherence and timing of cell cycle Start examined at single-cell resolution. Mol. Cell 21: 3–14. [DOI] [PubMed] [Google Scholar]
- Bi, E., P. Maddox, D. J. Lew, E. D. Salmon, J. N. McMillan et al., 1998. Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J. Cell Biol. 142: 1301–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, K. C., L. Calzone, A. Csikasz-Nagy, F. R. Cross, B. Novak et al., 2004. Integrative analysis of cell cycle control in budding yeast. Mol. Biol. Cell 15: 3841–3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross, F. R., 2003. Two redundant oscillatory mechanisms in the yeast cell cycle. Dev. Cell 4: 741–752. [DOI] [PubMed] [Google Scholar]
- Cross, F. R., V. Archambault, M. Miller and M. Klovstad, 2002. Testing a mathematical model of the yeast cell cycle. Mol. Biol. Cell 13: 52–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirick, L., T. Bohm and K. Nasmyth, 1995. Roles and regulation of Cln-Cdc28 kinases at the start of the cell cycle of Saccharomyces cerevisiae. EMBO J. 14: 4803–4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein, C. B., and F. R. Cross, 1992. CLB5: a novel B cyclin from budding yeast with a role in S phase. Genes Dev. 6: 1695–1706. [DOI] [PubMed] [Google Scholar]
- Fitch, I., C. Dahmann, U. Surana, A. Amon, K. Nasmyth et al., 1992. Characterization of four B-type cyclin genes of the budding yeast Saccharomyces cerevisiae. Mol. Biol. Cell 3: 805–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell, L. H., and M. W. Unger, 1977. Unequal division in Saccharomyces cerevisiae and its implications for the control of cell division. J. Cell Biol. 75: 422–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge, A., and M. Mendenhall, 1999. The cyclin-dependent kinase inhibitory domain of the yeast Sic1 protein is contained within the C-terminal 70 amino acids. Mol. Gen. Genet. 262: 55–64. [DOI] [PubMed] [Google Scholar]
- Jacobson, M. D., S. Gray, M. Yuste-Rojas and F. R. Cross, 2000. Testing cyclin specificity in the exit from mitosis. Mol. Cell. Biol. 20: 4483–4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp, D., L. Bhoite, D. J. Stillman and K. Nasmyth, 1996. The transcription factor Swi5 regulates expression of the cyclin kinase inhibitor p40SIC1. Mol. Cell. Biol. 16: 5701–5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib, K., J. F. Diffley and S. E. Kearsey, 1999. G1-phase and B-type cyclins exclude the DNA-replication factor Mcm4 from the nucleus. Nat. Cell Biol. 1: 415–422. [DOI] [PubMed] [Google Scholar]
- Lew, D. J., and S. I. Reed, 1993. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J. Cell Biol. 120: 1305–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malek, N. P., H. Sundberg, S. McGrew, K. Nakayama, T. R. Kyriakides et al., 2001. A mouse knock-in model exposes sequential proteolytic pathways that regulate p27Kip1 in G1 and S phase. Nature 413: 323–327. [DOI] [PubMed] [Google Scholar]
- Mateus, C., and S. V. Avery, 2000. Destabilized green fluorescent protein for monitoring dynamic changes in yeast gene expression with flow cytometry. Yeast 16: 1313–1323. [DOI] [PubMed] [Google Scholar]
- Mendenhall, M. D., 1993. An inhibitor of p34CDC28 protein kinase activity from Saccharomyces cerevisiae. Science 259: 216–219. [DOI] [PubMed] [Google Scholar]
- Miller, M. E., and F. R. Cross, 2001. Cyclin specificity: How many wheels do you need on a unicycle? J. Cell Sci. 114: 1811–1820. [DOI] [PubMed] [Google Scholar]
- Moffat, J., and B. Andrews, 2004. Late-G1 cyclin-CDK activity is essential for control of cell morphogenesis in budding yeast. Nat. Cell Biol. 6: 59–66. [DOI] [PubMed] [Google Scholar]
- Morgan, D. O., and J. M. Roberts, 2002. Oscillation sensation. Nature 418: 495–496. [DOI] [PubMed] [Google Scholar]
- Moriya, H., Y. Shimizu-Yoshida and H. Kitano, 2006. In vivo robustness analysis of cell division cycle genes in Saccharomyces cerevisiae. PLoS Genet. 2: e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg, D., R. Muller and M. Funk, 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156: 119–122. [DOI] [PubMed] [Google Scholar]
- Nash, P., X. Tang, S. Orlicky, Q. Chen, F. B. Gertler et al., 2001. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 414: 514–521. [DOI] [PubMed] [Google Scholar]
- Nasmyth, K., 1996. At the heart of the budding yeast cell cycle. Trends Genet. 12: 405–412. [DOI] [PubMed] [Google Scholar]
- Nasmyth, K., and T. Hunt, 1993. Cell cycle: dams and sluices. Nature 366: 634–635. [DOI] [PubMed] [Google Scholar]
- Newman, J. R., S. Ghaemmaghami, J. Ihmels, D. K. Breslow, M. Noble et al., 2006. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature 441: 840–846. [DOI] [PubMed] [Google Scholar]
- Nishizawa, M., M. Kawasumi, M. Fujino and A. Toh-e, 1998. Phosphorylation of sic1, a cyclin-dependent kinase (Cdk) inhibitor, by Cdk including Pho85 kinase is required for its prompt degradation. Mol. Biol. Cell 9: 2393–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlicky, S., X. Tang, A. Willems, M. Tyers and F. Sicheri, 2003. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell 112: 243–256. [DOI] [PubMed] [Google Scholar]
- Petroski, M. D., and R. J. Deshaies, 2003. a Context of multiubiquitin chain attachment influences the rate of Sic1 degradation. Mol. Cell 11: 1435–1444. [DOI] [PubMed] [Google Scholar]
- Petroski, M. D., and R. J. Deshaies, 2003. b Redundant degrons ensure the rapid destruction of Sic1 at the G1/S transition of the budding yeast cell cycle. Cell Cycle 2: 410–411. [PubMed] [Google Scholar]
- Schwab, M., A. S. Lutum and W. Seufert, 1997. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell 90: 683–693. [DOI] [PubMed] [Google Scholar]
- Schwob, E., T. Bohm, M. D. Mendenhall and K. Nasmyth, 1994. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell 79: 233–244. [DOI] [PubMed] [Google Scholar]
- Sherr, C. J., and J. M. Roberts, 1999. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 13: 1501–1512. [DOI] [PubMed] [Google Scholar]
- Thornton, B. R., and D. P. Toczyski, 2003. Securin and B-cyclin/CDK are the only essential targets of the APC. Nat. Cell Biol. 5: 1090–1094. [DOI] [PubMed] [Google Scholar]
- Tyers, M., 1996. The cyclin-dependent kinase inhibitor p40SIC1 imposes the requirement for Cln G1 cyclin function at Start. Proc. Natl. Acad. Sci. USA 93: 7772–7776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma, R., R. S. Annan, M. J. Huddleston, S. A. Carr, G. Reynard et al., 1997. a Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science 278: 455–460. [DOI] [PubMed] [Google Scholar]
- Verma, R., R. M. Feldman and R. J. Deshaies, 1997. b SIC1 is ubiquitinated in vitro by a pathway that requires CDC4, CDC34, and cyclin/CDK activities. Mol. Biol. Cell 8: 1427–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin, R., S. Prinz and A. Amon, 1997. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science 278: 460–463. [DOI] [PubMed] [Google Scholar]
- Wittenberg, C., and S. I. Reed, 2005. Cell cycle-dependent transcription in yeast: promoters, transcription factors, and transcriptomes. Oncogene 24: 2746–2755. [DOI] [PubMed] [Google Scholar]
- Zachariae, W., and K. Nasmyth, 1999. Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev. 13: 2039–2058. [DOI] [PubMed] [Google Scholar]
- Zachariae, W., M. Schwab, K. Nasmyth and W. Seufert, 1998. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science 282: 1721–1724. [DOI] [PubMed] [Google Scholar]