Abstract
A collection of nine salicylidene acylhydrazide compounds were tested for their ability to inhibit the activity of virulence-associated type III secretion systems (T3SSs) in Salmonella enterica serovar Typhimurium. The compounds strongly affected Salmonella pathogenicity island 1 (SPI1) T3SS-mediated invasion of epithelial cells and in vitro secretion of SPI1 invasion-associated effector proteins. The use of a SPI1 effector β-lactamase fusion protein implicated intracellular entrapment of the protein construct upon application of a salicylidene acylhydrazide, whereas the use of chromosomal transcriptional gene fusions revealed a compound-mediated transcriptional silencing of SPI1. Salicylidene acylhydrazides also affected intracellular bacterial replication in murine macrophage-like cells and blocked the transport of an epitope-tagged SPI2 effector protein. Two of the compounds significantly inhibited bacterial motility and expression of extracellular flagellin. We conclude that salicylidene acylhydrazides affect bacterial T3SS activity in S. enterica and hence could be used as lead substances when designing specific inhibitors of bacterial T3SSs in order to pharmaceutically intervene with bacterial virulence.
The prokaryotic type III secretion system (T3SS) constitutes a functionally conserved bacterial supramolecular apparatus that, like the structurally related flagellar multiprotein ensemble (32), traverses the cell walls of gram-negative bacteria (29) and allows transport of bacterial proteins from the bacterial cytoplasm into eukaryotic cells upon cell-to-cell contact (11, 21, 22, 23). Many of the translocated effector proteins interfere with intrinsic eukaryotic host cell functions to enable bacterial infection (3, 11). Concomitantly, T3SSs are known to play essential roles in the virulence of a large spectrum of gram-negative pathogens, including Chlamydia, Escherichia coli, Pseudomonas, Salmonella, Shigella, and Yersinia, as well as in the virulence of dedicated plant pathogens, such as Erwinia and Xanthomonas (21, 24).
Identification of specific bacterial virulence factors, such as adhesins, toxins, and T3SSs, and the dissection of their activities and functions at the molecular level have opened the possibility of using low-molecular-weight compounds to functionally interfere with selected virulence factors (25, 34). While still at an early experimental phase, this line of ambitions has identified selected small molecular compounds that do indeed affect adhesin and toxin expression in Vibrio cholerae (26), as well as collections of aromatic substances which inhibit T3SS activity in enteropathogenic E. coli (20). In parallel, salicylidene acylhydrazides have recently been shown to affect T3SSs in Yersinia pseudotuberculosis (27, 38) and Chlamydia trachomatis (37, 51).
Salmonella enterica is a gram-negative pathogen that causes diseases ranging from mild gastroenteritis to severe systemic infection both in humans and animals. Globally, S. enterica serovar Typhi, the causative agent of the most severe form of human salmonellosis, alone is estimated to infect around 22 million people annually, with a concomitant annual mortality of 220,000 individuals (4). Although S. enterica is intrinsically sensitive to many clinically applied antimicrobial agents, antibiotic resistance has become strongly established in S. enterica (40, 52). Since the beginning of the 1990s, strains of Salmonella that show multidrug resistance, including resistance to fluoroquinolone, have emerged to the extent that they today pose a serious public health problem (7, 40), emphasizing the requirement for new control and treatment regimens.
In the murine infection model for systemic salmonellosis, the virulence of S. enterica serovar Typhimurium depends on two separate T3SSs, encoded by Salmonella pathogenicity island 1 (SPI1) and 2 (SPI2), and their concomitant effector proteins (11, 21). SPI1 mediates invasion of the intestinal epithelium (10, 11) and induction of proinflammatory responses (22). At later stages of the infection, SPI2 supports intracellular replication inside professional phagocytic cells through secretion of effector proteins across the phagosomal vacuolar membrane into the cytosolic compartment of the phagocyte (3, 21, 23, 44, 49). Consequently, mutants defective in SPI1 activity are strongly attenuated in the mouse infection model when given orally but not when given intraperitoneally (19). Mutants that are defective in SPI2 activity are attenuated when given through the oral or intraperitoneal route of challenge (44), evidently reflecting their inability to replicate in the reticuloendothelial tissue. Accordingly, SPI2 deficiency results in impaired intracellular replication in cultured macrophage-like cells (3, 42).
We have studied whether salicylidene acylhydrazides are active against S. enterica by analyzing the effects of such compounds on the in vitro activities of the SPI1 and SPI2 T3SSs of S. enterica serovar Typhimurium. The results showed that the compounds efficiently affected SPI1 and SPI2 activity, as well as flagellum-based bacterial motility at micromolar concentrations. Thereby, our study implicates a common target among the various bacterial T3SSs that could be approached by pharmaceutical interference. The applied salicylidene acylhydrazides thus form a promising compound platform for generating broad-acting T3SS inhibitors.
MATERIALS AND METHODS
Bacterial strains and cultivation conditions.
This study was based on the S. enterica serovar Typhimurium strain TT16729 (5) that is deficient in the RpoS σ-factor (50) and on strain 14028 (American Type Culture Collection [ATCC], Manassas, VA). Escherichia coli strain TG1 was used as a host for amplification of plasmid DNA. Bacteria were generally grown in Luria-Bertani (LB) broth or on LB agar plates. To induce expression of SPI2, bacteria were taken from an overnight culture on a LB agar plate and suspended in a minimal medium of low pH (MM5.8 [16, 28]). When necessary, the culture media were supplemented with ampicillin (100 μg/ml; Sigma) or kanamycin sulfate (10 μg/ml; Sigma). The flagellar phase state was determined by agglutination of strain TT16729 with factor H1 or H2 antisera (Reagensia, Sweden).
The hilA::kan-334; hilA::Tn5lacZY, prgH::Tn5lacZY, and sipC::Tn5lacZY (1, 12) elements were kindly provided by Catherine A. Lee and Isabelle Hauteforth and introduced into S. enterica serovar Typhimurium TT16729 by P22 int transduction (6, 15).
Inhibitors.
Nine different salicylidene acylhydrazides were provided by Innate Pharmaceuticals AB, Umeå, Sweden, and their structures are shown in Fig. 1. Compounds D2, D3, D4, D5, D6, D8, and D9 have also been previously described for their effect on Yersinia pseudotuberculosis by Kauppi et al. (27) and Nordfelth et al. (38). The compounds were solubilized in dimethyl sulfoxide (DMSO; Sigma) to a concentration of 20 mM and, when indicated, added to bacterial or cell culture media at a final concentration of 1, 5, 20, 40, or 80 μM. The addition of a corresponding amount of plain DMSO was used as the negative control in all experiments. The solvent concentration never exceeded 0.2% (vol/vol).
FIG. 1.
Salicylidene acylhydrazides used in the study with reference to their D (derivative) and INP (Innate Pharmaceuticals) numbers.
Recombinant DNA technology and plasmids.
Isolation of plasmids and restriction endonuclease cleavages and ligations were carried out as described by Sambrook et al. (43) and following the instructions provided by the producer of the enzymes (New England Biolabs). For the isolation of chromosomal DNA, the Wizard Genomic DNA purification kit (Promega) was used. Plasmids pUC19 and pSU41 have been described previously by Yanisch-Perron et al. (53) and Bartolomé et al. (2), respectively. The plasmid sseJ2HA (31) was kindly provided by David W. Holden.
To generate the pAUN-1 plasmid for expression of a SipB-β-lactamase fusion protein, the open reading frame of the β-lactamase gene of pUC19 was PCR amplified such that the fragment coded for only the processed form of the enzyme. The β-lactamase fragment was subsequently inserted into pSU41 as a BamHI-XbaI fragment. Finally, a PCR fragment, amplified from Salmonella enterica serovar Typhimurium 14028 chromosomal DNA and containing a putative promoter region and the first 65 codons of sipB, was inserted at the 5′ end of the β-lactamase cassette as an EcoRI-BamHI fragment. Thereby, sipB was translationally fused to the β-lactamase reading frame (bla). The primers used for PCR amplification were as follows: sipB forward, 5′-GGAATTCTAAAAATGATTATCGCCC-3′; sipB reverse, 5′-CGGGATCCCGTATTAATAGCGCTCTC-3′; bla forward, 5′-CGGGATCCGTTTTTGCTCACCCAGAA-3′; and bla reverse, 5′-GCTCTAGATTACCAATGCTTAATCAG-3′. The implanted restriction sites are underlined.
Plasmid pHUB61 which carries an spvA-lacZ reporter was available from our previous work (48).
Cell culture and infection models.
For cell culture infections, we used either epithelial MDCK cells (ATCC, Rockville, MD) or murine macrophage-like RAW264.7 cells (TIB-71; American Type Culture Collection, Rockville, MD) cultivated in Dulbecco's modified Eagle medium (DMEM) and RPMI medium (Gibco, Paisley, United Kingdom), respectively. The cell culture media were supplemented with 10% fetal bovine serum (Gibco, Paisley, United Kingdom), l-glutamine (final concentration of 10 mM), HEPES (final concentration of 10 mM), and gentamicin (final concentration of 10 μg/ml). l-Glutamine, HEPES, and gentamicin were all from Sigma, St. Louis, MO.
To obtain phenotypically invasive bacteria for infection of MDCK cells, cultures were grown overnight (16 to 18 h at 37°C) in LB medium, diluted 1:10 into 4 ml of fresh medium, and grown in closed 15-ml Falcon tubes at 37°C on a Brunswick roller incubator for 3 h (6). Thereafter, the bacteria were diluted directly in HEPES-supplemented DMEM cell culture medium and seeded on the cells. RAW264.7 murine macrophage-like cells were infected with bacteria cultured on LB agar plates overnight at 37°C to ensure a noninvasive phenotype (6). Prior to infection, bacteria were suspended in phosphate-buffered saline (PBS) and opsonized in vitro for 30 min at 37°C using 10% fresh nonimmune serum from BALB/c mice. Thereafter, bacteria were diluted in RPMI medium supplemented with HEPES (final concentration of 10 mM) and subsequently seeded on semiconfluent cells grown in 24-well plates.
Cells were infected at a multiplicity of infection of 10:1 and centrifuged for 5 min at 1,500 rpm to facilitate infection. One hour postinfection, maintenance medium containing gentamicin at a final concentration of 50 μg/ml (killing medium) was applied to the plates, and cells were incubated for 45 min to kill extracellular bacteria. For continued incubations, the killing medium was replaced by maintenance medium containing 10 μg/ml of gentamicin. At the indicated time points, the cells were gently washed twice in PBS and disrupted either by using a solution of freshly prepared PBS containing 0.5% (vol/wt) sodium deoxycholate (Merck) (MDCK cells) or by hypotonic lysis (RAW264.7 cells). The number of intracellular bacteria was determined by CFU counts of viable bacteria. Invasion was measured as the ratio between the CFU counts at 2 h postinfection and CFU counts seeded on the cells, whereas bacterial growth yield is defined as CFU counts at 16 h postinfection/CFU counts at 2 h postinfection.
To probe for the effect of salicylidene acylhydrazides, the compounds were added at indicated concentrations either to the bacterial growth medium and the HEPES-buffered DMEM (MDCK cells) or to the killing and maintenance media (RAW264.7 cells).
Isolation and detection of SPI effector proteins.
SPI1-associated effector proteins were isolated as described by Eriksson et al. (15). In short, bacteria were grown in LB medium to promote invasiveness, and then the bacteria were recovered by centrifugation and resuspended in fresh DMEM supplemented with l-glutamine and HEPES. Thereafter, incubation was continued for another hour. The sample volumes were normalized according to the optical density at 600 nm (OD600), the bacteria were removed by repeated centrifugation (two centrifugations, each for 10 min at 14,000 × g), and the proteins were precipitated from the supernatant with the aid of trichloroacetic acid (TCA) at a final concentration of 10%. Pellets were washed in ice-cold acetone and dissolved in reducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. The enriched proteins were subsequently analyzed by 12% SDS-PAGE (30). Proteins were visualized either by staining with Coomassie blue or by immunoblotting as detailed below.
For isolation of SPI2 effector proteins, bacteria were taken from LB agar plates and grown overnight in MM5.8 (16, 28). The sample volumes were normalized according to the OD600, the bacteria were removed by repeated centrifugation as described above, and then the proteins were precipitated from the supernatant with TCA. Samples were also taken from the bacterial pellet and from the hexadecane-extractable fraction (35) obtained from the cellular pellet. The samples were solubilized in reducing SDS-PAGE sample buffer and separated on 12% SDS-polyacrylamide gels (30).
To detect β-lactamase or hemagglutinin (HA)-tagged fusion proteins, the SDS-polyacrylamide gels were blotted onto Hybond-P polyvinylidene difluoride membranes (Amersham Pharmacia Biotech). Fusion proteins were thereafter detected by immunostaining using either a polyclonal rabbit anti-β-lactamase serum (1:20,000; AB 3738; Chemicon International) or a mouse monoclonal anti-HA tag antibody (1:1,000; HA probe F-7; Santa Cruz Biotechnology). The Chemoluminescence Western Blotting kit (Roche) or the SuperSignal kit (Pierce) were used to detect bound antibody.
Bacterial cell fractionation.
Bacterial cultures of S. enterica serovar Typhimurium TT16729 carrying pAUN-1 were grown to invasiveness in kanamycin-supplemented LB medium (6), transferred to DMEM supplemented with kanamycin, l-glutamine, and HEPES, and incubated for another 2 h. When indicated, compound D9 was present in all cultivation steps at 20 μM. The cells were pelleted by centrifugation, washed once in PBS, dissolved in distilled water and finally frozen at −70°C. The subsequently defrosted cell suspensions were treated with lysozyme (Sigma) at a final concentration of 1 mg/ml for 1 hour at 4°C and sonicated until the opaque suspensions cleared. Next, the samples were centrifuged at low speed (5,000 × g, 5 min, 4°C) to remove intact bacteria and debris, and then the supernatants were fractionated by ultracentrifugation (50,000 × g, 1 h, 6°C). To separate cytoplasmic membrane-associated proteins from the cell wall-associated fraction, the samples were vigorously suspended in 20 mM Tris-HCl, 5 mM MgCl2, and 1% (vol/vol) Triton X-100 buffer, pH 7.5. This procedure allowed us to make a crude fractionation of the bacterial cell into a cytoplasmic Triton X-100-soluble fraction and into a fraction not soluble in a neutral detergent (41).
For enrichment of periplasmic proteins, the bacterial culture was grown as described above, yet the bacterial pellet was suspended in a 10 mM Tris-HCl and 2 mM EDTA buffer containing 20% (wt/vol) sucrose. The suspension was left standing for 20 min at room temperature, and then the bacteria were pelleted by centrifugation (14,000 × g, 15 min, room temperature). The pellet was dissolved in 5 mM Tris-HCl buffer to generate an osmotic shock. Bacteria were subsequently removed from the suspension by centrifugation (14,000 × g, 15 min, 4°C), and the supernatant was assayed for β-lactamase activity as detailed below.
Assaying β-lactamase activity.
The activity of β-lactamase was measured by probing the degradation of the chromogenic β-lactam nitrocefin (Calbiochem). The reactions were carried out in PBS supplemented with nitrocefin at a final concentration of 50 μg/ml. Two types of units were used to define activity: (i) “arbitrary units” defined as A486/reaction time (in minutes) × OD600 of the culture, or (ii) as “units” defined as OD486/reaction time (in minutes) × microgram of protein. Protein concentrations were measured by the Bradford method (kit provided by Bio-Rad) and using bovine serum albumin as the protein standard.
Determination of β-galactosidase activity.
The transcriptional activity of lacZ fusion constructs was measured by cleavage of ortho-nitrophenyl-β-d-galactopyranoside (ONPG; Sigma) by the method of Miller (36).
Detection of iNOS and production of nitric oxide.
Detection of inducible nitric oxide synthase (iNOS) was carried out by immunoblotting using anti-iNOS (BD Biosciences) as detailed by Bjur et al. (6). To enable comparisons of band intensities, actin was detected by immunoblotting in parallel as an internal standard. Rabbit anti-actin was from Sigma.
Production of nitric oxide was measured through the accumulation of nitrite in the cell culture medium. Nitrite was assayed with the Griess reagent (Merck), and the OD525 values were converted to micromolar concentrations using defined concentrations of sodium nitrite (Merck) as the standard (6, 15).
Motility assay and extraction of flagellin.
Bacterial motility (swimming) was tested on soft agar plates (tryptone [10 g/liter], NaCl [5 g/liter], and 0.3% agar) prepared 1 day ahead. Twenty microliters of an overnight culture was inoculated into the agar, and motility was read as the bacterial zone diameter after 8 h of incubation at 37°C. When indicated, salicylidene acylhydrazides were added to the bacterial swimming medium at a final concentration of 40 μM.
Flagellin was released from the bacterial cell surface using low pH denaturation (47). Briefly, bacteria were grown overnight (16 to 18 h) in LB medium in the absence or presence of salicylidene acylhydrazide D6 or D7. The bacteria were pelleted and suspended in 150 mM NaCl and 10 mM HCl and left standing for 5 min at room temperature. Thereafter, the bacteria were removed from the supernatant by repeated centrifugations (14,000 × g, 10 min), and then the proteins from the supernatant were precipitated with TCA as described above for SPI1 effector proteins and analyzed by SDS-PAGE on 9% gels (30).
Statistical analyses.
Each experiment was repeated at least three times, and a two-sided t test was used to determine statistical significance between the values for the different groups.
RESULTS AND DISCUSSION
Salicylidene acylhydrazides inhibit cellular invasion of S. enterica serovar Typhimurium.
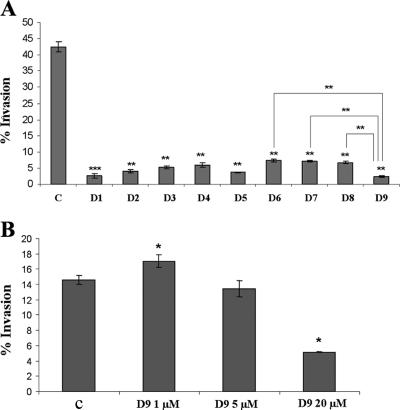
The ability to secrete effector proteins through the SPI1 T3SS strongly correlates with the ability of S. enterica serovar Typhimurium to invade cultured epithelial cells (10, 14, 24, 34). In this study we have analyzed salicylidene acylhydrazides (here designated D1 to D9) (Fig. 1), previously reported to affect the T3SS of Y. pseudotuberculosis (27, 38), for their ability to affect invasion of epithelial MDCK cells by S. enterica serovar Typhimurium TT16729. The rationale for choosing TT16729 was that this strain in our hands invades MDCK cells at a high frequency and that the strain expresses large amounts of SPI1 effector proteins in vitro. Thus, MDCK cells were infected with bacteria grown in the presence or absence of the different compounds under SPI1-inducing conditions. As would be expected from our previous study (6), up to 40% of the bacteria seeded on the cells were internalized within 2 hours postinfection in the absence of salicylidene acylhydrazides (Fig. 2A). Interestingly, all nine salicylidene acylhydrazides significantly inhibited bacterial invasion when the bacteria had been cultured in the presence of these compounds at a concentration of 20 μM (Fig. 2A).
FIG. 2.
Addition of salicylidene acylhydrazides to the bacterial growth medium inhibits the ability of bacteria to invade epithelial MDCK cells. (A) S. enterica serovar Typhimurium TT16729 was grown in LB medium in the presence of a solvent control (C) or in the presence of one of nine (D1 to D9) different derivatives of salicylidene acylhydrazides at a final concentration of 20 μM. Thereafter, the bacteria were assayed for their ability to invade epithelial MDCK cells in a gentamicin protection assay. The values are given as the percentage of invading bacteria as related to the initial input. (B) Dose dependency of salicylidene acylhydrazide D9 in inhibiting S. enterica serovar Typhimurium TT16729 from invading cultured MDCK cells. S. enterica serovar Typhimurium TT16729 was grown in LB medium in the presence of a solvent control (C) or in the presence of D9 at the indicated concentrations. Otherwise, the experiment was performed as described above for panel A. The levels of statistical significance are indicated as follows: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.005. Unless otherwise indicated, the P values were calculated by comparing the values to the value for the solvent-treated control.
Thereby, the salicylidene acylhydrazides showed a clear ability to block the invasiveness of S. enterica serovar Typhimurium at concentrations nearly identical to the concentrations required to affect either the Y. pseudotuberculosis or C. trachomatis T3SS activity (27, 37, 38, 51). One possible explanation for this broad antibacterial activity could be that salicylidene acylhydrazides exert a nonspecific inhibitory effect on the bacteria. However, several observations support a more specific mode of action. At 20 μM to 40 μM, the compounds did not affect the growth of S. enterica serovar Typhimurium TT16729 in LB medium (data not shown), and the compounds did not pose any apparent toxic effects on the MDCK cells at these concentrations (data not shown). Furthermore, not all compounds mediated inhibition to the same level; while compounds D6 to D8 inhibited invasion, the inhibition was significantly less effective compared to the most potent inhibitor D9 (Fig. 2A). Finally, when compound D9 was tested in the Yersinia model, it did not significantly block T3SS activity within the relevant concentration range (38). Yet, when the most potent inhibitor, compound D9, was tested at 5 μM, no significant inhibition of invasion of MDCK cells by S. enterica serovar Typhimurium was observed (Fig. 2B). At 1 μM we observed a small increase in invasion efficacy of D9-treated bacteria (Fig. 2B), which may reflect poor solubility of the compound in water. Therefore, we choose to use the compounds at concentrations ranging from 20 to 80 μM for the subsequent series of experiments.
Salicylidene acylhydrazides efficiently inhibit SPI1 effector protein secretion by S. enterica serovar Typhimurium in vitro.
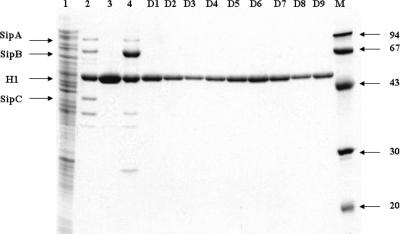
As Salmonella relies on the activity of SPI1 for invasion of epithelial cells (9, 10, 13, 21, 24, 39), the data presented in Fig. 2A indicated that the salicylidene acylhydrazides inhibited invasion by affecting SPI1 activity. To verify this, S. enterica serovar Typhimurium TT16729 wild-type bacteria were grown in the presence or absence of the different compounds (at 20 μM) under conditions well-known to induce secretion of SPI1 effector proteins into the culture medium (6, 9, 10, 39). When culture supernatants were analyzed by SDS-PAGE, a characteristic profile of SPI1 proteins accumulating in the culture supernatant was detected in the absence of the salicylidene acylhydrazides (Fig. 3, lane 2). This profile was distinct from that of a whole-cell bacterial lysate from the same culture (Fig. 3, lane 1) and not found in the culture supernatant of a hilA mutant defective in SPI1 gene expression (Fig. 3, lane 3), indicating that the proteins (apart from flagellin) were derived from SPI1. The SPI1 origin of the bands was further confirmed by analysis of the expression profile of a sipC mutant (Fig. 3, lane 4).
FIG. 3.
Salicylidene acylhydrazides inhibit secretion of S. enterica serovar Typhimurium invasion-associated SPI1 effector proteins in vitro. S. enterica serovar Typhimurium TT16729 was grown in LB medium in the presence of a solvent control or in the presence of one of nine different derivatives (D1 to D9) of salicylidene acylhydrazides at a final concentration of 20 μM. The bacteria were removed by repeated centrifugation, the proteins were precipitated from the culture supernatant (except for lane 1) with the use of TCA, and then the precipitates were analyzed by 12% SDS-PAGE followed by staining with Coomassie blue. Lane 1, whole-cell lysate prepared from a culture treated with the solvent alone; lane 2, the secreted protein profile obtained when treated only with the solvent control; lane 3, the protein profile obtained with a hilA mutant of S. enterica serovar Typhimurium; lane 4, the protein profile obtained with a sipC mutant of S. enterica serovar Typhimurium. The protein profiles obtained with compounds D1 to D9 are shown in the corresponding lanes. The positions of the molecular mass markers (in kilodaltons) are shown to the right of lane M, and the arrows to the left of the gel indicate the predicted mobility of Sip proteins presented in lane 2 and flagellin H1 proteins.
At 20 μM, the concentration defined as inhibiting invasion, all nine compounds inhibited SPI1 effector protein secretion to a level below detection (Fig. 3, lanes D1 to D9). This showed that salicylidene acylhydrazides affected the activity of the SPI1-associated T3SS and concomitantly provided an explanation for the ability of the compounds to inhibit T3SS-mediated invasion without compromising bacterial growth in vitro.
Salicylidene acylhydrazides prevent secretion of a SipB-β-lactamase fusion protein.
We next aimed to identify the level of inhibition of SPI1 effector protein secretion caused by salicylidene acylhydrazides. For this purpose, we generated a molecular reporter that would allow quantitative detection of a SPI1 effector protein in different bacterial cell fractions. To achieve this, a recombinant plasmid, pAUN-1, expressing the 65 N-terminal amino acids of the SPI1 effector protein SipB translationally fused to β-lactamase, was constructed (for details, see Materials and Methods). When bacteria carrying pAUN-1 were grown under SPI1-inducing conditions, the fusion protein was detected in the culture supernatant of S. enterica serovar Typhimurium, either by immunoblotting or by assaying the enzymatic activity of β-lactamase (Fig. 4A and D). Thereby, pAUN-1 proved suitable for analyzing the effects of the salicylidene acylhydrazides on SPI1 secretion in both a nonquantitative assay and a quantitative assay.
FIG. 4.
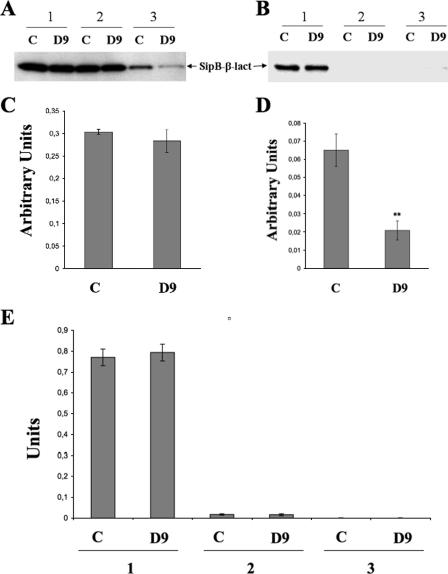
(A) Detection of β-lactamase by immunoblotting from S. enterica serovar Typhimurium TT16729 carrying pAUN-1 and corresponding culture fractions in the absence (solvent control [C]) or presence of D9 at 20 μM. Section 1, bacteria and the culture supernatant; section 2, whole bacterial cells only; 3, culture supernatant only. (B) Detection of β-lactamase by immunoblotting from subcellular fractions of S. enterica serovar Typhimurium TT16729 carrying pAUN-1. Section 1, cytoplasmic fraction; section 2, Triton X-100-soluble cell wall fraction; section 3, Triton X-100-insoluble cell wall fraction. SipB-β-lact., SipB-β-lactamase fusion protein. (C and D) Enzymatic detection of β-lactamase from S. enterica serovar Typhimurium TT16729 carrying pAUN-1 from whole-cell lysates (C) and soluble secreted culture fractions (D). (E) Enzymatic detection of β-lactamase from cytosolic fraction (section 1), Triton X-100-soluble fraction (section 2), and insoluble cell wall fraction (section 3) prepared from S. enterica serovar Typhimurium TT16729 carrying pAUN-1. Fractions obtained from solvent controls (C) and fractions obtained from D9 compound-treated cultures (D9) are indicated.
Next, the experiment was repeated with S. enterica serovar Typhimurium TT16729 carrying pAUN-1 grown in the absence or presence of the salicylidene acylhydrazide D9 (20 μM). Prior to analysis of the presence of SipB or β-lactamase activity, the cultures were fractionated into a soluble secreted fraction, a cytosolic fraction, and a cell wall-associated, Triton X-100-solubilized fraction and a detergent-insoluble fraction.
In untreated control cultures, the SipB-β-lactamase fusion protein was detected by immunoblotting both in the cytosolic and secreted fractions, whereas no significant signal could be detected in the Triton X-100-insoluble or -soluble, cell wall-associated fractions (Fig. 4A and B). When the bacterial fractions were assayed for β-lactamase activity, a high enzymatic activity was detected in the cytosolic fraction for both treated and untreated bacteria (Fig. 4E). In contrast, the soluble secreted fraction from D9-treated bacteria exhibited a substantially lower β-lactamase activity (Fig. 4D). No significant enzymatic activity could be detected in the cell wall-associated fractions. Thus, the data strongly indicated that compound D9 significantly affected transport, but not expression or intracellular stability of the SipB-β-lactamase fusion protein. The data also implied that D9-mediated inhibition of SipB-β-lactamase secretion did not involve trapping SipB-β-lactamase in the cell wall. Likewise, when osmotic shock samples were analyzed for periplasmic localization of the fusion protein, we could not demonstrate any detectable β-lactamase activity from either untreated or treated cultures (data not shown).
Combined, the experiments described in Fig. 4 showed that compound D9 did not affect the expression of the fusion protein but rather prevented its secretion from the bacterial cytoplasm.
Salicylidene acylhydrazides prevent expression of SPI1 lacZ transcriptional gene fusions.
The fact that salicylidene acylhydrazides blocked the secretion of T3SS effector proteins, but not the expression or translation of the SipB-β-lactamase effector fusion protein, suggested that the T3SS system itself was affected either through blockage in the expression of the T3SS machinery or through interference with T3SS activity. Therefore, we continued by analyzing the transcriptional activity of SPI1 in the absence or presence of salicylidene acylhydrazides.
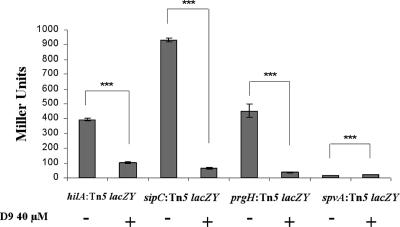
Usage of multicopy readout systems, such as the sipB-bla fusion construct, for assessment of gene expression may affect quantification of transcription. To exclude this possibility, we used single-copy chromosomal transcriptional lacZ fusion constructs that separately probed for the transcriptional activity of the genes encoding the SPI1 transcriptional activator HilA, the translocon protein PrgH, or the effector protein SipC. As expected, all three lacZ fusion constructs showed a clear activity when the bacteria were grown to invasiveness in the absence of the T3SS inhibitory compound (Fig. 5). In contrast, growth in the presence of compound D9 (40 μM) resulted in a significant decrease in transcription of the key SPI1 activator gene (hilA) and in an even more pronounced decrease in transcription of the prgH translocon gene and the sipC effector protein gene (Fig. 5).
FIG. 5.
Activities of chromosomal lacZ fusions probing for hilA, prgH, sipC, or spvA transcription in the absence (−) or presence (+) of compound D9. The level of statistical significance (P ≤ 0.001) for the values is indicated (***).
Next, we wanted to exclude the possibility that the salicylidene acylhydrazide compounds could have a specific inhibitory effect on β-galactosidase activity itself or on the expression of lacZ in general. Compound D9 or plain DMSO was therefore added to the reaction mixture at concentrations corresponding to 40 μM culturing conditions. However, this did not affect the ability of LacZ-driven cleavage of the chromogenic substrate used (data not shown). Furthermore, we tested the capacity of compound D9 to affect the LacZ activity of a lacZ reporter connected to the promoter of the Salmonella virulence gene spvA (48), which is poorly expressed in complex growth medium (16). In these experiments, neither DMSO nor compound D9 caused decreased spvA-lacZ expression when added to the growth medium of S. enterica serovar Typhimurium harboring the spvA-lacZ construct pHUB61. In contrast, we recorded a small but statistically significant increase in spvA-lacZ activity upon application of D9 (Fig. 5). Thus, treatment of S. enterica serovar Typhimurium with the salicylidene acylhydrazide D9 was clearly associated with a transcriptional down-regulation of the expression of SPI1-associated lacZ transcriptional gene fusions, yet the compound did not inhibit β-galactosidase activity per se or expression of the spvA-lacZ control. This clearly implied that the salicylidene acylhydrazide caused a transcriptional silencing of SPI1. The concomitant lack of SPI1-mediated secretion competence is therefore the most likely explanation for the observed lack of effector proteins in the presence of D9 (Fig. 3 and 4).
The Yersinia T3SS is transcriptionally down-regulated upon bacterial exposure to the compounds (38). For Y. pseudotuberculosis, this compound-mediated suppression of gene expression is envisioned to originate from an intracellular accumulation of the Yersinia T3SS repressor protein LcrQ (27, 38). In the absence of salicylidene acylhydrazides or Ca2+, LcrQ is secreted by the T3SS itself to create an autoactivating expression of the T3SS (27). Thus, the compounds have been suggested to mimic the classical Ca2+-mediated suppression of yersinial T3SS activity (38). In the present study, we could demonstrate a transcriptional silencing of the SPI1 T3SS in response to salicylidene acylhydrazides. However, Ca2+ does not suppress expression of SPI1, and SPI1 does not include any homologue for lcrQ (13). On the other hand, a number of insertion mutations have been reported in S. enterica serovar Typhimurium that relieve the SPI1 from its environmental regulation (17), which might implicate the presence of LcrQ-like proteins.
Salicylidene acylhydrazides affect bacterial intracellular replication.
Mutants of S. enterica serovar Typhimurium that are defective in SPI2 activity are affected for intracellular replication in macrophage-like cells (6, 21, 42). This prompted us to test whether salicylidene acylhydrazides could affect bacterial intracellular replication through inhibition of SPI2 T3SS activity. However, the S. enterica strain TT16729 is of LT2 origin and hence deficient in the expression of the virulence-associated alternative σ-factor RpoS (50). While RpoS deficiency is not expected to affect invasion (50), RpoS deficiency could potentially have an impact on intracellular replication through RpoS-dependent expression of stress responses (46) and the spv virulence genes (5, 16, 18). Therefore, we included the RpoS-producing S. enterica serovar Typhimurium strain 14028 in the following experiments.
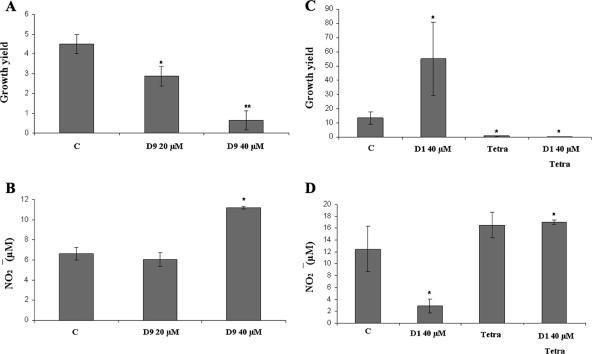
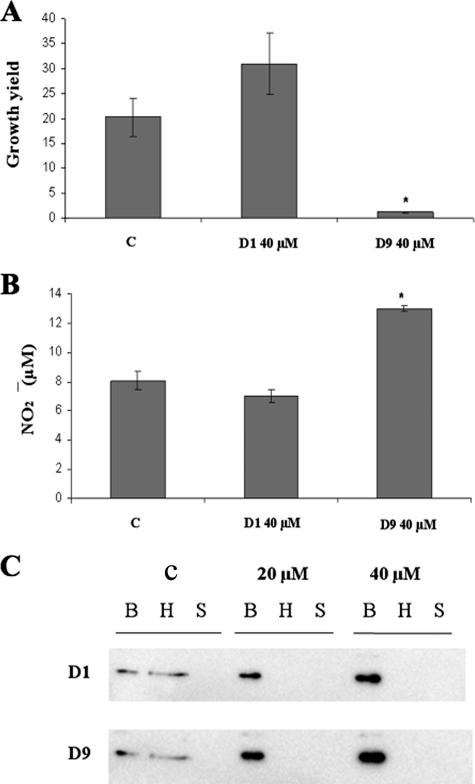
To probe for the effects of salicylidene acylhydrazides on SPI2 activity, we choose to use the two most efficient salicylidene acylhydrazide SPI1 inhibitors (D1 and D9). Thus, murine macrophage-like RAW264.7 cells were infected with either strain TT16729 or 14028 in the presence or absence of either D1 or D9 in the cell culture medium at a final concentration of 40 μM. Cells were lysed at 2 h and 16 h postinfection for CFU counts of viable bacteria. In this series of experiments, D9 indeed reduced the intracellular growth yields (CFU at 16 h) of both S. enterica serovar Typhimurium strains TT16729 and 14028 (Fig. 6A and 7A). Importantly, we could not observe any inhibition of bacterial intracellular replication if compound D9 was omitted from the cell culture medium and instead added to the bacterial growth medium prior to infection (data not shown). This suggested that D9 inhibited bacterial replication during the intracellular phase of infection. In contrast, when compound D1 was maintained in the cell culture medium during infection, a substantial increase in the bacterial intracellular replication was observed for TT16729 (Fig. 6C).
FIG. 6.
Effects of salicylidene acylhydrazides D1 and D9 on bacterial intracellular replication in murine macrophage-like RAW264.7 cells. Phenotypically noninvasive cultures of S. enterica serovar Typhimurium TT16729 were opsonized with complement and seeded on RAW264.7 macrophage-like cells. Compound D9 or D1 was added to the cell culture medium at the indicated concentrations 1 h postinfection and maintained throughout the experiment. Samples were taken at 2 and 16 h postinfection for CFU counts of intracellular bacteria. In parallel, the nitric oxide produced by the RAW264.7 macrophage-like cells was measured as the nitrite accumulated in the cell culture medium 16 h postinfection. (A and B) Growth yields (CFU at 16 h/CFU at 2 h) obtained with strain TT16729 (A) and with the corresponding nitric oxide levels (B). C, solvent-treated control. (C and D) Corresponding growth yields (C) and nitric oxide levels obtained with compound D1 at 40 μM (D). When indicated, the bacterial protein synthesis was blocked with the use of tetracycline (Tetra) at 10 μg/ml. The levels of statistical significance are indicated as follows: *, P ≤ 0.05; **, P ≤ 0.01. All P values were calculated by comparing the values to the value of the solvent-treated control (C).
FIG. 7.
Effects of salicylidene acylhydrazides D1 and D9 on bacterial intracellular replication in murine macrophage-like RAW264.7 cells and on the secretion of the SseJ-HA fusion protein in vitro. Phenotypically noninvasive cultures of S. enterica serovar Typhimurium 14028 were opsonized with complement and fed to RAW264.7 macrophage-like cells. Compound D9 or D1 was added to the cell culture medium at the indicated concentration 1 h postinfection and maintained throughout the experiment. Bacterial intracellular replication was measured 16 h postinfection. In parallel, the nitric oxide produced by the RAW264.7 macrophage-like cells was measured as the nitrite accumulated in the cell culture medium. (A and B) Growth yields (CFU at 16 h/CFU at 2 h) obtained with strain 14028 (A) and with the corresponding nitric oxide levels (B) when using compound D1 or D9 at 40 μM. C, solvent-treated control. (C) Detection of SseJ-HA fusion protein from S. enterica serovar Typhimurium 14028 by immunoblotting 12% SDS-polyacrylamide gels. Signals obtained with whole bacterial cells (B), hexadecane-extractable fraction (H), or cell-free supernatant (S) from cultures treated with solvent alone (C) or compound D1 or D9 are shown. The level of statistical significance (P ≤ 0.05) is indicated by an asterisk. All P values were calculated by comparing the values to the value for the solvent-treated control.
Salicylidene acylhydrazides, bacterial intracellular replication, and the production of nitric oxide.
The abilities of mice and of macrophage-like J774-A.1 and RAW264.7 cells to restrict intracellular replication of S. enterica serovar Typhimurium is largely dependent on nitric oxide (NO) produced through the innate induction and activation of iNOS in response to salmonella infection (6, 8, 15, 33). In parallel, SPI2 is known to interfere with the host cell NO response and thereby to promote bacterial intracellular replication in monocytic cells (6, 8, 33).
When RAW264.7 cell production of NO in response to infection was measured in parallel to bacterial growth yields, increased NO levels were observed when the infection was carried out in the presence of compound D9 (Fig. 6B and 7B). This observation is in line with the previously reported inverse correlation between the levels of NO produced by infected macrophage-like cells and the intracellular growth yields of S. enterica serovar Typhimurium (6, 8, 33) and could well explain the decreased bacterial intracellular replication observed upon treatment with compound D9. In contrast, the increased bacterial intracellular growth yield observed for strain TT16729 in the presence of D1 (Fig. 6C) was accompanied by a repression of host cell NO production (Fig. 6D).
Parallel immunoblotting of iNOS from infected cells did not show any compound-mediated decrease or increase in iNOS signals compared to infected cells treated with solvent control only (data not shown). This indicated that the compounds did not interfere with RAW264.7 cell protein synthesis and that the effect on NO production instead originated from a down-regulation of iNOS activity. As inhibition of iNOS activity has recently been associated with SPI2 activity (6), subsequent controls were carried out to establish whether the compounds affected iNOS activity of RAW264.7 cells directly or indirectly through interference with the activity of the infecting bacteria.
Exposure of uninfected RAW264.7 cells to compound D1 or D9 or to the solvent DMSO at 0.2% (vol/vol) did not affect the low background level of NO production in uninfected cells (data not shown). Thus, neither the solvent nor the salicylidene acylhydrazides per se caused activation of iNOS. To probe for the association between bacterial infection, decreased NO expression, and compound D1, bacterial protein synthesis was blocked at 2 h postinfection by the use of the bacteriostatic protein translation inhibitor tetracycline. In this experiment, RAW264.7 cells exhibited a strong NO response 16 h postinfection both in the absence and presence of D1 (Fig. 6D). Thus, the observed compound-mediated alterations in NO production were not caused directly by the compounds, but through the effect of the compounds on the infecting bacteria, potentially through interference with the bacterial SPI2 T3SS activity.
Effect of salicylidene acylhydrazide on SPI2 T3SS activity in S. enterica serovar Typhimurium in vitro.
Having demonstrated that salicylidene acylhydrazides affected bacterial intracellular replication, we proceeded to study the effect on the SPI2 T3SS in vitro. However, when either S. enterica serovar Typhimurium TT16729 or 14028 was grown under SPI2-inducing conditions, the yield of effector proteins remained too low to allow reproducible detection with Coomassie blue protein staining of SDS-polyacrylamide gels, even after extraction with hexadecane (data not shown), a procedure known to gently but specifically release T3SS effector proteins from the bacterial cell surface (35). An attempt to express a SPI2 effector protein fused to β-lactamase yielded β-lactamase signals of very low intensity as probed for by immunoblotting (data not shown). Evidently, SPI2 effector protein production remained relatively low even under the SPI2-inducing conditions applied. Furthermore, the use of large C-terminal extensions may in itself have affected SPI2 effector protein secretion.
To overcome the above-mentioned problems, we continued by using an sseJ2HA construct that expressed the entire SseJ SPI2 effector protein with a short C-terminal HA tag (31). Thus, S. enterica serovar Typhimurium 14028 containing the sseJ2HA construct was grown under SPI2-inducing conditions and analyzed by immunoblotting. In this experiment, a clear signal was detected in the expected region of the fusion protein (approximately 65 kDa) in the whole-cell fraction, as well as in the hexadecane-extractable fraction (Fig. 7C). In the presence of either compound D1 or D9, however, a significant reduction in signal intensity was observed in the hexadecane-extracted fraction accompanied by a corresponding increase in signal intensity in the bacterial cell-containing fraction (Fig. 7C). This implied that the compounds D1 and D9 affected the secretion of HA-tagged SseJ (SseJ-HA).
We could not detect any soluble secreted form of the SseJ-HA protein from S. enterica serovar Typhimurium strain 14028 or strain TT16729. One explanation for this could be that the protein was transported but not released from the bacterial cell surface. Another possible explanation for the absence of soluble SseJ-HA may be proteolytic degradation of soluble SseJ-HA. Our previous microarray analyses show that growth of S. enterica serovar Typhimurium in MM5.8 strongly induces the expression of the gene encoding the outer membrane protease PgtE (16). Pla, the PgtE homologue of Yersinia pestis, is known to mediate degradation of T3SS effector proteins (45). Hence, under the growth conditions used, the SPI2 effector proteins may have been degraded by PgtE. However, we did not record any pattern of SseJ-HA degradation in the hexadecane-extractable fraction in the absence of the salicylidene acylhydrazides (Fig. 7C); therefore, the latter explanation seems highly unlikely. Thus, the data in Fig. 4 and Fig. 7 indicated that the salicylidene acylhydrazides tested prevented secretion, but not expression or stability of the tagged SseJ-HA protein and the SipB-β-lactamase fusion proteins.
Salicylidene acylhydrazides affect motility and flagellin expression in S. enterica serovar Typhimurium.
The enterobacterial flagellar motility system is related to the SPI1 and SPI2 T3SSs and could in essence be regarded to constitute an additional T3SS in S. enterica (32). Therefore, we also tested the potential of salicylidene acylhydrazides to affect motility. Indeed, two of the salicylidene acylhydrazide compounds (D5 and D6 at 40 μM) significantly reduced bacterial motility on soft agar plates (Table 1).
TABLE 1.
Swimming zones obtained with S. enterica serovar Typhimurium TT16729 in the presence of salicylidene acylhydrazides or the solvent control
| Compounda | Diameter (cm) of swimming zoneb |
|---|---|
| C | 3.1 (0.3) |
| D1 | 3.1 (0.2) |
| D2 | 3.3 (0.2) |
| D3 | 2.9 (0.1) |
| D4 | 2.7 (0.2) |
| D5 | 2.4 (0.2)* |
| D6 | 1.7 (0.2)** |
| D7 | 2.7 (0.1) |
| D8 | 2.9 (0.2) |
| D9 | 2.7 (0.3) |
The compounds are salicylidene acylhydrazides (D1 to D9) or the solvent control (C).
Values represent means and standard deviations (shown in parentheses) from three separate experiments. Values that are significantly different from the value for the solvent control are indicated as follows: *, P ≤ 0.05; **, P ≤ 0.01.
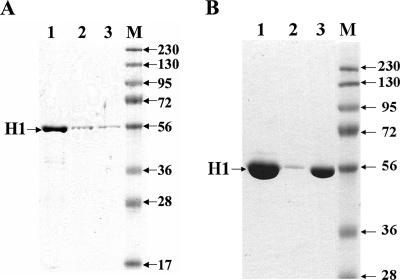
As the above experiments on SPI1 and SPI2 implied that the compounds affected effector protein secretion, we isolated the flagellar fraction from S. enterica serovar Typhimurium TT16729 when grown in the absence or presence of compound D5 or D6 at 40 μM. When analyzed by SDS-PAGE, the flagellin signal was significantly lower in the sample obtained from compound-treated cultures (Fig. 8A). We next analyzed flagellin expression by growing the bacteria in the presence of compound D6 or in the presence of the less potent motility inhibitor D7 at 80 μM. At this concentration, the flagellin signal was barely detectable in the sample from the D6-treated culture, and a larger amount of the sample had to be loaded on the gel to enable detection (Fig. 8B). At 80 μM, the bacteria treated with compound D7 still exhibited surface expression of flagellin but at a lower level compared to that of the solvent-treated control (Fig. 8B). Thus, salicylidene acylhydrazides had the ability to inhibit motility, evidently due to preventing surface expression of flagellae.
FIG. 8.
(A) Detection of bacterial cell-bound flagellin on 9% SDS-polyacrylamide gels from cultures treated with solvent alone (lane 1) or with compound D5 (lane 2) or D6 (lane 3) at 40 μM. (B) Detection of flagellin on SDS-polyacrylamide gels from cultures treated with solvent alone (lane 1), or with compound D6 (lane 2) or 7 (lane 3) at 80 μM. M indicates The positions of molecular mass markers (M) (in kilodaltons) are shown to the right of the gels. The OD600 values of the cultures were used to normalize the sample volume loaded.
Conclusions.
With the aim of specifically targeting bacterial virulence determinants by the use of small molecular compounds, a screen was recently performed that identified salicylidene acylhydrazides affecting the T3SS of Yersinia pseudotuberculosis (27, 38). Here, we have applied a subset of the salicylidene acylhydrazide collection (Fig. 1) to scrutinize their effects on the SPI1 and SPI2 T3SS systems and the related flagellar motility system of S. enterica serovar Typhimurium. Our results showed that the compounds affected all three T3SSs yet with somewhat different efficiencies and consequences.
Although the components of the bacterial T3SSs have been conserved, they possess substantial differences in their primary structures (21, 23, 32). Therefore, it may come as no surprise that different, structurally related, small molecular compounds act differently upon encountering separate T3SSs, yet they merge on a common target or target mechanism. Since this class of compound targets T3SS in Chlamydia, Salmonella, and Yersinia, we are now undertaking additional studies which aim to identify the molecular target of these compounds in each organism. Moreover, based on the activity profiles, more target-focused libraries of salicylidene acylhydrazides will be designed. Subsequent synthesis of such compounds and evaluation in all three organisms will allow us to determine the structural features required for activity against all three organisms and features resulting in selectivity for one over the others. This strategy will result in valuable research tools and chemical starting points for development of novel antibacterial drugs.
Acknowledgments
This study was supported by the Swedish Research Council, Cancerfonden, and the EU-founded Marie Curie IMO-TRAIN Graduate Education Program.
We are also grateful to David W. Holden, Catherine A. Lee, and Isabelle Hautefort for providing genetic constructs and to Innate Pharmaceuticals AB for providing salicylidene acylhydrazides.
Footnotes
Published ahead of print on 4 June 2007.
REFERENCES
- 1.Bajaj, V., C. Hwang, and C. A. Lee. 1995. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol. Microbiol. 18:715-727. [DOI] [PubMed] [Google Scholar]
- 2.Bartolomé, B., Y. Jubete, E. Martínez, and F. de la Cruz. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75-78. [DOI] [PubMed] [Google Scholar]
- 3.Beuzon, C. R., S. P. Salcedo, and D. W. Holden. 2002. Growth and killing of a Salmonella enterica serovar Typhimurium sifA mutant strain in the cytosol of different host cell lines. Microbiology 148:2705-2715. [DOI] [PubMed] [Google Scholar]
- 4.Bhan, M. K., R. Bahl, and S. Bhatnagar. 2005. Typhoid and paratyphoid fever. Lancet 366:749-762. [DOI] [PubMed] [Google Scholar]
- 5.Björkman, J., M. Rhen, and D. I. Andersson. 1996. Salmonella typhimurium cob mutants are not hypervirulent. FEMS Microbiol. Lett. 139:121-126. [DOI] [PubMed] [Google Scholar]
- 6.Bjur, E., S. Eriksson Ygberg, and M. Rhen. 2006. The O-antigen affects replication of Salmonella enterica serovar Typhimurium in murine macrophage-like J774-A.1 cells through modulation of host cell nitric oxide production. Microbes Infect. 8:1826-1838. [DOI] [PubMed] [Google Scholar]
- 7.Butaye, P., G. B. Michael, S. Schwarz, T. J. Barrett, A. Brisabois, and D. G. White. 2006. The clonal spread of multidrug-resistant non-typhi Salmonella serotypes. Microbes Infect. 8:1891-1897. [DOI] [PubMed] [Google Scholar]
- 8.Chakravottry, D., I. Hansen-Wester, and M. Hensel. 2002. Salmonella pathogenicity island 2 mediates protection of intracellular Salmonella from reactive nitrogen species. J. Exp. Med. 195:1155-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clements, M. O., S. Eriksson, A. Thompson, S. Lucchini, J. C. Hinton, S. Normark, and M. Rhen. Polynucleotide phosphorylase is a global regulator of virulence and persistency in Salmonella enterica. Proc. Natl. Acad. Sci. USA 99:8784-8789. [DOI] [PMC free article] [PubMed]
- 10.Collazo, C. M., and J. E. Galan. 1996. Requirement for exported proteins in secretion through the invasion-associated type III system of Salmonella typhimurium. Infect. Immun. 64:3524-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collazo, C. M., and J. E. Galan. 1997. The invasion-associated type-III protein secretion system in Salmonella—a review. Gene 192:51-59. [DOI] [PubMed] [Google Scholar]
- 12.Darwin, K. H., and V. I. Miller. 1999. InvF is required for expression of genes encoding proteins secreted by the SPI1 type III secretion apparatus in Salmonella typhimurium. J. Bacteriol. 181:4949-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Day, J. B., and C. A. Lee. 2003. Secretion of the orgC gene product by Salmonella enterica serovar Typhimurium. Infect. Immun. 71:6680-6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eichelberg, K., and J. E. Galán. 1999. Differential regulation of Salmonella typhimurium type III secreted proteins by pathogenicity island 1 (SPI-1)-encoded transcriptional activators InvF and HilA. Infect. Immun. 67:4099-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eriksson, S., J. Björkman, S. Borg, A. Syk, S. Pettersson, D. I. Andersson, and M. Rhen. 2000. Salmonella typhimurium mutants downregulating phagocyte nitric oxide production. Cell. Microbiol. 2:239-250. [DOI] [PubMed] [Google Scholar]
- 16.Eriksson Ygberg, S., M. O. Clements, A. Rytkönen, A. Thompson, D. W. Holden, J. C. Hinton, and M. Rhen. 2006. Polynucleotide phosphorylase negatively controls spv virulence gene expression in Salmonella enterica. Infect. Immun. 74:1243-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fahlen, T. F., N. Mathur, and B. D. Jones. 2000. Identification and characterization of mutants with increased expression of hilA, the invasion gene transcriptional activator of Salmonella typhimurium. FEMS Immunol. Med. Microbiol. 28:25-35. [DOI] [PubMed] [Google Scholar]
- 18.Fang, F. C., S. Libby, N. A. Buchmeier, P. C. Loewen, J. Switala, J. Harwood, and D. G. Guiney. 1992. The alternative sigma factor katF (rpoS) regulates Salmonella virulence. Proc. Natl. Acad. Sci. USA 89:11978-11982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galan, J. E., and R. Curtiss III. 1989. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc. Natl. Acad. Sci. USA 86:6383-6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gauthier, A., M. L. Robertson, M. Lowden, J. A. Ibarra, J. L. Puente, and B. B. Finlay. 2005. Transcriptional inhibitor of virulence factors in enteropathogenic Escherichia coli. Antimicrob. Agents. Chemother. 49:4101-4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansen-Wester, I., and M. Hensel. 2001. Salmonella pathogenicity islands encoding type III secretion systems. Microbes Infect. 3:549-559. [DOI] [PubMed] [Google Scholar]
- 22.Hobbie, S., L. M. Chen, R. J. Davis, and J. E. Galan. 1997. Involvement of mitogen-activated protein kinase pathways in the nuclear responses and cytokine production induced by Salmonella typhimurium in cultured intestinal epithelial cells. J. Immunol. 159:5550-5559. [PubMed] [Google Scholar]
- 23.Holden, D. W. 2002. Trafficking of the Salmonella vacuole in macrophages. Traffic 3:161-169. [DOI] [PubMed] [Google Scholar]
- 24.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hung, D. T., and E. J. Rubin. 2006. Chemical biology and bacteria: not simply a matter of life or death. Curr. Opin. Chem. Biol. 10:321-326. [DOI] [PubMed] [Google Scholar]
- 26.Hung, D. T., E. A. Shakhnovich, E. Pierson, and J. J. Mekalanos. 2005. Small-molecule inhibitor of Vibrio cholerae virulence and intestinal colonization. Science 310:670-674. [DOI] [PubMed] [Google Scholar]
- 27.Kauppi, A. M., R. Nordfelth, H. Uvell, H. Wolf-Watz, and M. Elofsson. 2003. Targeting bacterial virulence: inhibitors of type III secretion in Yersinia. Chem. Biol. 10:241-249. [DOI] [PubMed] [Google Scholar]
- 28.Kox, L. F., M. M. Wosten, and E. A. Groisman. 2000. A small protein that mediates the activation of a two-component system by another two-component system. EMBO J. 19:1861-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kubori, T., Y. Matsushima, D. Nakamura, J. Uralil, M. Lara-Tejero, A. Sukhan, J. E. Galan, and S. I. Aizawa. 1998. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280:602-605. [DOI] [PubMed] [Google Scholar]
- 30.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of the bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 31.Linehan, S. A., A. Rytkönen, X.-J. Yu, M. Liu, and D. W. Holden. 2005. SlyA regulates function of Salmonella pathogenicity island 2 (SPI-2) and expression of SPI-2-associated genes. Infect. Immun. 73:4354-4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCarter, L. L. 2006. Regulation of flagella. Curr. Opin. Microbiol. 9:180-186. [DOI] [PubMed] [Google Scholar]
- 33.McCollister, B. D., T. J. Bourret, R. Gill, J. Jones-Carson, and A. Vazquez-Torres. 2005. Repression of SPI2 transcription by nitric oxide-producing IFN-γ-activated macrophages promotes maturation of Salmonella phagosomes. J. Exp. Med. 202:625-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKinney, J. S., H. Zhang, T. Kubori, J. E. Galan, and S. Altman. 2004. Disruption of type III secretion in Salmonella enterica serovar Typhimurium by external guide sequences. Nucleic Acids Res. 32:848-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michiels, T., P. Wattiau, R. Brasseur, J. M. Ruysschaert, and G. Cornelis. 1990. Secretion of Yop proteins by yersiniae. Infect. Immun. 58:2840-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller, J. H. 1972. Experiments in molecular genetics, p. 325-355. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 37.Muschiol, S., L. Bailey, Å. Gylfe, C. Sundin, K. Hultenby, S. Bergström, M. Eloffson, H. Wolf-Watz, S. Normark, and B. Henriques-Normark. 2006. A small-molecule inhibitor of type III secretion inhibits different stages of the infectious cycle of Chlamydia trachomatis. Proc. Natl. Acad. Sci. USA 103:14566-14571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nordfelth, R., A. M. Kauppi, H. A. Norberg, H. Wolf-Watz, and M. Elofsson. 2005. Small-molecule inhibitors specifically targeting type III secretion. Infect. Immun. 73:3104-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pegues, D. A., M. J. Hantman, I. Behlau, and S. I. Miller. 1995. PhoP/PhoQ transcriptional repression of Salmonella typhimurium invasion genes: evidence for a role in protein secretion. Mol. Microbiol. 17:169-181. [DOI] [PubMed] [Google Scholar]
- 40.Quinn, T., R. O'Mahony, A. W. Baird, D. Drudy, P. Whyte, and S. Fanning. 2006. Multi-drug resistance in Salmonella enterica: efflux mechanisms and their relationships with the development of chromosomal resistance gene clusters. Curr. Drug Targets 7:849-860. [DOI] [PubMed] [Google Scholar]
- 41.Rhen, M., D. O'Connor, and S. Sukupolvi. 1987. The outer membrane permeability mutation of the virulence-associated plasmid of Salmonella typhimurium is located in a traT-like gene. FEMS Microbiol. Lett. 52:145-154. [Google Scholar]
- 42.Ruiz-Albert, J., X. J. Yu, C. R. Beuzon, A. N. Blakey, E. E. Galyov, and D. W. Holden. 2002. Complementary activities of SseJ and SifA regulate dynamics of the Salmonella typhimurium vacuolar membrane. Mol. Microbiol. 44:645-661. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 44.Shea, J. E., M. Hensel, C. Gleeson, and D. W. Holden. 1996. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 93:2593-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sodeinde, O. A., A. K. Sample, R. R. Brubaker, and J. D. Goguen. 1988. Plasminogen activator/coagulase gene of Yersinia pestis is responsible for degradation of plasmid-encoded outer membrane proteins. Infect. Immun. 56:2749-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spector, M. P., F. Garcia del Portillo, S. M. Bearson, A. Mahmud, M. Magut, B. B. Finlay, G. Dougan, J. W. Foster, and P. J. Pallen. 1999. The rpoS-dependent starvation-stress response locus stiA encodes a nitrate reductase (narZYWV) required for carbon-starvation-inducible thermotolerance and acid tolerance in Salmonella typhimurium. Microbiology 145:3035-3045. [DOI] [PubMed] [Google Scholar]
- 47.Steiner, T. S., J. P. Nataro, C. E. Poteet-Smith, J. A. Smith, and R. L. Guerrant. 2000. Enteroaggregative Escherichia coli expresses a novel flagellin that causes IL-8 release from intestinal epithelial cells. J. Clin. Investig. 105:1769-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taira, S., P. Heiskanen, H. Heikkilä, P. Riikonen, and M. Rhen. 1995. Evidence for functional polymorphism of the spvR gene regulating virulence gene expression in Salmonella. Mol. Gen. Genet. 246:437-444. [DOI] [PubMed] [Google Scholar]
- 49.Vazquez-Torres, A., Y. Xu, J. Jones-Carson, D. W. Holden, S. M. Lucia, M. C. Dinauer, P. Mastroeni, and F. C. Fang. 2000. Salmonella pathogenicity island-dependent evasion of the phagocyte NADPH oxidase. Science 287:1655-1658. [DOI] [PubMed] [Google Scholar]
- 50.Wilmes-Riesenberg, M. R., J. W. Foster, and R. Curtiss III. 1997. An altered rpoS allele contributes to the avirulence of Salmonella typhimurium LT2. Infect. Immun. 65:203-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wolf, K., H. J. Betts, B. Chellas-Géry, S. Hower, C. N. Linton, and K. A. Fields. 2006. Treatment of Chlamydia trachomatis with a small molecule inhibitor of the Yersinia type III secretion system disrupts progression of the chlamydial development cycle. Mol. Microbiol. 61:1543-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woo, Y. K., and S. H. Lee. 2006. Genetic diversity of multi-resistant Salmonella enterica serovar Typhimurium from animals and humans. J. Microbiol. 44:106-112. [PubMed] [Google Scholar]
- 53.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]