Abstract
The leukemic fusion protein AML1-ETO occurs frequently in human acute myeloid leukemia (AML) and has received much attention over the past decade. An initial model for its pathogenetic effects emphasized the conversion of a hematopoietic transcriptional activator, RUNX1 (or AML1), into a leukemogenic repressor which blocked myeloid differentiation at the level of target gene regulation. This view has been absorbed into a larger picture of AML1-ETO pathogenesis, encompassing dysregulation of hematopoietic stem cell homeostasis at several mechanistic levels. Recent reports have highlighted a multifaceted capacity of AML1-ETO directly to inhibit key hematopoietic transcription factors that function as tumor suppressors at several nodal points during hematopoietic differentiation. A new model is presented in which AML1-ETO coordinates expansion of the stem cell compartment with diminished lineage commitment and with genome instability.
1. Introduction
AML1-ETO, the product of the chromosomal translocation t(8;21)(q22;q22) usually occurs in acute myeloid leukemia (AML) with granulocytic differentiation, FAB M2 subtype [1]. The fusion protein comprises the conserved runt homology (rhd) from the hematopoietic transcription factor RUNX1 (originally known as AML1), encoded on chromosome 21, joined to the majority of the ETO repressor protein, encoded on chromosome 8. ETO possesses four phylogenetically conserved segments homologous to the Drosophila Nervy protein and appears to function as a scaffolding factor for the assembly of protein complexes. Two of these ETO segments, Nervy Homology Region 2 (NHR2) and Nervy Homolgy Region 4 (NHR4) serve as docking sites for corepressors including mSin3A, SMRT and histone deacetylases (HDAC) [2]. AML1-ETO, through its rhd, retains the ability to bind RUNX1 DNA binding sites and through the ETO moiety gains the capacity to recruit additional cofactors, mostly corepressors. Thus AML1-ETO has been predicted to act as a transcriptional repressor for RUNX1 target genes.
From a clinical standpoint, AML with t(8;21) tends to occur in early adulthood and has a better prognosis than most other types of AML. A consistent pathological feature consists of enhanced marrow granulopoiesis with inhibition of erythropoiesis. The bone marrow morphology consists of blasts with solitary Auer rods and abnormal myelocytes with increased cytoplasmic granulation. As will be discussed below, the impaired erythropoiesis seen in clinical cases of t(8;21)-positive AML has been recapitulated both in animal and in ex vivo model systems [3]
Experimental and clinical studies underscore the importance of secondary mutations or events in AML1-ETO mediated leukemogenesis. For example, mice expressing AML1-ETO require exposure to a mutagen like N-ethyl-N-nitrosourea or coexpression of a constitutively active tyrosine kinase such as the TEL-PDGFRβ fusion in order to develop AML [4,5]. Similarly, the clinical data show a high incidence of secondary genetic alterations affecting tyrosine kinase signal transduction pathways in patients with t(8;21)-positive AML. Wang et al. observed that ∼50% of their patients with t(8;21)-positive AML had point mutations in the receptor tyrosine kinase gene C-KIT [6]. The mutations caused constitutive activation and frequently affected residues N822 or D816. They also found the majority of t(8;21)-positive leukemias specifically to overexpress c-Kit regardless of mutational status. In another cohort, Schessl and colleagues found more than one third of their patients with t(8;21)-positive AML to harbour activating mutations in either FLT3, C-KIT or NRAS [7]. In addition, this study demonstrated that AML1-ETO could collaborate with mutant FLT3 to induce leukemia in transplanted mice. These observations highlight the critical synergy between AML1-ETO expression and aberrant tyrosine kinase signaling in leukemic progression. The secondary mutations associated with t(8;21)-positive AML have therapeutic implications, as leukemias carrying the c-Kit N822K mutation may respond to tyrosine kinase inhibitors such as Imatinib [6]. In contrast, those leukemias bearing c-Kit D816 mutations are resistant to Imatinib [6], but may respond to the SRC/ABL inhibitor Dasatinib (BMS-354825) or to the tyrosine kinase inhibitor Nilotinib (AMN 107) [8,9].
2. The classical model of leukemogenesis by AML1-ETO
Initial models for the pathogenetic role of AML1-ETO drew from landmark discoveries made on another leukemia-associated transcription factor fusion, PML-RARα, involved in acute promyelocytic leukemia (APL). As a result of the t(15;17) chromosomal translocation associated with APL, a transcriptional activator normally involved in programming granulocytic differentiation (RARα: Retinoic Acid Receptor α) underwent conversion to a repressor by fusion to a heterologous sequence (PML). In an analogous model, RUNX1 normally functioned as a trancriptional activator, promoting granulocytic differentiation through the coordinated upregulation of lineage-specific target genes such as myeloperoxidase. Through fusion with ETO, as a consequence of the t(8;21) chromosomal translocation, RUNX1 underwent a metamorphosis from activator to repressor, now downregulating all of its target genes involved in granulocytic differentiation. Multiple findings supported this model: 1) wild type RUNX1 bound transcriptional coactivators, such as p300/CBP, leading to enhanced target gene transactivation [10]; 2) ETO recruited corepressors including HDACs1-3, NCoR, SMRT and mSin3A [11-13]; 3) AML1-ETO repressed tumor suppressor genes, such as p14ARF, which were normally activated by RUNX1 [14]; 4) treatment of cell lines with HDAC (deacetylase) and DNMT (DNA methyl transferase) inhibitory compounds relieved the repressive effects of AML1-ETO on chromatin, gene transcription, and cell differentiation [15-17]; and 5) in an elegant, genetically-based model of Drosophila eye development, AML1-ETO behaved as a classical, direct transcriptional repressor [18]. In this fascinating study of Wildonger and Mann, AML1-ETO expression produced an abnormal eye phenotype that was notably distinct from that associated with loss of Lozenge (Lz), a Drosophila homolog of RUNX1. Fusion of the rhd with ETO was necessary to obtain this phenotype. Overexpression of the CBFβ homologs, Bro or Bgb, enhanced the effects of AML1-ETO, while their downregulation suppressed the inhibitory effect of AML1-ETO on eye development. As expected of a classical, direct transcriptional repressor, AML1-ETO silenced the expression of Lz target genes regardless of whether they were positively or negatively regulated by Lz, e.g. PAX2 and dpn. In addition, fusion of Lz to the constitutive repressor Engrailed produced an eye phenotype similar to that obtained with AML1-ETO, further supporting a classical model of direct target gene repression.
3. Challenges to the classical model: gene expression profiles, physiologic model systems, potential dispensability of corepressor recruitment
Categorization of transcripts affected by AML1-ETO has provided new layers of mechanistic complexity which challenge the sufficiency of direct target gene repression. In an initial study using differential display analysis, 24 AML1-ETO target genes were identified, only 10 of which displayed regulation by wild type RUNX1, thus providing the first evidence that AML1-ETO could affect expression of genes not normally under the control of RUNX1 [19]. Subsequent microarray profiles found AML1-ETO to activate as many genes as it repressed. Many genes activated by AML1-ETO were involved in stem cell self-renewal. Using inducible expression in the U937 cell line, Alcalay et al. discovered AML1-ETO induction of Jagged1, a Notch ligand known to drive stem cell proliferation [20]. Jagged upregulation triggered activation of the Notch pathway, and primary AML blasts with t(8;21) showed expression of the Notch target gene HES1. In a similar U937-based inducible system, Muller-Tidow and colleagues identified Plakoglobin as a target gene activated by AML1-ETO [21]. This upregulation occurred through direct transcriptional activation of the Plakoglobin promoter by AML1-ETO, a unique example of promoter activation by a putative transcriptional repressor. Plakoglobin mediates Wnt signalling through its ability to bind TCF/LEF and activate the Wnt target genes C-MYC and CYCLIN D1. Notably, Wnt signalling, like that of Notch, promotes stem cell self-renewal. Muller-Tidow et al. found that AML1-ETO also activated expression of β-catenin, another Wnt mediator that upregulates c-Myc and cyclin D1. In a separate study using retrovirally transduced human CD34+ hematopoietic progenitors, microarray analysis identified the nerve growth factor receptor TRKA as yet another positive target of AML1-ETO [22]. Real time qRT-PCR confirmed a significant increase in TRKA transcripts in AML cases with t(8;21). Enforced AML1-ETO expression rendered human CD34+ cells responsive to the growth promoting effects of exogenous nerve growth factor combined with IL-3. Thus a common theme emerging from the gene expression profiles is that AML1-ETO may drive expansion of early multipotent progenitors through the paradoxical upregulation of target genes.
Assays of AML1-ETO function employing either transduction of primary hematopoietic progenitors or inducible transgenic mice have confirmed its role in early multipotent progenitor expansion [4,23-26]. This role is more subtle than the differentiation blockade observed in transfected cell lines and argues against a simple unimodal model of transcriptional repression of RUNX1 target genes. Furthermore, in vivo models of AML1-ETO leukemogenesis have shown no disease responsiveness to HDAC inhibitors [5], raising questions regarding the relevance of HDAC recruitment, at least in the latter stages of the disease.
Additional evidence challenging the importance of corepressor recruitment has come from two recent studies dissecting domains of AML1-ETO involved in transformation of primary murine HSC. Yan and colleagues identified an incidental mutant of AML1-ETO which had acquired potent leukemogenic activity in HSC transduction/transplantation [27]. Suprisingly, this mutant, a single base expansion within a coding polyA tract, truncated AML1-ETO at amino acid 552 and added 4 novel carboxy terminal amino acids. The resultant fusion retained NHR1 and NHR2 of ETO but lost NHR3 and NHR4, the last of which serves as a critical docking site for NCoR/SMRT. A similar C-terminal truncation, resulting from alternative splicing, has been identified in many patients with t(8;21)-positive AML [28]. These results suggest that corepressor recruitment could actually prevent rather than promote AML1-ETO-mediated leukemogenesis. In the second study, the Bushweller and Speck labs applied detailed structure-function analyses to the NHR2 domain and concluded that the oligomerization of NHR2, rather than its ability to recruit mSin3A and HDACs, correlated most closely with the capacity of AML1-ETO to transform primary murine hematopoietic progenitors [29]. While both studies do not categorically rule out a role for RUNX1 target gene repression by copreressor recruitment, they emphasize that AML1-ETO leukemogenesis clearly depends on alternative pathways in primary cell systems.
4. Launching a multipronged attack on hematopoietic tumor suppressor proteins: AML1-ETO directly targets E2A, PU.1, C/EBPα, and GATA-1
An emerging theme in the pathogenesis of acute leukemias is the loss of function of lineage-programming transcription factors. Complete loss of function precludes cellular survival and subsequent transformation, but partial loss of function, as recently highlighted by Rosenbauer et al., perturbs stem cell homeostasis enough to promote leukemogenesis [30]. In particular, lineage commitment defects induce expansion of a self-renewing progenitor compartment, which may then be poised for leukemic transformation through additional mutations. Through direct protein-protein interactions repressing several of the key hematopoietic transcription factors, AML1-ETO may thereby expand stem cell pools. And through repression of genes involved in DNA base excision repair [20], AML1-ETO may promote genetic instability within this expanded stem cell pool. Below are listed four hematopoietic transcription factors with the following critical features: 1) expressed at early stages in hematopoietic development, 2) implicated in early lineage commitment events, 3) known to function as tumor suppressors in human leukemia, 4) involved in positive autoregulation during normal differentiation and 5) functionally repressed by AML1-ETO through protein-protein interactions.
E proteins, consisting of E2A, HEB, and E2-2, constitute a subset of basic helix-loop-helix (bHLH) transcription factors characterized by broad tissue expression and heterodimerization with a wide array of tissue-specific bHLH factors. Homodimeric E proteins participate in the programming of B cell ontogeny and T cell maturation. E2A−/− mice display an early arrest in B cell development and a high incidence of T cell malignancies [31]. In human disease, ectopic thymocyte expression of the bHLH oncoprotein SCL/tal promotes T cell acute lymphoblastic leukemia through a mechanism involving E protein sequestration [31]. Recent work from the Roeder lab identified an interaction of AML1-ETO with the AD1 activation domain of E proteins, leading to their repression through displacement of p300/CBP coactivators [32]. The interacting domain within ETO consisted of the NHR1, which bears homology to the general transcription factor TAF4. Whether NHR1 actually contributes to AML1-ETO transformation of primary cells remains untested. Paradoxically, human AML with t(8;21) is characterized by the aberrant expression of the lymphoid markers TdT and CD19, both of which are positively regulated by E2A [33-35].
PU.1, an ets domain transcription factor, programs development of neutrophil, monocyte, and B cell lineages, all of which are deficient in PU.1−/− mice. Mice with deletion of the PU.1 upstream regulatory element (URE) manifest an 80% reduction in PU.1 expression accompanied by a high incidence of rapid-onset AML, a striking contrast to the non-leukemic phenotype of PU.1 null mice [36]. In one series of human AMLs, ∼7% of cases (9/126) displayed heterozygous PU.1 coding mutations, many of which disrupted transcriptional function [37]. Notably, none of the PU.1 mutant cases in this series possessed t(8;21). In subsequent studies of AML cases, mutations of PU.1 have not been identified, suggesting that this event may be rare [38]. In vitro, Vangala et al. found AML1-ETO to bind directly to the β3β4 domain of PU.1 causing displacement of the coactivator c-Jun [39]. The resultant blockade of PU.1 transactivation prevented PU.1-mediated autoregulation, growth arrest, and monocytic differentiation in transduced murine marrow cells. Furthermore, overexpression of PU.1 in the t(8;21)-positive Kasumi cell line induced growth arrest and monocytic differentiation.
C/EBPα, a 42 kd leucine zipper transcriptional activator, programs development of neutrophils, which undergo maturation arrest in C/EBPα−/− mice [40]. Knock-in mice expressing an amino terminal truncated 30 kd isoform (p30) lacking TAD1 develop myeloid leukemia, again in contrast to the non-malignant phenotype of their null counterparts [30]. Nevertheless, C/EBPα−/− mice do display an expanded HSC compartment reminiscent of that seen in AML1-ETO transduced progenitors [30]. In a series of 137 patients with AML, 10 samples (∼7%) displayed heterozygous coding mutations which in 5 cases led to the expression of the p30 dominant negative isoform [41]. Functional assays confirmed repressive effects of the AML-associated C/EBPα mutations on DNA binding, transactivation of granulocytic target genes, and induction of G-CSF receptor expression in U937 cells. Subsequent surveys have confirmed a C/EBPA mutation rate of ∼9% in AML with mutations either leading to p30 expression or causing disruption of the carboxy terminal basic leucine zipper region [42-46]. Notably, throughout all of these studies, none of the AML cases expressing mutant C/EBPα possessed the t(8;21) abnormality. Conversely, t(8;21)-positive AMLs uniformly expressed >5-fold lower levels of C/EBPα transcripts and protein, as compared with other types of AML [47]. This effect resulted from an interruption of C/EBPα autoregulation caused by physical interaction with AML1-ETO. As with PU.1, enforced expression in Kasumi cells induced differentiation, along the neutrophil lineage in the case of C/EBPα.
GATA-1, an X chromosome encoded zinc finger transcription factor, programs development along erythroid, megakaryocytic, mast cell, and eosinophil lineages. The most prominent feature of GATA-1 −/− mice is the embryonic lethality caused by early erythroblast developmental arrest and apoptosis [48]. A knockdown allele expressing 5% of normal GATA-1 levels, in the context of a female heterozygote (in order to bypass embryonic lethality), is associated with development of acute leukemias [49]. These leukemias do not express erythroid markers but rather consist of primitive c-Kit+ myeloid cells or of CD19+ B lineage cells. In human AMLs, GATA-1 mutations have been found almost exclusively in association with Down syndrome acute megakaryoblastic leukemias (DS-AMKL). These mutations consistently lead to the production of an amino terminally truncated 40 kd isoform lacking an ill-defined transcriptional activation domain [50]. We have found AML1-ETO to function as a potent inhibitor of GATA-1 transcriptional activation [3]. Correspondingly, transfection of AML1-ETO into primary human CD34+ hematopoietic progenitors efficiently blocked erythroid lineage commitment, normally denoted by CD36 and GPA upregulation coupled with CD34 and CD13 downregulation [3]. This effect of AML1-ETO was reminiscent of the erythroid blockade uniquely associated with t(8;21)-positive human AMLs and of the erythroid inhibition seen in several murine models of AML1-ETO transformation [51-54]. AML1-ETO employed a novel mechanism in its inhibition of erythropoiesis, interfering with the acetylation of GATA-1 mediated by p300/CBP. Thus, mutants of GATA-1 with enforced docking and acetylation by p300/CBP overrode the inhibitory effects of AML1-ETO on primary erythropoiesis [3].
5. An integrated mechanism of AML1-ETO action on HSC: filling the pool while closing the overflow valves
Given the multifaceted effects of AML1-ETO on transcriptional regulation in hematopoiesis, we propose a leukemogenic mechanism that couples induction of HSC expansion (“filling the pool”) with closure of the escape routes of lineage commitment (“closing the overflow valves”). This combination of effects will circumvent homeostatic mechanisms to restrict the HSC pool and will cooperate with the loss of DNA base excision repair to promote eventual generation of fully transformed leukemic stem cells (Figure 1). RUNX1 haploinsufficiency causes expansion of multilineage progenitors both in human disease as well as in murine model systems [55,56]. Therefore the antagonism of wild type RUNX1 by AML1-ETO, in part through the classical gene repression pathway, could provide an initial impetus for HSC expansion (Figure 2). However, this effect could be counterbalanced by homeostatic mechanisms that accelerate HSC lineage commitment in order to rectify pool size. Recent models of the hematopoietic hierarchy suggest that lymphoid (LMPP), myeloid (CMP), and megakaryocytic/erythroid (MEP) progenitors may all emerge directly from a primitive highly proliferative progenitor/HSC compartment [57]. As an example, the upregulation of GATA-1 within HSC promotes direct transition to MEP and offers one potential valve for lowering the HSC pool. Critical for this valve function is the autoregulatory aspect of GATA-1, creating a self-amplifying circuit that locks cells into their MEP fates, i.e. propping the valve open. However, mathematical modeling has shown an exquisite sensitivity of self-amplifying transcriptional circuits to inhibitors, “circuit destabilization” [58]. Thus AML1-ETO repression of GATA-1 may initially disrupt its positive autoregulation and thereby undermine its entire transcriptional program. Notably, a critical component of the GATA-1 transcriptional program consists of downregulating c-Kit expression [59]. Abnormally sustained c-Kit expression in GATA-1-deficient cells may then predispose these genetically unstable cells to positive selection for receptor-activating mutations, accounting for the high frequency of C-KIT mutation in t(8;21)-positive AMLs (Figure 1). One can envision analogous consequences accruing from repression of PU.1 and C/EBPα, blocking the transition to CMP, and from the repression of E proteins, blocking the transition to LMPP (Figure 2). The inhibitory effects of AML1-ETO on commitment to the various lineages most likely are incomplete and the rising of the HSC or early progenitor pool is most likely very gradual, explaining the lack of a striking phenotype in many murine models. Nevertheless, this relentless expansion in the setting of genetic instability provides a platform for acquisition of secondary cooperating mutations, such as receptor tyrosine kinase activation.
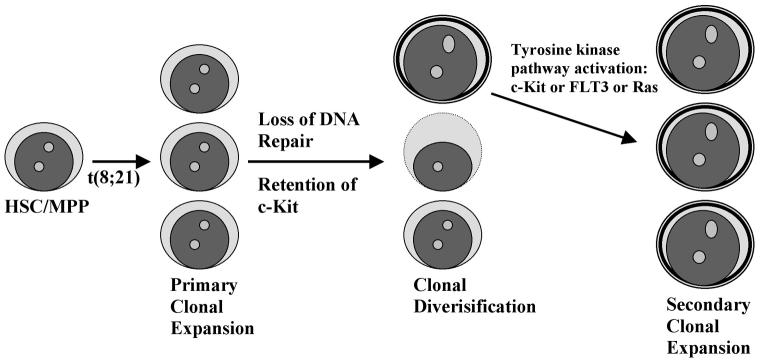
Fig. 1.
Diagram of the multistep clonal progression associated with t(8;21)-positive leukemogenesis. In this model, t(8;21) occurs as the initial event within a cellular compartment encompassing the hematopoietic stem cell (HSC) or a multipotent progenitor (MPP). The resultant expression of the AML1-ETO fusion protein causes an initial expansion of this HSC/MPP compartment. The mechanism for this expansion is depicted below in Fig. 2. Additional consequences of AML1-ETO expression consist of failure to downregulate c-Kit and impairment of DNA base excision repair. Defects in DNA repair ultimately produce mutagenic clonal diversification, and the continued high level expression of c-Kit provides a target for tyrosine kinase activating mutations. Other components of tyrosine kinase signaling may also undergo selection for activating mutations. As a next step, the combination of AML1-ETO expression and constitutively active tyrosine kinase pathway signaling induces a secondary clonal expansion, which may eventuate in acute myeloid leukemia (AML).
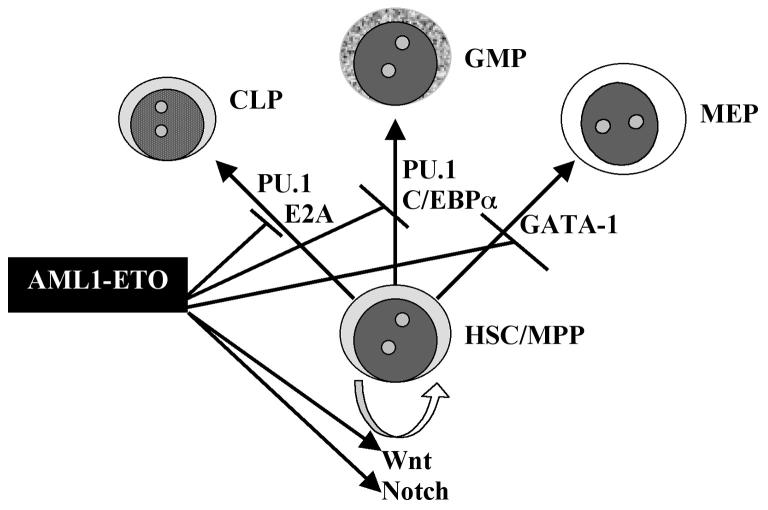
Fig. 2.
A postulated mechanism for AML1-ETO expansion of HSC/MPP. Homeostasis of the HSC/MPP compartment depends on a balance between self-renewal, programmed by Wnt and Notch signalling, and lineage commitment programmed by the indicated transcription factors. In particular, GATA-1 functions as an inducer of megakaryocyteerythroid progenitors (MEP); PU.1 and C/EBPα promote the development of granulocyte-monocyte progenitors (GMP); and E2A and PU.1 contribute to common lymphocyte progenitor (CLP) commitment. AML1-ETO acts at all of the indicated steps, promoting HSC/MPP self-renewal through activation of Wnt and Notch pathways and blocking commitment to the various lineages through inhibitory interactions with differentiation-promoting hematopoietic transcription factors, including GATA-1, PU.1, C/EBPα, and E2A. Each of these transcription factors has previously been demonstrated to function as a tumor suppressor, both in human disease and in murine models.
Acknowledgments
Kamal Elagib is supported by the NIH grant, T32 CA009109 (“Cancer Research Training in Molecular Biology”), and Adam Goldfarb is supported in part by NIH grants CA100057 and CA93735.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brunning RD, McKenna RW. Tumors of the Bone Marrow. 1 ed. Armed Forces Institute of Pathology; Washington, D. C.: 1994. Acute Myeloid Leukemias; pp. 22–100. [Google Scholar]
- 2.Peterson LF, Zhang D-E. The 8;21 translocation in leukemogenesis. Oncogene. 2004;23:4255–4262. doi: 10.1038/sj.onc.1207727. [DOI] [PubMed] [Google Scholar]
- 3.Choi Y, Elagib KE, Delehanty LL, Goldfarb AN. Erythroid inhibition by the leukemic fusion AML1-ETO is associated with impaired acetylation of the major erythroid transcription factor GATA-1. Cancer Res. 2006;66:2990–2996. doi: 10.1158/0008-5472.CAN-05-2944. [DOI] [PubMed] [Google Scholar]
- 4.Higuchi M, O'Brien D, Kumaravelu P, Lenny N, Yeoh E-J, Downing JR. Expression of a conditional AML1-ETO oncogene bypasses embryonic lethality and establishes a murine model of human t(8;21) acute myeloid leukemia. Cancer Cell. 2002;1:63–74. doi: 10.1016/s1535-6108(02)00016-8. [DOI] [PubMed] [Google Scholar]
- 5.Grisolano JL, O'Neal J, Cain J, Tomasson MH. An activated receptor tyrosine kinase, TEL/PDGFβR, cooperates with AML1/ETO to induce acute myeloid leukemia in mice. Proc. Natl. Acad. Sci. USA. 2003;100:9506–9511. doi: 10.1073/pnas.1531730100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y-Y, Zhou G-B, Yin T, Chen B, Shi J-Y, Liang W-X, et al. AML1-ETO and C-KIT mutation/overexpression in t(8;21) leukemia: Implication in stepwise leukemogenesis and response to Gleevec. Proc. Natl. Acad. Sci. USA. 2005;102:1104–1109. doi: 10.1073/pnas.0408831102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schessl C, Rawat VPS, Cusan M, Deshpande A, Kohl TM, Rosten PM, et al. The AML1-ETO fusion gene and the FLT3 length mutation collaborate in inducing acute leukemia in mice. J. Clin. Invest. 2005;115:2159–2168. doi: 10.1172/JCI24225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shah NP, Lee FY, Luo R, Jiang Y, Donker M, Akin C. Dasatinib (BMS-354825) inhibits KITD816V, an imatinib-resistant activating mutation that triggers neoplastic growth in most patients with systemic mastocytosis. Blood. 2006;108:286–291. doi: 10.1182/blood-2005-10-3969. [DOI] [PubMed] [Google Scholar]
- 9.von Bubnoff N, Gorantla SH, Kancha RK, Lordick F, Peschel C, Duyster J. The systemic mastocytosis-specific activating cKit mutation D816V can be inhibited by the tyrosine kinase inhibitor AMN 107. Leukemia. 2005;19:1670–1671. doi: 10.1038/sj.leu.2403887. [DOI] [PubMed] [Google Scholar]
- 10.Kitabayashi I, Yokoyama A, Shimizu K, Ohki M. Interaction and functional cooperation of the leukemia-associated factors AML-1 and p300 in myeloid cell differentiation. EMBO J. 1998;17:2994–3004. doi: 10.1093/emboj/17.11.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amann JM, Nip J, Strom DK, Lutterbach B, Harada H, Lenny N, et al. ETO, a target of t(8;21) in acute leukemia, makes distinct contacts with muliple histone deacetylases and binds mSin3A through its oligomerization domain. Mol. Cell. Biol. 2001;21:6470–6483. doi: 10.1128/MCB.21.19.6470-6483.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hildebrand D, Tiefenbach J, Heinzel T, Grez M, Maurer AB. Multiple regions of ETO cooperate in transcriptional repression. J. Biol. Chem. 2001;276:9889–9895. doi: 10.1074/jbc.M010582200. [DOI] [PubMed] [Google Scholar]
- 13.Zhang J, Hug BA, Huang EY, Chen CW, Gelmetti V, Maccarana M, et al. Oligomerization of ETO is obligatory for corepressor interaction. Mol. Cell. Biol. 2001;21:156–163. doi: 10.1128/MCB.21.1.156-163.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Linggi B, Muller-Tidow C, Van de Locht L, Hu M, Nip J, Serve H, et al. The t(8;21) fusion protein, AML1-ETO, specifically represses the transcription of the p14ARF tumor suppressor in acute myeloid leukemia. Nature Med. 2002;8:743–750. doi: 10.1038/nm726. [DOI] [PubMed] [Google Scholar]
- 15.Wang J, Saunthararajah Y, Redner RL, Liu JM. Inhibitors of histone deacetylase relieve ETO-mediated repression and induce differentiation of AML1-ETO leukemia cells. Cancer Res. 1999;59:2766–2769. [PubMed] [Google Scholar]
- 16.Klisovic ML, Maghraby EA, Parthun MR, Guimond M, Sklenar AR, Whitman SP, et al. Depsipeptide (FR 901228) promotes histone acetylation, gene transcription, apoptosis and its activity is enhanced by DNA methyltransferase inhibitors in AML1/ETO-positive leukemic cells. Leukemia. 2003;17:350–358. doi: 10.1038/sj.leu.2402776. [DOI] [PubMed] [Google Scholar]
- 17.Liu S, Shen T, Huynh L, Klisovic ML, Rush LJ, Ford JL, et al. Interplay of RUNX1/MTG8 and DNA methyltransferase 1 in acute myeloid leukemia. Cancer Res. 2005;65:1277–1284. doi: 10.1158/0008-5472.CAN-04-4532. [DOI] [PubMed] [Google Scholar]
- 18.Wildonger J, Mann RS. The t(8;21) translocation converts AML1 into a constitutive transcriptional repressor. Dev. 2005;132:2263–2272. doi: 10.1242/dev.01824. [DOI] [PubMed] [Google Scholar]
- 19.Shimada H, Ichikawa H, Nakamura S, Katsu R, Iwasa M, Kitabayashi I, Ohki M. Analysis of genes under the downstream control of the t(8;21) fusion protein AML1-MTG8: overexpression of the TIS11b (ERF-1, cMG1) gene induces myeloid cell proliferation in response to G-CSF. Blood. 2000;96:655–663. [PubMed] [Google Scholar]
- 20.Alcalay M, Meani N, Gelmetti V, Fantozzi A, Fagioli M, Orleth A, et al. Acute myeloid leukemia fusion proteins deregulate genes involved in stem cell maintenance and DNA repair. J. Clin. Invest. 2003;112:1751–1761. doi: 10.1172/JCI17595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muller-Tidow C, Steffen B, Cauvet T, Tickenbrock L, Ji P, Diederichs S, et al. Translocation products in acute myeloid leukemia activate the Wnt signaling pathway in hematopoietic cells. Mol. Cell. Biol. 2004;24:2890–2904. doi: 10.1128/MCB.24.7.2890-2904.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mulloy JC, Jankovic V, Wunderlich M, Delwel R, Cammenga J, Krejci O, et al. AML1-ETO fusion protein up-regulates TRKA mRNA expression in human CD34+ cells, allowing nerve growth factor-induced expansion. Proc. Natl. Acad. Sci. USA. 2005;102:4016–4021. doi: 10.1073/pnas.0404701102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mulloy JC, Cammenga J, MacKenzie KL, Berguido FJ, Moore MAS, Nimer SD. The AML1-ETO fusion protein promotes the expansion of human hematopoietic stem cells. Blood. 2002;99:15–23. doi: 10.1182/blood.v99.1.15. [DOI] [PubMed] [Google Scholar]
- 24.Basecke J, Schwieger M, Griesinger F, Schiedlmeier B, Wulf G, Trumper L, Stocking C. AML1/ETO promotes the maintenance of early hematopoietic progenitors in NOD/SCID mice but does not abrogate their lineage specific differentiation. Leukemia & Lymphoma. 2005;46:265–272. doi: 10.1080/10428190400010767. [DOI] [PubMed] [Google Scholar]
- 25.Tonks A, Pearn L, Tonks AJ, Pearce L, Hoy T, Phillips S, et al. The AML1-ETO fusion gene promotes extensive self-renewal of human primary erythroid cells. Blood. 2002;101:624–632. doi: 10.1182/blood-2002-06-1732. [DOI] [PubMed] [Google Scholar]
- 26.de Guzman CG, Warren AJ, Zhang Z, Gartland L, Erickson P, Drabkin H, et al. Hematopoietic stem cell expansion and distinct myeloid developmental abnormalities in a murine model of the AML1-ETO translocation. Mol. Cell. Biol. 2002;22:5506–5517. doi: 10.1128/MCB.22.15.5506-5517.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan M, Burel SA, Peterson LF, Kanbe E, Iwasaki H, Boyapati A, et al. Deletion of an AML1-ETO C-terminal NcoR/SMRT-interacting region strongly induces leukemia development. Proc. Natl. Acad. Sci. USA. 2004;101:17186–17191. doi: 10.1073/pnas.0406702101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan M, Kanbe E, Peterson LF, Boyapati A, Miao Y, Wang Y, et al. A previously unidentified alternatively spliced isoform of t(8;21) transcript promotes leukemogenesis. Nat. Med. 2006;12:945–949. doi: 10.1038/nm1443. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y, Cheney MD, Gaudet JJ, Chruszcz M, Lukasik SM, Sugiyama D, et al. The tetramer structure of the Nervy homology two domain, NHR2, is critical for AML1/ETO's activity. Cancer Cell. 2006;9:249–260. doi: 10.1016/j.ccr.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 30.Rosenbauer F, Koschmieder S, Steidl U, Tenen DG. Effect of transcription-factor concentrations on leukemic stem cells. Blood. 2005;106:1519–1524. doi: 10.1182/blood-2005-02-0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Massari ME, Murre C. Helix-Loop-Helix Proteins: Regulators of Transcription in Eucaryotic Organisms. Mol. Cell. Biol. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang J, Kalkum M, Yamamura S, Chait BT, Roeder RG. E protein silencing by the leukemogenic AML1-ETO fusion protein. Science. 2004;305:1286–1289. doi: 10.1126/science.1097937. [DOI] [PubMed] [Google Scholar]
- 33.Brunning RD, Matutes E, Flandrin G, Vardiman J, Bennett J, Head D, Harris NL. Acute myeloid leukaemia with recurrent genetic abnormalities. In: Jaffe ES, Harris NL, Stein H, Vardiman JW, editors. World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues. 1st ed. IARC Press; Lyon, France: 2001. pp. 81–82. [Google Scholar]
- 34.Bain G, Robanus Maandag EC, Izon DJ, Amsen D, Kruisbeek AM, Weintraub BC, et al. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell. 1994;79:885–892. doi: 10.1016/0092-8674(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 35.Choi JK, Shen C-P, Radomska HS, Eckhardt LA, Kadesch T. E47 activates the Ig-heavy chain and TdT loci in non-B cells. EMBO J. 1996;15:5014–5021. [PMC free article] [PubMed] [Google Scholar]
- 36.Rosenbauer F, Wagner K, Kutok JL, Iwasaki H, Le Beau MM, Okuno Y, et al. Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nat. Genet. 2004;36:624–630. doi: 10.1038/ng1361. [DOI] [PubMed] [Google Scholar]
- 37.Mueller BU, Pabst T, Osato M, Asou N, Johansen LM, Minden MD, et al. Heterozygous PU.1 mutations are associated with acute myeloid leukemia. Blood. 2002;100:998–1007. doi: 10.1182/blood.v100.3.998. [DOI] [PubMed] [Google Scholar]
- 38.Dohner K, Tobis K, Bischof T, Hein S, Schlenk RF, Frohling S, Dohner H. Mutation analysis of the transcription factor PU.1 in youger adults (16-60 years) with acute myeloid leukemia: a study of the AML Study Group Ulm (AMLSG ULM) Blood. 2003;102:3850. doi: 10.1182/blood-2003-08-2654. [DOI] [PubMed] [Google Scholar]
- 39.Vangala RK, Heiss-Neumann MS, Rangatia JS, Singh SM, Schoch C, Tenen DG, et al. The myeloid master regulator transcription factor PU.1 is inactivated by AML1-ETO in t(8;21) myeloid leukemia. Blood. 2003;101:270–277. doi: 10.1182/blood-2002-04-1288. [DOI] [PubMed] [Google Scholar]
- 40.Zhang D-E, Zhang P, Wang ND, Hetherington CJ, Darlington GJ, Tenen DG. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha-deficient mice. Proc. Natl. Acad. Sci. USA. 1997;94:569–574. doi: 10.1073/pnas.94.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pabst T, Mueller BU, Zhang P, Radomska HS, Narravula S, Schnittger S, et al. Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-α (C/EBPα), in acute myeloid leukemia. Nat. Genet. 2001;27:263–270. doi: 10.1038/85820. [DOI] [PubMed] [Google Scholar]
- 42.Gombart AF, Hofmann W-K, Kawano S, Takeuchi S, Krug U, Kwok SH, et al. Mutations in the gene encoding the transcription factor CCAAT/enhancer binding protein α in myelodysplastic syndromes and acute myeloid leukemias. Blood. 2002;99:1332–1340. doi: 10.1182/blood.v99.4.1332. [DOI] [PubMed] [Google Scholar]
- 43.Preudhomme C, Sagot C, Boissel N, Cayuela J-M, Tigaud I, de Botton S, et al. Favorable prognostic significance of CEBPA mutations in patients with de novo acute myeloid leukemia: a study from the Acute Leukemia French Association (ALFA) Blood. 2002;100:2717–2723. doi: 10.1182/blood-2002-03-0990. [DOI] [PubMed] [Google Scholar]
- 44.Snaddon J, Smith ML, Neat M, Cambal-Parrales M, Dixon-McIver A, Arch R, et al. Mutations of CEBPA in acute myeloid leukemia FAB types M1 and M2. Genes Chrom. Cancer. 2003;37:72–78. doi: 10.1002/gcc.10185. [DOI] [PubMed] [Google Scholar]
- 45.Smith ML, Cavenagh JD, Lister A, Fitzgibbon J. Mutation of CEBPA in familial acute myeloid leukemia. N. Engl. J. Med. 2004;351:2403–2407. doi: 10.1056/NEJMoa041331. [DOI] [PubMed] [Google Scholar]
- 46.Nerlov C. C/EBPα mutations in acute myeloid leukemias. Nat. Rev. Cancer. 2004;4:394–400. doi: 10.1038/nrc1363. [DOI] [PubMed] [Google Scholar]
- 47.Pabst T, Mueller BU, Harakawa N, Schoch C, Haferlach T, Behre G, et al. AML1-ETO downregulates the granulocytic differentiation factor C/EBPα in t(8;21) myeloid leukemia. Nat. Med. 2001;7:444–451. doi: 10.1038/86515. [DOI] [PubMed] [Google Scholar]
- 48.Pevny L, Simon MC, Robertson E, Klein WH, Tsai S-F, D'Agati V, et al. Erythroid differentiation in chimeric mice blocked by a targeted mutation of the gene for transcription factor GATA-1. Nature. 1991;349:257–260. doi: 10.1038/349257a0. [DOI] [PubMed] [Google Scholar]
- 49.Shimizu R, Kuroha T, Ohneda O, Pan X, Ohneda K, Takahashi S, et al. Leukemogenesis caused by incapacitated GATA-1 function. Mol. Cell. Biol. 2004;24:10814–10825. doi: 10.1128/MCB.24.24.10814-10825.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wechsler J, Greene M, McDevitt MA, Anastasi J, Karp JE, Le Beau MM, Crispino JD. Acquired mutations in GATA1 in the megakaryoblastic leukemia of Down syndrome. Nat. Gen. 2002;32:148–152. doi: 10.1038/ng955. [DOI] [PubMed] [Google Scholar]
- 51.Yamasaki H, Era T, Asou N, Sanada I, Matutes E, Yamaguchi K, Takatsuki K. High degree of myeloid differentiation and granulocytosis is associated with t(8;21) smoldering leukemia. Leukemia. 1995;9:1147–1153. [PubMed] [Google Scholar]
- 52.Kojima K, Omoto E, Hara M, Sasaki K, Katayama Y, Nawa Y, et al. Myelodysplastic syndrome with translocation (8;21): a distinct myelodysplastic syndrome entity or M2-acute myeloid leukemia with extensive myeloid maturation? Ann. Hematol. 1998;76:279–282. doi: 10.1007/s002770050402. [DOI] [PubMed] [Google Scholar]
- 53.Schwieger M, Lohler J, Friel J, Scheller M, Horak I, Stocking C. AML1-ETO inhibits maturation of multiple lymphohematopoietic lineages and induces myeloblast transformation in synergy with ICSBP deficiency. J. Exp. Med. 2002;196:1227–1240. doi: 10.1084/jem.20020824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fenske TS, Pengue G, Mathews V, Hanson PT, Hamm SE, Riaz N, Graubert TA. Stem cell expression of the AML1/ETO fusion protein induces a myeloproliferative disorder in mice. Proc. Natl. Acad. Sci. USA. 2004;101:15184–15189. doi: 10.1073/pnas.0400751101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Song W-J, Sullivan MG, Legare RD, Hutchings S, Tan X, Kufrin D, et al. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukemia. Nature Genet. 1999;23:166–175. doi: 10.1038/13793. [DOI] [PubMed] [Google Scholar]
- 56.Sun W, Downing JR. Haploinsufficiency of AML1 results in a decrease in the number of LTR-HSC while simultaneously inducing an increase in more mature progenitors. Blood. 2004;104:3565–3572. doi: 10.1182/blood-2003-12-4349. [DOI] [PubMed] [Google Scholar]
- 57.Adolfsson J, Mansson R, Buza-Vidas N, Hultquist A, Liuba K, Jensen CT, et al. Identification of FLt3+ lympho-myeloid stem cells lacking erythromegakaryocytic potential: a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 58.Arron JR, Winslow MM, Polleri A, Chang C-P, Wu H, Gao X, et al. NFAT dysregulation by increased dosage of DSCR1 and DYRK!A on chromosome 21. Nature. 2006;441:595–600. doi: 10.1038/nature04678. [DOI] [PubMed] [Google Scholar]
- 59.Munugalavadla V, Dore LC, Tan BL, Hong L, Vishnu M, Weiss MJ, Kapur R. Repression of c-Kit and its downstream substrates by GATA-1 inhibits cell proliferation during erythroid maturation. Mol. Cell. Biol. 2005;25:6747–6759. doi: 10.1128/MCB.25.15.6747-6759.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]