Abstract
Treatment of mouse or human cells with the protein kinase C (PKC) inhibitors H7 or bisindolylmaleimide I induced an increase in the lifetime of p53, leading to its accumulation. In inhibitor-treated cells, p53 translocated to the nuclei and bound to DNA but was not competent to induce transcription. However, transactivation could be induced by subsequent DNA damage. Phorbol ester, a potent activator of PKC, significantly inhibited the accumulation of p53 after DNA damage. Therefore, constitutive PKC-dependent phosphorylation of p53 itself, or of a protein that interacts with p53, is required for the rapid degradation of p53 in untreated cells. Furthermore, an increase in the lifetime of p53 is not accompanied necessarily by its activation. Treatment with the PKC inhibitors decreased the overall level of p53 phosphorylation but led to the appearance of a phosphopeptide not seen in tryptic digests of p53 from untreated cells. Therefore, the lifetime and activities of p53 are likely to be regulated by distinct alterations of the phosphorylation pattern of p53, probably caused by the actions of different kinases.
The tumor suppressor protein p53 plays an important role in maintaining genetic integrity in mammalian cells (1), and the gene encoding p53 is inactivated in human tumors (2). p53 is induced in response to DNA damage (3, 4) or stresses such as hypoxia (5) or nucleotide deprivation (6). The induction of p53 leads either to arrest at different stages of cell cycle [reviewed by Agarwal et al. (7)] or to activation of apoptosis [reviewed by Gottlieb and Oren (8), Ko and Prives (9), and Smith and Fornace (10)], thus helping to prevent DNA damage and blocking the propagation of cells that have experienced damage. The two major aspects of the p53 response, activation and accumulation, are regulated by different mechanisms (11, 12), so that transactivation by p53 of genes required for cell cycle arrest is not accompanied necessarily by an increase in the amount of p53 protein (11).
The accumulation of p53 is caused primarily by an increase in its half-life (3). p53 degradation is mediated by ubiquitin-dependent proteolysis (13), but the signals targeting p53 for ubiquitination and the changes in the protein after DNA damage that prevent degradation have been obscure until recently. However, we now know that the binding of mdm2 to p53 is important not only in regulating transactivation by p53 but also in determining its rate of degradation (14, 15). DNA damage induces phosphorylation of the N-terminal domain of p53, thus preventing mdm2 from binding to and stabilizing the protein (16).
Posttranslational modification is the major mechanism of p53 regulation. Several different protein kinases can phosphorylate distinct domains of p53 in vitro. For example, the DNA-activated protein kinase mitogen-activated protein kinase, Raf-1, and the casein kinase I-like protein kinase each phosphorylate different sites in the N-terminal transactivation domain of p53 [reviewed by Steegenga et al. (17)]. These phosphorylation events affect the transactivation function of p53. For example, mutation of three N-terminal serine residues that are kinase targets to alanines in mouse p53 causes a significant decrease in suppressor function and reduces transactivation (18). Mutation of various serine residues has a differential effect on the ability of rat p53 to bind to and induce transcription from various promoters (19), indicating that specific patterns of serine phosphorylation are likely to occur in response to different types of stress, leading in turn to stress-specific patterns of gene expression.
The C-terminal regulatory domain of p53 is a target for phosphorylation by casein kinase II (20), protein kinase C (PKC) (21), cdk (22) and p34cdc2 (23). p34cdc2 can bind to p53 in vivo (23) and phosphorylate it in vitro (22). Phosphorylation by cell cycle-dependent protein kinases suggests the possibility that the activity of p53 is regulated differentially during the cell cycle. Phosphorylation by PKC and casein kinase II in vitro stimulates p53 to bind to DNA (24, 25), probably by changing the conformation of the protein. However, the activation of PKC by phorbol ester does not cause a change in phosphorylation of the C-terminal domain of mouse p53 (26), indicating that the PKC site may be phosphorylated constitutively. Experiments with the human p53 mutant S392A revealed that phosphorylation of the C-terminal domain by casein kinase II is not required for p53 to transactivate target genes (27). Taken together, the data suggest that, in vivo, the C-terminal domain of p53 may have functions in addition to regulating DNA binding and that constitutive phosphorylation of this domain may be required to elicit such functions.
In the current work, we examined the role played by PKC in the rapid turnover of p53 in untreated cells. PKC inhibitors slow the degradation of p53 and lead to changes in its phosphorylation, whereas a PKC activator inhibits the DNA damage-induced accumulation of p53. The most likely possibility is that PKC-dependent phosphorylation of one or more sites of p53 is required for its rapid degradation.
MATERIALS AND METHODS
Cells and Inhibitors.
Mouse (12)1/CA cells carrying a p53-responsive β-galactosidase reporter construct have been described (12). All cells were grown in DMEM (GIBCO/BRL) supplemented by 10% fetal calf serum (GIBCO/BRL). H7, H8, and phorbol ester were obtained from Sigma, and A3 and bisindolylmaleimide I (Bis) were obtained from Alexis Biochemicals (San Diego). The human cell lines were WI-38, from the American Type Culture Collection; HT1080, from Peter Goodfellow (Imperial Cancer Research Fund, London); and HCT116 and RKO from Bert Vogelstein (Johns Hopkins Oncology Center, Baltimore).
RNA and Protein Assays.
Total RNA was extracted by using the Trizol reagent (GIBCO/BRL) according to the manufacturer’s protocol. Western analyses were performed as described by Chernov and Stark (12), and p53 was detected with the mAb PAb421 (a gift of A. Levine, Princeton University, Princeton). Horseradish peroxidase-conjugated goat anti-mouse secondary antibody (Bio-Rad) was detected with the Renaissance chemoluminescence reagent (DuPont/NEN). Quantitation was obtained by using the nih image program.
Electrophoretic Mobility Shift Assays.
Nuclear extracts were prepared, and electrophoretic mobility shift assays were performed as described by Chernov and Stark (12), using as the probe an oligonucleotide containing a 26-mer p53 binding site [AGCTTAGACATGCCTAGACATGCCTA (28)].
Immunostaining.
Immunostaining was performed as described by Harlow and Lane (29). In brief, cells grown on coverslips were fixed with methanol:acetone∷1:1 for 2 min at room temperature, followed by incubation for 2 hr at room temperature with primary PAb421 antibody and then for 30 min with secondary fluorescein-conjugated anti-mouse Ig (Sigma). Images were obtained with a Nikon Optiphot epifluorescence microscope coupled to a cooled computer-controlled charge-coupled device camera (Oncor).
Protein Kinase Assays and Phosphopeptide Analysis.
Analysis of PKC activity using histone H1 as a substrate and using a partially purified cell extract was performed as described by Gopalakrishna et al. (30). Phosphopeptide analyses were performed as described by Adler et al. (31). After a 1-hr preincubation in phosphate-free medium containing 10% dialyzed fetal calf serum, cells at ≈80% confluency were labeled in the same medium for 7 hr, using 200 μCi/ml 32P-labeled orthophosphate/150-mm plate. Proteins were extracted in PBS containing 1% Nonidet P-40, 0.5% sodium deoxycholate, 1 mM Na2VO3, 0.1 phenylmethane sulfonyl fluoride, and 1 μg/ml each of the protease inhibitors pepstatin, leupeptin, and aprotinin. p53 was immunoprecipitated with the PAb421 antibody, and the proteins, separated by SDS/PAGE, were transferred to a polyvinylidene difluoride membrane. After exposure of the membrane to film, the radioactive p53 band was cut out with a razor blade. After oxidation with performate, the p53 was digested overnight with tosylamide-2-phenylethyl chloromethyl ketone-treated trypsin. The peptides were concentrated by freeze drying, dissolved in loading buffer, and separated in an isoelectric focusing polyacrylamide gel (pH 3.5–10) for 2 hr at 600 V and 4°C. The gel was dried and exposed to film to localize the peptides.
RESULTS
PKC Inhibitors Induce the Accumulation of p53.
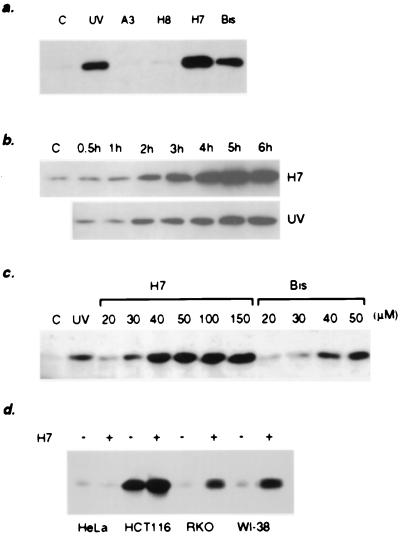
To investigate the roles of specific kinases in determining the level and activity of p53 in vivo, we used the PKC inhibitors H7 (32) and Bis (33) and the protein kinase A and G inhibitors H8 and A3 (32, 34). We used both mouse (12)1/CA cells, which have wild-type p53 and have been transfected with a LacZ gene driven by a p53-dependent promoter (12), and human HT1080 cells, which also have wild-type p53 (unpublished data). The PKC inhibitors H7 and Bis, but not the protein kinase A and G inhibitors H8 and A3, induced p53 to a very high level, comparable to or even higher than (in the case of H7) the level of p53 in UV-irradiated cells (Fig. 1a). The PKC activity present in partially purified cell extracts, analyzed by the in vitro phosphorylation of histone H1, was eliminated after treating the cells with H7 or Bis for 5 hr (data not shown). The time course of p53 accumulation was similar for H7-treated and UV-irradiated cells, but the amount of p53 after 6 hr was higher in the case of H7 (Fig. 1b). The induction was dose-dependent (Fig. 1c). We also analyzed the effect of H7 on the level of p53 in the human cell lines HT1080, HeLa, RKO, and WI-38 (Fig. 1d) and in the rat fibroblast cell line REF52 (data not shown). In all of the cells except HeLa, we observed p53 accumulation. The lack of induction in HeLa cells may be caused by the presence of the papilloma viral protein E6, which destabilizes p53 (35).
Figure 1.
The effect of kinase inhibitors on the level and activity of p53 in mouse and human cells. (a) The level of p53 in (12)1/CA cells treated for 6 hr with different kinase inhibitors. (b) Time course of p53 accumulation in mouse (12)1/CA cells treated with 50 μM H7 or irradiated with 25 J/m2 UV light. (c) The induction of p53 in mouse cells treated for 6 hr with different concentrations of H7 or Bis. (d) The induction of p53 by H7 in human cells.
The Accumulation of p53 in Cells Treated with Inhibitors of PKC Is Caused by Protein Stabilization.
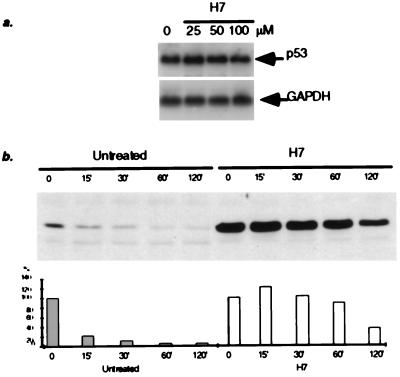
Previous studies have shown that transcriptional activation of the p53 gene is not involved in the accumulation of p53 induced by DNA damage (36). We found no increase in the levels of p53 mRNA in H7- or Bis-treated cells, compared with the untreated controls (Fig. 2a; data not shown). These data indicate that, as in the case of p53 induced by DNA damage, transcription of the p53 gene is not induced by H7 or Bis. Because the most likely cause of p53 accumulation is protein stabilization, we compared the half-lives of p53 in untreated cells and in cells treated with H7. Mouse (12)1/CA cells were treated with H7 for 5 hr, and cycloheximide then was added to inhibit new protein synthesis. Extracts were prepared at the indicated times, and the levels of p53 were determined. The half-life of p53 in untreated cells, ≈15 min (37), was increased to 1–2 hr in cells treated with H7 (Fig. 2b).
Figure 2.
Effect of H7 on the level of p53 mRNA and the stability of the p53 protein. (a) Analysis of p53 mRNA from human cells treated with different concentrations of H7. Total RNA was extracted 5 hr after treatment. (b) The effect of H7 on the lifetime of p53 in mouse cells. (12)1/CA cells were treated with H7 (50 μM) for 6 hr, followed by incubation in the presence of cycloheximide (CHX). The levels of p53 protein were analyzed at the times indicated. The amount of protein at zero time was scored as 100%.
Pretreatment with Phorbol Ester Inhibits the Accumulation of p53 after DNA Damage.
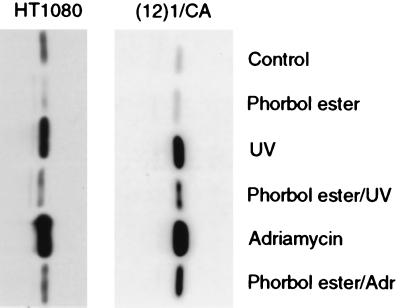
Because treatment with inhibitors of PKC leads to the stabilization and accumulation of p53, we also investigated the effects of PKC activation. The activity of PKC in cells treated with 80 ng/ml phorbol ester increases gradually for 6–8 hr and then decreases (38). Therefore, we pretreated cells with 80 ng/ml phorbol ester for 6 hr and then irradiated them with UV light, or treated them with Adriamycin to damage their DNA. In mouse cells, phorbol ester did not affect the basal level of p53, whereas in HT1080 cells it caused a small decrease (Fig. 3). However, in both cases, pretreatment with phorbol ester significantly inhibited the accumulation of p53 after exposure to UV light or Adriamycin (Fig. 3). In accordance with the small effect of phorbol ester on the basal level of p53 in untreated cells, this compound did not cause any change in the half-life of p53 (data not shown).
Figure 3.
The effect of phorbol ester on the basal and induced levels of p53 in mouse and human cells. (12)1/CA or HT1080 cells were treated with phorbol ester (80 ng/ml), UV light (25 J/m2), or Adriamycin (200 ng/ml) or were pretreated with phorbol ester for 6 hr followed by UV light or Adriamycin. Protein extracts were prepared 6 hr after the DNA-damaging treatment.
Location and DNA Binding Activity of p53 Induced by Inhibiting PKC.
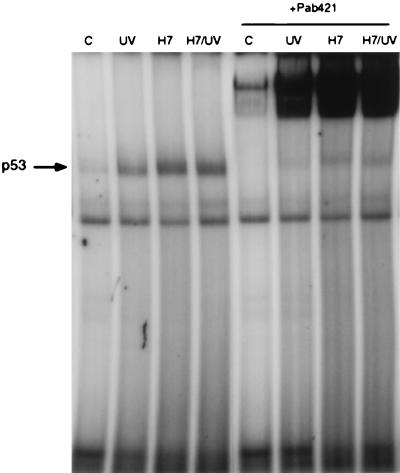
Translocation of p53 from the cytoplasm to the nucleus and induction of its DNA binding activity [reviewed by Gottlieb and Oren (8)] are required for the transactivation of target genes in response to DNA damage. In situ immunostaining of H7-treated cells, using the p53-specific antibody PAb421, revealed that the accumulated p53 is present in nuclei (Fig. 4). In contrast to UV-irradiated cells, the nuclear accumulation of p53 in H7-treated cells can be seen in almost all of the cells (Fig. 4). We tested the DNA binding activity of p53 in electrophoretic mobility shift assays with a labeled p53-specific consensus binding element (28) by using nuclear extracts of H7- and UV-treated cells 6 hr after treatment. DNA binding was induced in H7-treated cells, and the induced band could be super-shifted by the PAb421 antibody (Fig. 5). In accord with the higher level of p53, the induction of DNA binding was also higher in H7-treated cells, compared with UV-irradiated cells (Fig. 5).
Figure 4.
Nuclear accumulation of p53 in mouse cells treated with H7. Cells were irradiated with 25 J/m2 UV light or treated with H7 (50 μM). After 6 hr, the cells were fixed and probed with the p53-specific antibody PAb421 and with fluorescein-conjugated second antibody. Staining with 4′,6′-diamidino-2-phenylindole (DAPI) was used to reveal the nuclei.
Figure 5.
DNA binding activity of p53 in cells treated with H7 or irradiated with UV light. p53-specific DNA binding activity in nuclear extracts was analyzed 5 hr after treatment. The last four lanes show the effect of the p53-specific antibody PAb421.
The p53 Induced by Inhibitors of PKC Does Not Activate Transcription From p53-Dependent Promoters.
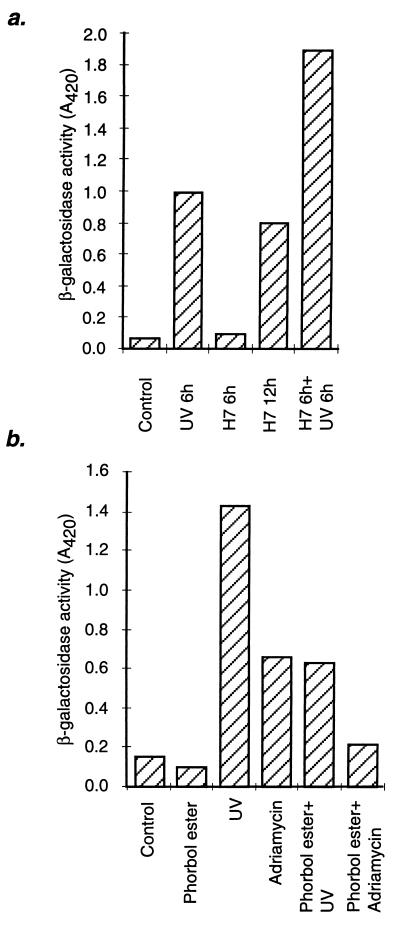
To explore the transcriptional activation of p53-responsive genes, we used LacZ, whose expression in (12)1/CA cells is regulated by a p53-dependent promoter (12, 39). The cells were treated with H7 or Bis for 6 hr, a time sufficient to induce substantial accumulation of p53, followed by analysis of β-galactosidase activity. As a control, parallel plates were irradiated with UV light. No increase in β-galactosidase activity was detected in the inhibitor-treated cells, whereas at the same time, induction by more than 10-fold was detected in the UV-irradiated cells (Fig. 6a; data not shown). The same was true for expression of the p53-responsive gene waf1 (data not shown). To determine whether the accumulated p53 in cells treated for 6 hr with PKC inhibitors could be activated, we irradiated the treated cells with UV light. Significantly more β-galactosidase activity was present 6 hr after irradiation of inhibitor-treated cells than in irradiated untreated cells, as expected from the higher levels of p53 present in the treated cells before irradiation (Fig. 6a) if all of the accumulated protein could be activated. Longer exposure to H7 or Bis (12 hr) caused induction of β-galactosidase activity to a level lower than that obtained after UV irradiation (Fig. 6a; data not shown), probably because of a secondary stress response or to a nonspecific effect of the inhibitors. Cotreatment with phorbol ester inhibited the induction of β-galactosidase by UV light or Adriamycin (Fig. 6b), correlating with a smaller increase in the accumulation of p53 protein, indicating that phorbol ester does not have a direct effect on p53 activation.
Figure 6.
Effect of H7 or phorbol ester on p53-dependent expression of β-galactosidase in mouse cells, with or without DNA damage. (a) (12)1/CA cells were treated with 50 μM H7 or irradiated with UV light (25 J/m2), and β-galactosidase activity was measured at the times shown. Alternatively, the cells were treated with H7 for 6 hr and then irradiated with UV light before analysis 6 hr later. (b) (12)1/CA cells were treated with phorbol ester (80 ng/ml), UV light (25 J/m2), or Adriamycin (200 ng/ml) or pretreated with phorbol ester for 6 hr, followed by treatment with UV light or Adriamycin. β-galactosidase activity was analyzed 6 hr after the DNA damaging treatment.
Alteration of p53 Phosphorylation in Cells Treated with Inhibitors of PKC.
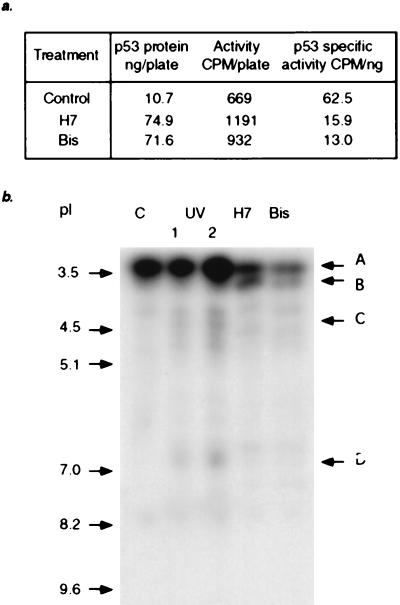
To determine the mechanism involved in stabilizing p53 in cells treated with inhibitors of PKC, we analyzed the phosphopeptides obtained after tryptic digestion of p53 labeled with 32P in vivo. (12)1/CA cells were treated with H7 or Bis and then labeled with [32P]-orthophosphate. p53 was immunoprecipitated and, after electrophoresis and transfer to a membrane, its specific activity was determined (Fig. 7a). As expected, more labeled p53 was precipitated from the treated cells, and its degree of phosphorylation was lower. Moreover, when we determined the endogenous kinase activity in total cell extracts by using in vitro phosphorylation of histidine-tagged human p53, treatment of the cells with H7 or Bis led to inhibition of p53 phosphorylation (data not shown).
Figure 7.
Phosphorylation of p53 in mouse cells treated with H7 or Bis or irradiated with UV light. The treated cells were labeled for 5 hr with [32P]-orthophosphate, and p53 was immunoprecipitated with PAb421. (a) Specific activities of p53 from untreated or treated cells. (b) Tryptic peptide analysis of the labeled p53. The phosphopeptides were separated by isoelectric focusing. Equal amounts of radioactivity were loaded in each lane.
The immunoprecipitated mouse p53 was digested with trypsin, and the labeled peptides were separated by isoelectric focusing in a polyacrylamide gel. Six tryptic peptides, representing the N- and C-terminal domains (Table 1), contain in vitro phosphorylation sites for several protein kinases [reviewed by Steegenga et al. (17)]. A tryptic digest of p53 from untreated (12)1/CA cells contains a single major band (A) in the pI region 3–3.5 (Fig. 7b). Based on the relative intensity, more than one phosphopeptide may migrate in this region. Treatment with H7 or Bis resulted in a significant decrease in the intensity of band A, with the simultaneous appearance of phosphopeptide B, pI ≈ 3.7 (Fig. 7b). Irradiation with UV light induced the appearance of phosphopeptide C, pI between 4 and 4.5, and phosphopeptide D, pI between 6.5 and 7.0 (Fig. 7b), which are not found in inhibitor-treated cells. On the other hand, phosphopeptide B was not detected in the UV-irradiated samples. The data demonstrate that treatment of cells with H7 or Bis causes rearrangement of the phosphorylated sites of p53, with the appearance of a new phosphopeptide (B), and that these effects correlate with the stabilization of p53. Furthermore, the activation of p53 in UV-treated cells involves the phosphorylation of sites that are distinct from those affected by H7 or Bis.
Table 1.
The sequence of mouse p53 (50) and tryptic peptides that contain phosphorylation sites from the N- or C-terminal domain
| MTAMEESQSD | ISLELPLSQE | TFSGLWKLLP | PEDILPSPHC | MDDLLLPQDV | 50 |
| EEFFEGPSEA | LRVSGAPAAQ | DPVTETPGPV | APAPATPWPL | SSFVPSQKTY | 100 |
| QGNYGFHLGF | LQSGTAKSVM | CTYSPPLNKL | FCQLAKTCPV | QLWVSATPPA | 150 |
| GSRVRAMAIY | KKSQHMTEVV | RRCPHHERCS | DGDGLAPPQH | LIRVEGNLYP | 200 |
| EYLEDRQTFR | HSVVVPYEPP | EAGSEYTTIH | YKYMCNSSCM | GGMNRRPILT | 250 |
| IITLEDSSGN | LLGRDSFEVR | VCACPGRDRR | TEEENFRKKE | VLCPELPPGS | 300 |
| AKRALPTCTS | ASPPQKKKPL | DGEYFTLKIR | GRKRFEMFRE | LNEALELKDA | 350 |
| HATEESGDSR | AHSSYLKTKK | GQSTSRHKKT | MVKKVGPDSD | 390 |
| Peptide | pI |
|---|---|
| 1–27 | 3.74 |
| 28–62 | 3.60 |
| 63–98 | 4.18 |
| 304–316 | 8.23 |
| 371–376 | 10.04 |
| 385–390 | 3.23 |
DISCUSSION
Our data demonstrate that phosphorylation plays an important role not only in activating p53, but also in regulating its stability, increasing its level after DNA damage or other stress. p53 is degraded through ubiquitination and proteosome-mediated proteolysis (13, 40), but the signals that induce or inhibit this process have been obscure until recently. We now know that the interaction of p53 with mdm2 not only regulates its ability to transactivate target genes (41, 42) but also is required to induce degradation of the protein (14–16). Hupp et al. (11) have shown that the induction of p53-dependent transcription by low doses of UV radiation does not require accumulation of the protein, and we found previously that transcriptional induction is inhibited by salicylate without inhibiting accumulation of the protein (12). The effect of salicylate is likely due to its ability to inhibit protein phosphorylation (43). In the experiments presented here, we show that the stability of p53 can be regulated by phosphorylation, independently of transcriptional activation. Treatment of mouse or human cells with the PKC inhibitors H7 or Bis induces the accumulation of a latent form of p53, which, despite its nuclear localization and efficient DNA binding, does not activate the transcription of target genes. The mechanism of the accumulation of p53 is protein stabilization, which correlates with a decrease in overall phosphorylation, but with the appearance of a new phosphopeptide. Based on these observations, we propose that phosphorylation, mediated by PKC or by another kinase that can be inhibited by H7 or Bis, stimulates p53 degradation. The most likely target of PKC is p53 itself, which is consistent with the observed changes in the phosphorylation of p53. However, we cannot completely rule out the possibility that the stabilization of p53 caused by PKC inhibitors is due to decreased phosphorylation of a separate protein that regulates p53 stability. Other correlations of p53 stability and its state of phosphorylation have tended to suggest that decreased phosphorylation increases the rate of p53 degradation. For example, a serine-to-alanine mutation at position 15 of human p53, which prevents phosphorylation at this site, increases the rate of degradation (44), and N-terminal phosphorylation may interfere with the p53–mdm2 interaction, leading to stabilization (16). Because the effects we observe with PKC inhibitors are in the opposite direction, we conclude that the situation is complex, with phosphorylation at different p53 sites, catalyzed by different kinases, leading to opposite effects on stability.
The p53 that accumulates in cells treated with H7 or Bis is localized in the nucleus and can bind to DNA, indicating that the absence of p53-dependent transcriptional activation does not depend on inhibition of DNA binding activity or on cytoplasmic retention. Activation of the transcriptional function of p53 probably requires additional modification of p53 or the activation of cofactors (45–47). Consistent with this idea, additional treatment with UV light led to the induction of p53-dependent transcription to an extent related to the amount of p53 protein. The effect of H7 on the nuclear accumulation of p53 is also different from the effect of UV irradiation. Approximately half of the UV-irradiated cells accumulated p53 in the nucleus, but almost all of the H7-treated cells did so (Fig. 4). The nuclear accumulation of p53 after DNA damage may depend on the position of each individual cell in the cell cycle at the moment of damage (48).
We have found different changes in the phosphorylation of p53 in UV-irradiated cells compared with cells treated with PKC inhibitors, in agreement with the different activities of p53 in these cells. Although the accumulation of p53 is caused by stabilization in both cases, we did not detect any common change in the phosphorylation pattern, again indicating that more than one mechanism may be involved in regulating p53 stability. Consistent with this hypothesis, UV irradiation and γ irradiation have different effects on p53 ubiquitination (49).
Acknowledgments
This work was supported by Grant GM-49345 from the National Institutes of Health.
ABBREVIATIONS
- Bis
bisindolymaleimide
- PKC
protein kinase C
References
- 1.Yin Y, Tainsky M A, Bischoff F Z, Strong L C, Wahl G M. Cell. 1992;70:937–948. doi: 10.1016/0092-8674(92)90244-7. [DOI] [PubMed] [Google Scholar]
- 2.Hollstein M, Sidransky D, Vogelstein B, Harris C C. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 3.Kastan M B, Onyekwere O, Sidransky D, Vogelstein B, Craig R W. Cancer Res. 1991;51:6304–6311. [PubMed] [Google Scholar]
- 4.Fritsche M, Haessler C, Brandner G. Oncogene. 1993;8:307–318. [PubMed] [Google Scholar]
- 5.Graeber T G, Peterson J F, Tsai M, Monica K, Fornace A, Jr, Giaccia A J. Mol Cell Biol. 1994;14:6264–6277. doi: 10.1128/mcb.14.9.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Linke S P, Clarkin K C, DiLeonardo A, Tsou A, Wahl G M. Genes Dev. 1996;10:934–947. doi: 10.1101/gad.10.8.934. [DOI] [PubMed] [Google Scholar]
- 7.Agarwal M L, Taylor W R, Chernov M V, Chernova O B, Stark G R. J Biol Chem. 1998;273:1–4. doi: 10.1074/jbc.273.1.1. [DOI] [PubMed] [Google Scholar]
- 8.Gottlieb T M, Oren M. Biochim Biophys Acta. 1996;1287:77–102. doi: 10.1016/0304-419x(95)00019-c. [DOI] [PubMed] [Google Scholar]
- 9.Ko L J, Prives C. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 10.Smith M L, Fornace A J., Jr Proc Natl Acad Sci USA. 1997;94:12255–12257. doi: 10.1073/pnas.94.23.12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hupp T R, Sparks A, Lane D P. Cell. 1995;83:237–245. doi: 10.1016/0092-8674(95)90165-5. [DOI] [PubMed] [Google Scholar]
- 12.Chernov M V, Stark G R. Oncogene. 1997;14:2503–2510. doi: 10.1038/sj.onc.1201104. [DOI] [PubMed] [Google Scholar]
- 13.Chowdary D R, Dermody J J, Jha K K, Ozer H L. Mol Cell Biol. 1994;14:1997–2003. doi: 10.1128/mcb.14.3.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haupt Y, Maya R, Kazaz A, Oren M. Nature (London) 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 15.Kubbutat M H, Jones S N, Vousden K H. Nature (London) 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 16.Shieh S-Y, Ikeda M, Taya Y, Prives C. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 17.Steegenga W T, Van der Eb A J, Jochemsen A G. J Mol Biol. 1996;263:103–113. doi: 10.1006/jmbi.1996.0560. [DOI] [PubMed] [Google Scholar]
- 18.Mayr G A, Reed M, Wang P, Wang Y, Schwedes J F, Tegtmeyer P. Cancer Res. 1995;55:2410–2417. [PubMed] [Google Scholar]
- 19.Lohrum M, Scheidtmann K H. Oncogene. 1996;13:2527–2539. [PubMed] [Google Scholar]
- 20.Herrmann C P, Kraiss S, Montenarh M. Oncogene. 1991;6:877–884. [PubMed] [Google Scholar]
- 21.Baudier J, Delphin C, Grunwald D, Khochbin S, Lawrence J J. Proc Natl Acad Sci USA. 1992;89:11627–11631. doi: 10.1073/pnas.89.23.11627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Prives C. Nature (London) 1995;376:88–91. doi: 10.1038/376088a0. [DOI] [PubMed] [Google Scholar]
- 23.Milner J, Cook A, Mason J. EMBO J. 1990;9:2885–2889. doi: 10.1002/j.1460-2075.1990.tb07478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hupp T R, Meek D W, Midgley C A, Lane D P. Nucleic Acids Res. 1993;21:3167–3174. doi: 10.1093/nar/21.14.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hupp T R, Lane D P. Cold Spring Harb Symp Quant Biol. 1994;59:195–206. doi: 10.1101/sqb.1994.059.01.024. [DOI] [PubMed] [Google Scholar]
- 26.Milne D M, McKendrick L, Jardine L J, Deacon E, Lord J M, Meek D W. Oncogene. 1996;13:205–211. [PubMed] [Google Scholar]
- 27.Fiscella M, Zambrano N, Ullrich S J, Unger T, Lin D, Cho B, Mercer W E, Anderson C W, Appella E. Oncogene. 1994;9:3249–3257. [PubMed] [Google Scholar]
- 28.El-Deiry W S, Kern S E, Pietenpol J A, Kinzler K W, Vogelstein B. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 29.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 30.Gopalakrishna R, Chen Z H, Gundimeda U, Wilson J C, Anderson W B. Anal Biochem. 1992;206:24–35. doi: 10.1016/s0003-2697(05)80006-5. [DOI] [PubMed] [Google Scholar]
- 31.Adler V, Pincus M R, Minamoto T, Fuchs S Y, Bluth M J, Brandt-Rauf P W, Friedman F K, Robinson R C, Chen J M, Wang X W, Harris C C, Ronai Z. Proc Natl Acad Sci USA. 1997;94:1686–1691. doi: 10.1073/pnas.94.5.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hidaka H, Inagaki M, Kawamoto S, Sasaki Y. Biochemistry. 1984;23:5036–5041. doi: 10.1021/bi00316a032. [DOI] [PubMed] [Google Scholar]
- 33.Toullec D, Pianetti P, Coste H, Bellevergue P, Grand-Perret T, Ajakane M, Baudet V, Boissin P, Boursier E, Loriolle F, et al. J Biol Chem. 1991;266:15771–15781. [PubMed] [Google Scholar]
- 34.Inagaki M, Kawamoto S, Itoh H, Saitoh M, Hagiwara M, Takahashi J, Hidaka H. Mol Pharmacol. 1986;29:577–581. [PubMed] [Google Scholar]
- 35.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 36.Liu M, Dhanwada K R, Birt D F, Hecht S, Pelling J C. Carcinogenesis. 1994;15:1089–1092. doi: 10.1093/carcin/15.6.1089. [DOI] [PubMed] [Google Scholar]
- 37.Oren M, Maltzman W, Levine A J. Mol Cell Biol. 1981;1:101–110. doi: 10.1128/mcb.1.2.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Imbra R J, Karin M. Mol Cell Biol. 1987;7:1358–1363. doi: 10.1128/mcb.7.4.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Funk W D, Pak D T, Karas R H, Wright W E, Shay J W. Mol Cell Biol. 1992;12:2866–2871. doi: 10.1128/mcb.12.6.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ciechanover A, Shkedy D, Oren M, Bercovich B. J Biol Chem. 1994;269:9582–9589. [PubMed] [Google Scholar]
- 41.Oliner J D, Pietenpol J A, Thiagalingam S, Gyuris J, Kinzler K W, Vogelstein B. Nature (London) 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 42.Perry M E, Piette J, Zawadzki J A, Harvey D, Levine A J. Proc Natl Acad Sci USA. 1993;90:11623–11627. doi: 10.1073/pnas.90.24.11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frantz B, O’Neill E A. Science. 1995;270:2017–2019. doi: 10.1126/science.270.5244.2017. [DOI] [PubMed] [Google Scholar]
- 44.Fiscella M, Ullrich S J, Zambrano N, Shields M T, Lin D, Lees-Miller S P, Anderson C W, Mercer W E, Appella E. Oncogene. 1993;8:1519–1528. [PubMed] [Google Scholar]
- 45.Avantaggiati M L, Ogryzko V, Gardner K, Giordano A, Levine A S, Kelly K. Cell. 1997;89:1175–1185. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- 46.Garkavtsev I, Grigorian I A, Ossovskaya V S, Chernov M V, Chumakov P M, Gudkov A V. Nature (London) 1998;391:295–298. doi: 10.1038/34675. [DOI] [PubMed] [Google Scholar]
- 47.Lill N L, Grossman S R, Ginsberg D, DeCaprio J, Livingston D M. Nature (London) 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 48.Komarova E A, Zelnick C R, Chin D, Zeremski M, Gleiberman A S, Bacus S S, Gudkov A V. Cancer Res. 1997;57:5217–5220. [PubMed] [Google Scholar]
- 49.Maki C G, Howley P M. Mol Cell Biol. 1997;17:355–363. doi: 10.1128/mcb.17.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zakut-Houri R, Oren M, Bienz B, Lavie V, Hazum S, Givol D. Nature (London) 1983;306:594–597. doi: 10.1038/306594a0. [DOI] [PubMed] [Google Scholar]