Abstract
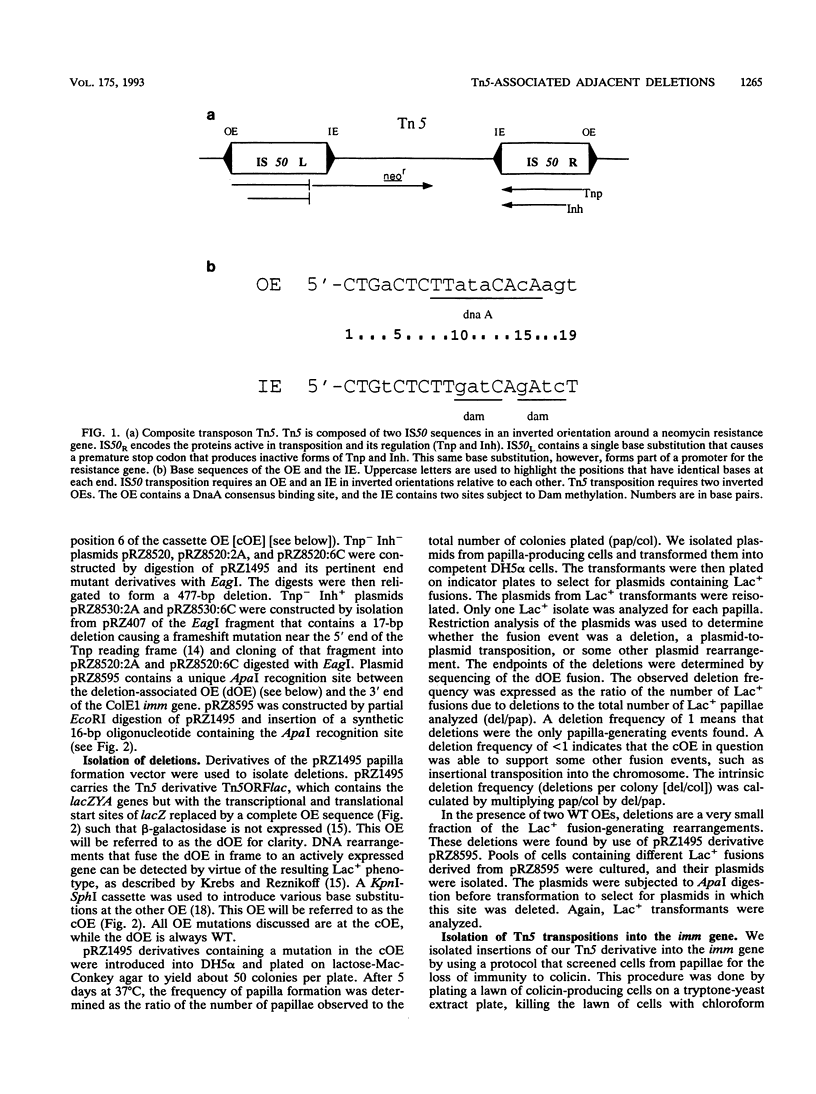
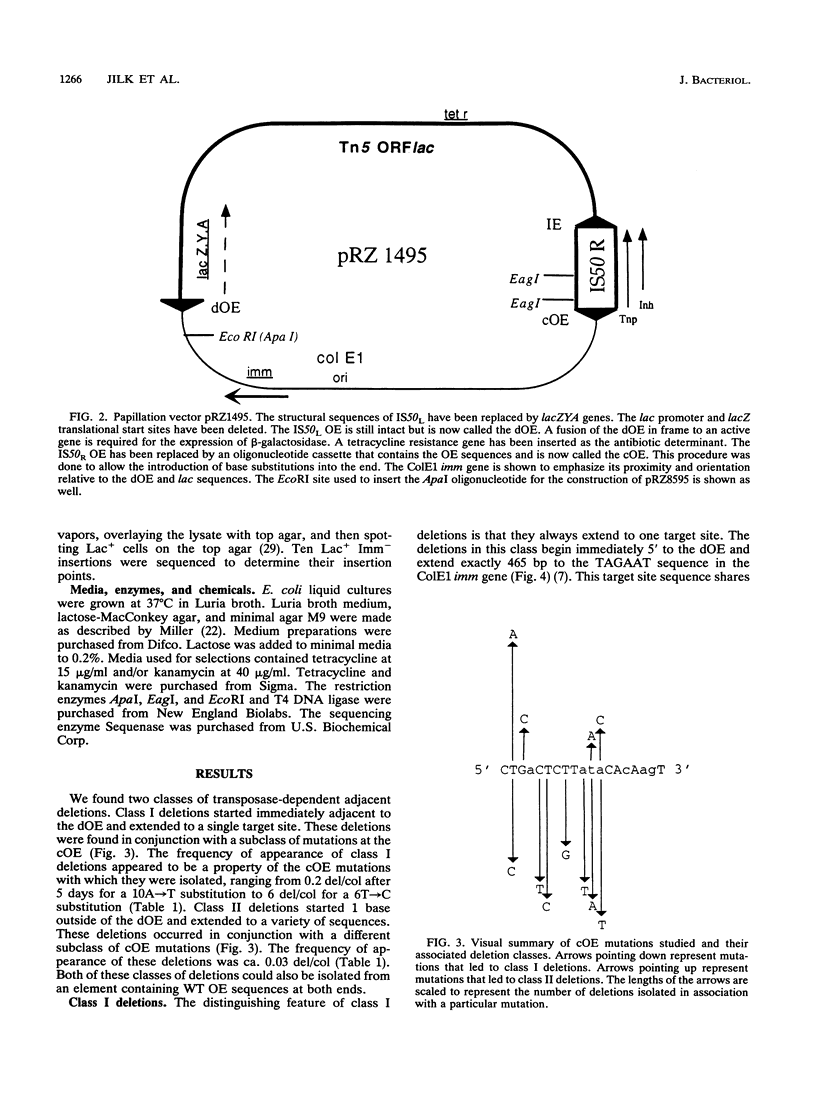
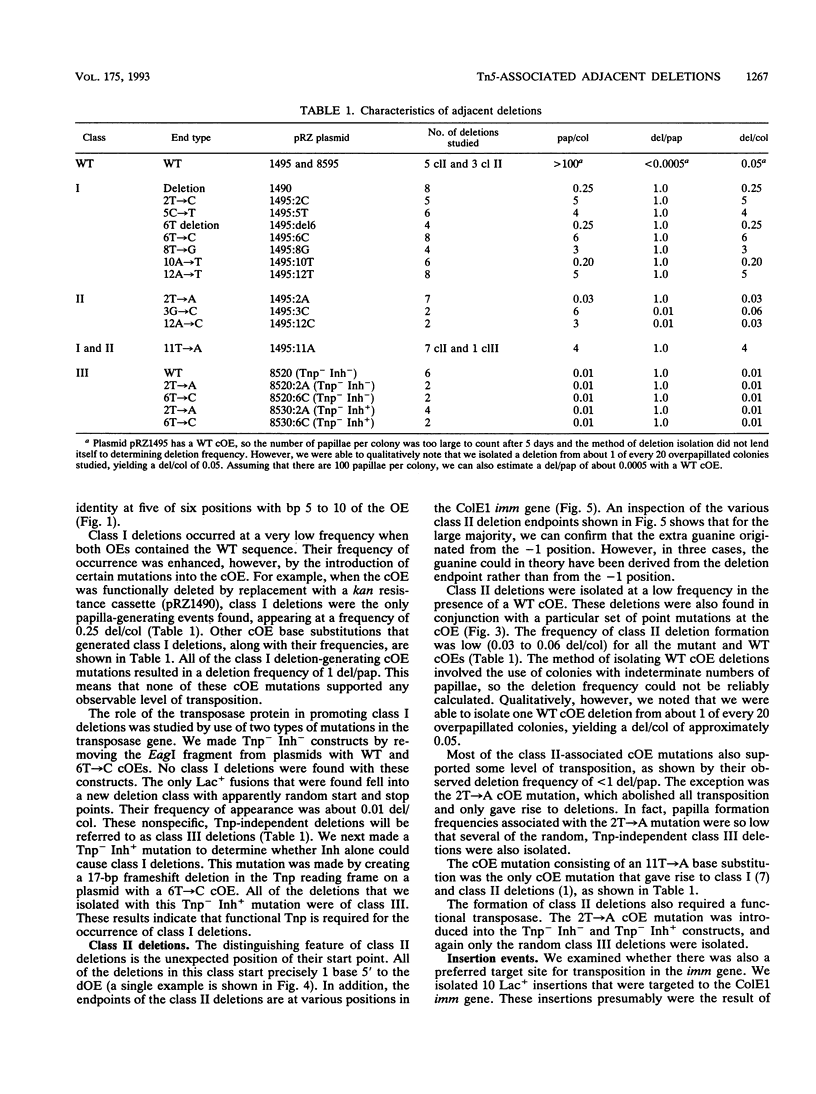
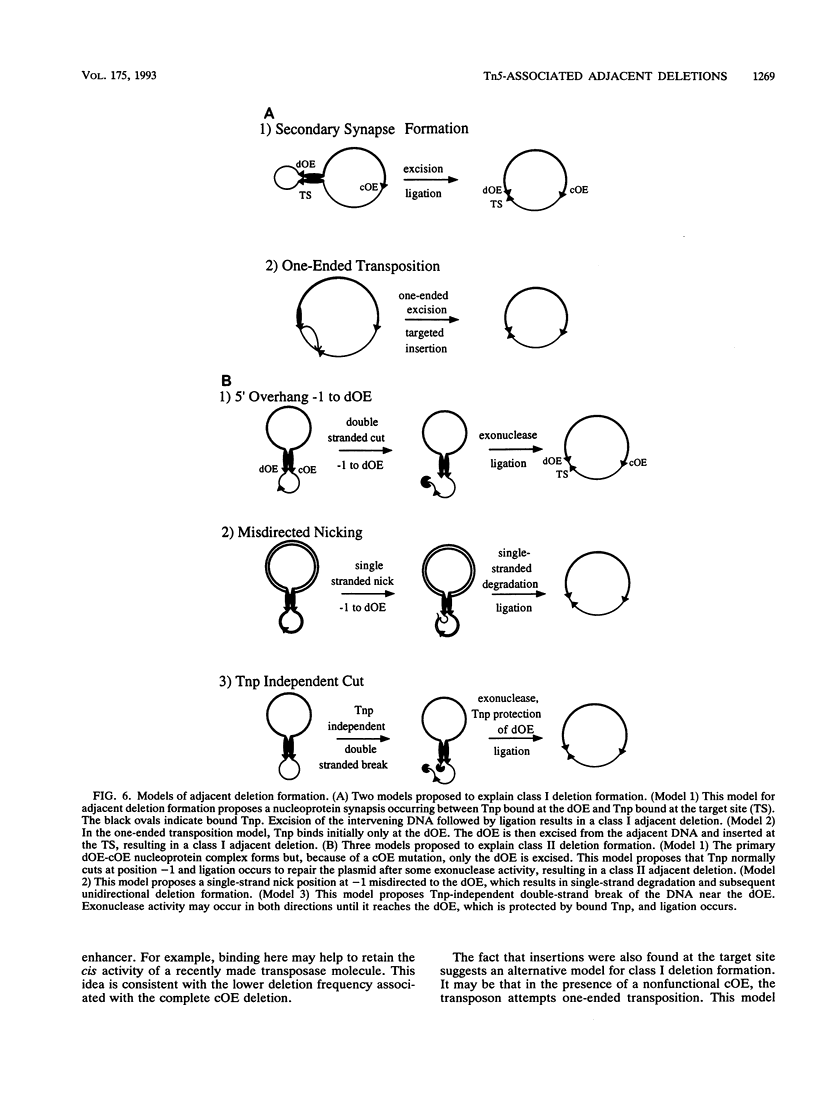
The prokaryotic transposable element Tn5 has been found to promote the formation of adjacent deletions. The frequency of adjacent deletion formation is much lower than that of normal transposition events. Like normal transposition, however, adjacent deletion formation requires the activity of the transposase protein. The deletions can be divided into two classes, as distinguished by their endpoints. The occurrence of one of the two deletion classes is increased when the frequency of normal transposition is reduced by the introduction of a deletion or a certain base substitution at one of the two outside ends (OEs). The nature of the base substitution at the mutant OE influences the class of deletion found adjacent to the wild-type OE, even though these two ends are about 12 kbp apart. By studying the formation of these deletions, we have gained some insight into the way in which the transposase interacts with the OEs. Our observations suggest that there is a protein-mediated interaction between the two ends, that different end base pairs are involved in different transposition-related processes, and that the adjacent deletions are the result of nonproductive attempts at transposition.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adzuma K., Mizuuchi K. Interaction of proteins located at a distance along DNA: mechanism of target immunity in the Mu DNA strand-transfer reaction. Cell. 1989 Apr 7;57(1):41–47. doi: 10.1016/0092-8674(89)90170-0. [DOI] [PubMed] [Google Scholar]
- Arthur A., Sherratt D. Dissection of the transposition process: a transposon-encoded site-specific recombination system. Mol Gen Genet. 1979 Oct 1;175(3):267–274. doi: 10.1007/BF00397226. [DOI] [PubMed] [Google Scholar]
- Bainton R., Gamas P., Craig N. L. Tn7 transposition in vitro proceeds through an excised transposon intermediate generated by staggered breaks in DNA. Cell. 1991 May 31;65(5):805–816. doi: 10.1016/0092-8674(91)90388-f. [DOI] [PubMed] [Google Scholar]
- Berg D. E., Schmandt M. A., Lowe J. B. Specificity of transposon Tn5 insertion. Genetics. 1983 Dec;105(4):813–828. doi: 10.1093/genetics/105.4.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan P. T., Ohmori H., Tomizawa J., Lebowitz J. Nucleotide sequence and gene organization of ColE1 DNA. J Biol Chem. 1985 Jul 25;260(15):8925–8935. [PubMed] [Google Scholar]
- Derbyshire K. M., Hwang L., Grindley N. D. Genetic analysis of the interaction of the insertion sequence IS903 transposase with its terminal inverted repeats. Proc Natl Acad Sci U S A. 1987 Nov;84(22):8049–8053. doi: 10.1073/pnas.84.22.8049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichinger D. J., Boeke J. D. A specific terminal structure is required for Ty1 transposition. Genes Dev. 1990 Mar;4(3):324–330. doi: 10.1101/gad.4.3.324. [DOI] [PubMed] [Google Scholar]
- Huisman O., Errada P. R., Signon L., Kleckner N. Mutational analysis of IS10's outside end. EMBO J. 1989 Jul;8(7):2101–2109. doi: 10.1002/j.1460-2075.1989.tb03619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isberg R. R., Syvanen M. Tn5 transposes independently of cointegrate resolution. Evidence for an alternative model for transposition. J Mol Biol. 1985 Mar 5;182(1):69–78. doi: 10.1016/0022-2836(85)90028-2. [DOI] [PubMed] [Google Scholar]
- Johnson R. C., Bruist M. F. Intermediates in Hin-mediated DNA inversion: a role for Fis and the recombinational enhancer in the strand exchange reaction. EMBO J. 1989 May;8(5):1581–1590. doi: 10.1002/j.1460-2075.1989.tb03542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. C., Reznikoff W. S. Role of the IS50 R proteins in the promotion and control of Tn5 transposition. J Mol Biol. 1984 Aug 25;177(4):645–661. doi: 10.1016/0022-2836(84)90042-1. [DOI] [PubMed] [Google Scholar]
- Krebs M. P., Reznikoff W. S. Use of a Tn5 derivative that creates lacZ translational fusions to obtain a transposition mutant. Gene. 1988 Mar 31;63(2):277–285. doi: 10.1016/0378-1119(88)90531-8. [DOI] [PubMed] [Google Scholar]
- Lavoie B. D., Chaconas G. Immunoelectron microscopic analysis of the A, B, and HU protein content of bacteriophage Mu transpososomes. J Biol Chem. 1990 Jan 25;265(3):1623–1627. [PubMed] [Google Scholar]
- Lichens-Park A., Syvanen M. Cointegrate formation by IS50 requires multiple donor molecules. Mol Gen Genet. 1988 Feb;211(2):244–251. doi: 10.1007/BF00330600. [DOI] [PubMed] [Google Scholar]
- Makris J. C., Nordmann P. L., Reznikoff W. S. Mutational analysis of insertion sequence 50 (IS50) and transposon 5 (Tn5) ends. Proc Natl Acad Sci U S A. 1988 Apr;85(7):2224–2228. doi: 10.1073/pnas.85.7.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinus M. G., Morris N. R. Isolation of deoxyribonucleic acid methylase mutants of Escherichia coli K-12. J Bacteriol. 1973 Jun;114(3):1143–1150. doi: 10.1128/jb.114.3.1143-1150.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B., D'Alençon E., Ehrlich S. D. Deletion hot spots in chimeric Escherichia coli plasmids. J Bacteriol. 1989 Apr;171(4):1846–1853. doi: 10.1128/jb.171.4.1846-1853.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B., Ehrlich S. D. Illegitimate recombination at the replication origin of bacteriophage M13. Proc Natl Acad Sci U S A. 1986 May;83(10):3386–3390. doi: 10.1073/pnas.83.10.3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordmann P. L., Makris J. C., Reznikoff W. S. Inosine induced mutations. Mol Gen Genet. 1988 Sep;214(1):62–67. doi: 10.1007/BF00340180. [DOI] [PubMed] [Google Scholar]
- Phadnis S. H., Berg D. E. Identification of base pairs in the outside end of insertion sequence IS50 that are needed for IS50 and Tn5 transposition. Proc Natl Acad Sci U S A. 1987 Dec;84(24):9118–9122. doi: 10.1073/pnas.84.24.9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts D. E., Ascherman D., Kleckner N. IS10 promotes adjacent deletions at low frequency. Genetics. 1991 May;128(1):37–43. doi: 10.1093/genetics/128.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J. A. Molecular model for the transposition and replication of bacteriophage Mu and other transposable elements. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1933–1937. doi: 10.1073/pnas.76.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surette M. G., Harkness T., Chaconas G. Stimulation of the Mu A protein-mediated strand cleavage reaction by the Mu B protein, and the requirement of DNA nicking for stable type 1 transpososome formation. In vitro transposition characteristics of mini-Mu plasmids carrying terminal base pair mutations. J Biol Chem. 1991 Feb 15;266(5):3118–3124. [PubMed] [Google Scholar]
- Tomcsanyi T., Berg C. M., Phadnis S. H., Berg D. E. Intramolecular transposition by a synthetic IS50 (Tn5) derivative. J Bacteriol. 1990 Nov;172(11):6348–6354. doi: 10.1128/jb.172.11.6348-6354.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren G. J., Saul M. W., Sherratt D. J. ColE1 plasmid mobility: essential and conditional functions. Mol Gen Genet. 1979 Feb 16;170(1):103–107. doi: 10.1007/BF00268585. [DOI] [PubMed] [Google Scholar]
- Weinreich M. D., Reznikoff W. S. Fis plays a role in Tn5 and IS50 transposition. J Bacteriol. 1992 Jul;174(14):4530–4537. doi: 10.1128/jb.174.14.4530-4537.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J. C., Krebs M. P., Reznikoff W. S. Effect of dam methylation on Tn5 transposition. J Mol Biol. 1988 Jan 5;199(1):35–45. doi: 10.1016/0022-2836(88)90377-4. [DOI] [PubMed] [Google Scholar]
- Yin J. C., Reznikoff W. S. dnaA, an essential host gene, and Tn5 transposition. J Bacteriol. 1987 Oct;169(10):4637–4645. doi: 10.1128/jb.169.10.4637-4645.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]


