Abstract
Mycobacteria contain genes for several DNA ligases, including ligA, which encodes a NAD+-dependent enzyme that has been postulated to be a target for novel antibacterial compounds. Using a homologous recombination system, direct evidence is presented that wild-type ligA cannot be deleted from the chromosome of Mycobacterium smegmatis. Deletions of native ligA in M. smegmatis could be obtained only after the integration of an extra copy of M. smegmatis or Mycobacterium tuberculosis ligA into the attB site of the chromosome, with expression controlled by chemically inducible promoters. The four ATP-dependent DNA ligases encoded by the M. smegmatis chromosome were unable to replace the function of LigA. Interestingly, the LigA protein from M. smegmatis could be substituted with the NAD+-dependent DNA ligase of Escherichia coli or the ATP-dependent ligase of bacteriophage T4. The conditional mutant strains allowed the analysis of the effect of LigA depletion on the growth of M. smegmatis. The protein level of the conditional mutants was estimated by Western blot analysis using antibodies raised against LigA of M. tuberculosis. This revealed that a strong overproduction or depletion of LigA did not affect the growth or survival of mycobacteria under standard laboratory conditions. In conclusion, although NAD+-dependent DNA ligase is essential for mycobacterial viability, only low levels of protein are required for growth. These findings suggest that very efficient inhibition of enzyme activity would be required if NAD+-dependent DNA ligase is to be useful as an antibiotic target in mycobacteria. The strains developed here will provide useful tools for the evaluation of the efficacy of any appropriate compounds in mycobacteria.
Tuberculosis (TB) remains a major threat to public health. It is an important infectious disease, causing high morbidity and mortality worldwide, and the situation is made even worse by the emergence of drug-resistant strains of Mycobacterium tuberculosis (4). For example, multidrug-resistant (MDR) TB (resistant at least to rifampin and isoniazid) takes longer to treat (up to 2 years) with second-line drugs, which are expensive and have side effects. Even worse is extensively drug-resistant TB, which is resistant to first- and second-line drugs (MDR plus resistance to any fluoroquinolone and at least one of three injectable second-line drugs: capreomycin, kanamycin, or amikacin). Thus, options for treatment are becoming seriously limited, returning TB control to the preantibiotic era (3, 20). Drug resistance in M. tuberculosis is not caused by a universal mechanism for all drugs but can be caused by mutations of various chromosomal genes, as identified for MDR occurrence due to the sequential accumulation of mutations in different genes that provide resistance to individual drugs. The mutations connected to resistance can appear in targets of current drugs (e.g., inhA and kasA for isoniazid, rpoB for rifampin, and embCAB for ethambutol) or enzymes required for the intracellular activation of current drugs (katG for isoniazid, pncA for pyrazinamide, and etaA for ethionamide) (34). These concerns lead to the conclusion that the identification of novel, sensitive targets or new drugs is necessary for the control of drug-resistant forms of TB.
A requirement for an antibacterial enzyme target is that it be essential for the organism and not present in the host. One such candidate has been proposed to be NAD+-dependent DNA ligase (5, 32). DNA ligases are essential constituents of all organisms due to their critical roles in DNA replication and repair. The mechanism of DNA ligation shares common features regardless of the cellular origin of the enzyme, with a key step being the formation of a covalent DNA ligase-adenylate intermediate. Importantly, two classes of DNA ligase that are categorized by whether NAD+ or ATP is used as the source of adenylate have been identified. While the essential DNA ligases of bacteria are NAD+ dependent, those used in eukaryotes, archaea, and viruses are ATP dependent. It is this distribution of cofactor specificity that has led to the suggestion that NAD+-dependent DNA ligases may be exploited as useful new targets for broad-spectrum antibacterial compounds (5, 24, 29, 32). Indeed, recent studies have begun to make important progress in identifying small molecules that have some specificity towards the inhibition of NAD+-dependent DNA ligases (2, 26-28).
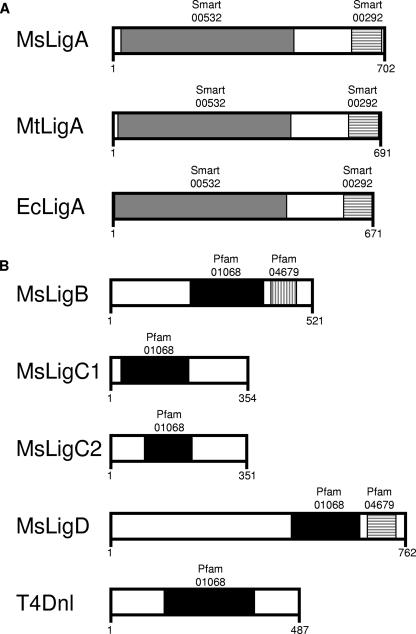
Although NAD+-dependent DNA ligases appear to be produced in all bacteria, some bacteria encode additional ATP-dependent versions of the proteins (5, 24, 29, 32). This complicates potential strategies to target NAD+-dependent DNA ligases with antibiotics, as it is not clear whether the ATP-dependent enzymes would influence the efficacy of any compound. Such factors are particularly relevant to mycobacteria, because multiple DNA ligases are encoded within their genomes (Fig. 1). Mycobacterial genomes carry a single gene, ligA, that is homologous to NAD+-dependent DNA ligases, with the activity of its expressed product confirmed by in vitro studies (10, 26-28, 33). Three different types of ATP-dependent DNA ligases are encoded in the genomes of mycobacteria, and the biochemical activities of these proteins have also been confirmed (9, 10, 31).
FIG. 1.
DNA ligases of mycobacteria, E. coli, and bacteriophage T4. A schematic diagram of conserved domains within the various DNA ligases used in this study is shown. The number of amino acids in each specific protein is indicated. Approximate relative positions of domains conserved within more than one of the proteins are shown by the shaded regions. Domains were identified from their reference within the Pfam (8) or SMART (15) database, as indicated. (A) NAD+-dependent ligases from M. smegmatis (MsLigA), M. tuberculosis (MtLigA), and E. coli (EcLigA). (B) ATP-dependent DNA ligases from M. smegmatis (MsLigB, MsLigC1, MsLigC2, and MsLigD) and bacteriophage T4 (T4Dnl).
The potential for antibiotics to target NAD+-dependent DNA ligases relies on the fact that these enzymes are believed to be essential for all bacteria due to their participation in DNA replication. However, it is difficult to establish this indispensability in a definitive manner, which is a fundamental requirement if these enzymes are going to be assessed as antibiotic targets. In this report, we undertake a series of experiments that prove directly that ligA is essential in Mycobacterium smegmatis. We demonstrate that this gene can be complemented by non-host NAD+-dependent DNA ligases and the ATP-dependent DNA ligase from bacteriophage T4. A detailed analysis of the amount of DNA ligase in various strains identifies that the level of protein can vary by around 10-fold, with little effect on growth under standard laboratory conditions. Strains produced during this study will be useful in any detailed evaluation of antibiotics targeting NAD+-dependent DNA ligases.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All strains used in this study were based on M. smegmatis mc2155 (25) and were cultured in Middlebrook 7H9 broth supplemented with albumin-dextrose-sodium chloride or NB broth (8.0 g/liter nutrient broth [Difco], 10.0 g/liter glucose). As required, further additions included 0.2% Tween 80 (pH 6.0 to 6.2), 50 μg/ml hygromycin, 7.5 μg/ml gentamicin, and 25 μg/ml kanamycin. Mycobacterial transformants were selected on Middlebrook 7H10 agar plates enriched with albumin-dextrose-sodium chloride containing kanamycin (25 μg/ml), gentamicin (7.5 μg/ml), or hygromycin (50 μg/ml).
Gene cloning strategies.
Standard molecular biology protocols were used for all cloning protocols (22). All PCR products were obtained using thermostable ExTaq polymerase (Takara, Japan), cloned initially into a TA vector (pGEM-T Easy; Promega), and then released by digestion with appropriate restriction enzymes before cloning into the final vectors. To facilitate subcloning into expression vectors, restriction enzyme recognition sites were incorporated into the sequence of the primers. The plasmids used in this work are listed in Table 1.
TABLE 1.
Plasmids used in this study
| Plasmid | Description | Source or reference |
|---|---|---|
| Cloning vectors | ||
| pGemTEasy | T/A cloning | Promega |
| pMV306 | Mycobacterial integrating vector, Hygr | Med-Immune Inc. |
| p2NIL | Recombination vector, nonreplicating in mycobacteria, Kanr | 17 |
| pGoal17 | Source of PacI cassette, Ampr | 17 |
| pJam2 | Shuttle vector carrying inducible Pami promoter, Kanr | 30 |
| PMV306GM | pMV306 with defective Hygr gene (digested with EcoRI, filled-in resultant cohesive ends, and self-ligated) carrying Gmr gene in KpnI site | This study |
| pMVPami | pMV306 with Pami promoter of pJam2 cloned into HindIII-XbaI restriction sites | This study |
| pMV306tetR | Mycobacterial integrating vector carrying tetracycline repressor, Kmr | 7 |
| pSE100 | Shuttle vector carrying inducible Ptet promoter, Hygr | 7 |
| Vectors used for gene replacement | ||
| pMK104 | M. smegmatis 5′ ΔligA PstI-HindIII fragment including 5′ end and its upstream region (1,754 bp) in p2NIL, Kanr | This study |
| pMK106 | M. smegmatis ΔligA HindIII-BamHI fragment including 3′ end and its downstream region (1,741 bp) in pMK104, Kanr | This study |
| pMK107 | pMK106 with PacI cassette from pGoal17, Kanr | This study |
| pMK140 | pMK104 with Hygr gene cloned between 5′ and 3′ fragments of ligA (ΔligA::Hygr), Kanr Hygr | This study |
| pMK141 | pMK140 with PacI cassette from pGoal17, Kanr Hygr | This study |
| Overproduction vectors | ||
| pMK114 | ligAMs under Pami promoter in pJam2, Kanr | This study |
| pMK129 | ligAMs under Pami promoter in pJam2, Kanr | This study |
| pMKB144 | ligBMs under Pami promoter in pJam2, Kanr | This study |
| pMKC146 | ligC1Ms under Pami promoter in pJam2, Kanr | This study |
| pMKC148 | ligC2Ms under Pami promoter in pJam2, Kanr | This study |
| pMK113 | ligDMs under Pami promoter in pJam2, Kanr | This study |
| pJamligT4 | bacteriophage T4 ligase under Pami promoter in pJam2, Kanr | This study |
| pMK124 | ligAMs under Pami promoter of pMK114 cloned in pMV306, Hygr | This study |
| pMK130 | ligAMt under Pami promoter of pMK129 cloned in pMV306, Hygr | This study |
| pMK155 | ligBMs under Pami promoter of pMKB144 cloned in pMV306GM, Gmr | This study |
| pMK152 | ligC1Ms under Pami promoter of pMKC146 cloned in pMV306GM, Gmr | This study |
| pMK154 | ligC2Ms under Pami promoter of pMKC148 cloned in pMV306GM, Gmr | This study |
| pMK150 | ligDMs under Pami promoter of pMK113 cloned in pMV306GM, Gmr | This study |
| pMVligAEc | ligAEc cloned under control of Pami promoter in pMVPami, Hygr | This study |
| pMVligT4 | Bacteriophage T4 ligase under Pami promoter of pJamligT4 cloned in pMV306, Hygr | This study |
Construction of ligA gene replacement vector.
Following strategies reported previously that deleted DNA ligase genes in M. smegmatis (12), a suicidal recombination delivery vector was constructed to perform unmarked deletions in the ligA gene (MSMEG2361) of M. smegmatis. The vector carried the 5′ end of ligA (68 bp) with the upstream region amplified with primers A-GR1 and A-GR2 (Table 2) connected to the 3′ end of the gene (936 bp) and with the downstream region amplified with A-GR3 and A-GR4 primers (Table 2). Note that this cloning results in the 5′ and 3′ fragments of the gene being ligated out of frame, allowing expression of nonfunctional protein only.
TABLE 2.
Primer sequences used for PCR amplification of gene sequences
| Amplified region | Primer | Sequence | Product size (bp) |
|---|---|---|---|
| Primers used to amplify DNA for targeted gene replacement | |||
| ligA 5′ flanking region, sense | A-GR1 | 5′-CTGCAGTCTCGCGCGTCGCGCGCTCG-3′ | 1,754 |
| ligA 5′ flanking region, reverse | A-GR2 | 5′-AAGCTTCGCCGCTCGTCGGCGTCAGG-3′ | |
| ligA 3′ flanking region, sense | A-GR3 | 5′-AAGCTTGGCGACACCGTGGTGATCCGC-3′ | 1,741 |
| ligA 3′ flanking region, reverse | A-GR4 | 5′-GGATCCAGCCGAGACCACAGGGCGGG-3′ | |
| Primers used to clone genes for complementation experiments | |||
| ligAMs gene, sense | MsA-s | 5′-GGATCCGTGAGCGAGAAGGCAACCG-3′ | 2,106 |
| ligAMs gene, reverse | MsA-r | 5′-TCTAGACTACTCGGCCGGCTCGACC-3′ | |
| ligBMs gene, sense | MsB-s | 5′-AGATCTATGGATGTCCGACCCGCG-3′ | 1,566 |
| ligBMs gene, reverse | MsB-r | 5′-TCTAGATCAGTCGCGCTCGTAGAAC3′ | |
| ligC1Ms gene, sense | MsC1-s | 5′-GGATCCATGGACCTGCCCGTGCTGC-3′ | 1,065 |
| ligC1Ms gene, reverse | MsC1-r | 5′-TCTAGACTAAACCCGGCACGATGTC-3′ | |
| ligC2Ms gene, sense | MsC2-s | 5′-AGATCTATGGGAAGGATGGACTTGC-3′ | 1,056 |
| ligC2Ms gene, reverse | MsC2-r | 5′-TCTAGATCACTGTTCCTCCAGCACG-3′ | |
| ligDMs gene, sense | MsD-s | 5′-GGATCCGTGGCGAGGCATCCTTGGG-3′ | 2,289 |
| ligDMs gene, reverse | MsD-r | 5′-TCTAGACTATTCCCACACAACCTC-3′ | |
| ligAMt gene, sense | MtbA-s | 5′-AGATCTGTGAGCTCCCCAGACGCCG-3′ | 2,076 |
| ligAMt gene, reverse | MtbA-r | 5′-TCTAGATTACGTTCGTGACGCGGG-3′ | |
| ligAEc gene, sense | EcA-s | 5′-TCTAGAATGGAATCAATCGAACAAC-3′ | 2,016 |
| ligAEc gene, reverse | EcA-r | 5′-TCTAGATTAGCTACCCAGCAAACGC-3′ | |
| T4Dnl gene, sense | T4-s | 5′-GGATCCATGATTCTTAAAATTCTGAACG-3′ | 1,464 |
| T4Dnl gene, reverse | T4-r | 5′-GGATCCTCATAGACCAGTTACCTCATG-3′ |
Construction of complementation plasmids.
M. smegmatis genes (ligA, ligB [MSMEG2280], ligC1 [MSMEG6264], ligC2 [MSMEG6263], and ligD [MSMEG5550]), M. tuberculosis ligA (ligAMt) (Rv3014), Escherichia coli ligA (ligAEc) (EC2345), and DNA ligase of bacteriophage T4 (M32518) were amplified by PCR and cloned downstream from the Pami promoter (Tables 1 and 2). Mycobacterial ligases were cloned first into the BamHI-XbaI sites of pJam2, and the cloned genes and Pami promoter were then excised with HindIII and XbaI and cloned into the integration vector pMV306HygR or pMV306GmR. ligAEc was introduced into the single XbaI site of pMV306-Pami under the control of the acetamidase promoter. The gene for T4 DNA ligase was introduced into the BamHI site of pJam2, excised from this vector with HindIII, and cloned into pMV306. Finally, ligA of M. smegmatis was cloned into pSE100 downstream from the Ptet promoter (Tables 1 and 2).
Testing of ligA essentiality and engineering of conditional mutants.
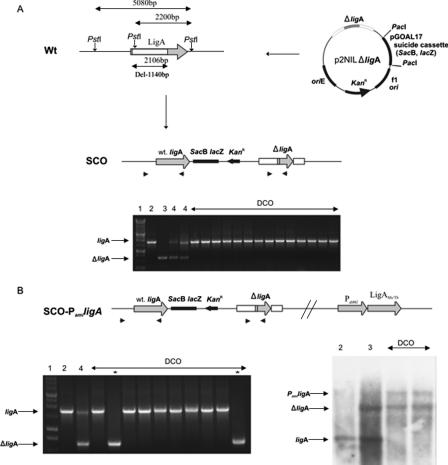
A two-step recombination protocol (12, 17) was used to obtain single-crossover (SCO) strains carrying wild-type (wt) and disrupted ligA (ΔligA) at its native locus on the chromosome. Using the strategy outlined in Fig. 2A, the suicidal recombination plasmid DNA (p2NILΔligA) was treated with NaOH (0.2 mM) and integrated into the M. smegmatis mc2155 chromosome by homologous recombination. The resulting SCO recombinant mutant colonies were blue, Kanr, and sensitive to sucrose. Additionally, the suicidal recombination plasmid (p2NILΔligA) was enriched with the Hygr gene cloned in a single HindIII site of ΔligA (ΔligA::hyg) and introduced into M. smegmatis. The resulting SCO-2 mutant colonies were blue, Kanr, Hygr, and sensitive to sucrose. The SCO strains were further processed to select for double-crossover (DCO) mutants that were white, Kans, and resistant to sucrose (2%). PCR was used to identify the presence of ligA or ΔligA in resultant DCOs.
FIG. 2.
NAD+-dependent ligA is essential for viability of M. smegmatis. (A) A deleted version of ligA (ΔligA) of M. smegmatis was constructed by PCR and introduced into the suicide recombination vector p2NIL. SCO mutants carrying both ligA and ΔligA were obtained by the integration of plasmid DNA (p2NILΔligA) into regions of the chromosome that flank the gene of interest. SCO mutant strains were processed directly for DCO mutant strains. The genotype of selected strains (>50) was confirmed by PCR, indicating that all DCO strains carried wt ligA exclusively. (B) SCO strains from A were enriched with intact ligA from M. smegmatis or M. tuberculosis controlled by an inducible promoter (Pami ligAMs/ligAMt). The genotype of selected strains was confirmed by PCR and Southern hybridization analysis, indicating that DCO strains were both wt (ligA) and mutant type (ΔligA). Numbers above the lanes of each gel represent the following samples: 1, 1-kb DNA ladder; 2, M. smegmatis wt control; 3, p2NILΔligA plasmid; 4, SCO mutant strains. DCO mutant strains are indicated in the figure, with those containing ΔligA highlighted by an asterisk (*). Note that the middle PstI recognition site (A) was present within the deleted region of ligA (ΔligA), thus causing the ΔligA band detected by Southern hybridization to be larger than the wt ligA band. The Southern hybridization probe was amplified using the 3′ undeleted end of ligA.
Further experiments used the strategies outlined in Fig. 2B and Fig. 3 to introduce the complementation plasmids into SCO strains. The resultant strains SCO-Pami ligAMs, SCO-Ptet ligAMs, SCO-Pami ligAMt, SCO-Pami ligAEc, SCO-Pami ligB, SCO-Pami ligC1, SCO-Pami ligC2, SCO-Pami ligD, and SCO-Pami T4Dnl were processed to select for DCO mutant strains in the presence of inducers (0.2% acetamide and 25 ng/ml anhydrotetracycline for Pami and Ptet, respectively). If DCOs carrying ΔligA were not identified during the processing of SCO strains with complementing genes, the complemented SCO-2 strains (SCO-2 Pami ligB, SCO-2 Pami ligC1, SCO-2 Pami ligC2, and SCO-2 Pami ligD) were processed for DCO strains. This approach allowed the attempted selection of DCOs exclusively carrying ΔligA::hyg at the native locus, with the resultant resistance to hygromycin allowing the detection of mutated strains even if they appeared at very low efficiencies.
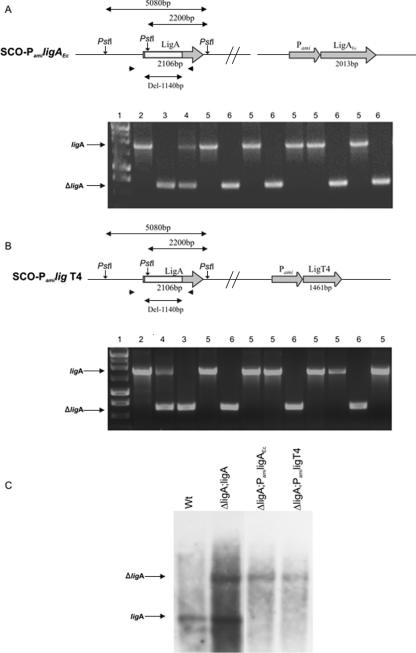
FIG. 3.
NAD+-dependent ligA of M. smegmatis can be substituted with NAD+-dependent ligA of E. coli or ATP-dependent ligase of enterobacteriophage T4. SCO strains carrying both ligA and ΔligA (Fig. 2) were enriched with ligAEc (A) or ligT4 (B) controlled by a Pami promoter. DCO mutant strains resulting from the processing of these SCO strains were both wt (ligA) and mutant type (ΔligA). The genotype of selected DCO strains was confirmed by PCR (A and B) and Southern hybridization (C) analyses. Numbers above the lanes of each gel represent the following samples: 1, 1-kb DNA ladder; 2, M. smegmatis wt control; 3, p2NILΔligA plasmid; 4, SCO mutant strains; 5, DCO mutant strains carrying wt ligA; 6, DCO mutant strains carrying ΔligA. In the Southern hybridization shown in C, 6A and 6B represent the complementation by ligAEc and ligT4, respectively. Note that the middle PstI recognition site (A) was present within the deleted region of ligA (ΔligA), thus causing the ΔligA band detected by Southern hybridization to be larger than the wt ligA band. The Southern hybridization probe was amplified using the 3′ undeleted end of ligA.
For all SCO and DCO strains, PCR and Southern hybridization were used to identify the presence of ligA or ΔligA at the native locus on the chromosome. A hybridization probe (to the 3′ end of ligA) was generated by PCR (using primers A-GR3 and MsA-r) (Table 2) and labeled by a nonradioactive primer extension system (DIG-labeling system; Amersham).
Preparation of antibodies against LigAMt.
Cloning and purification of a recombinant form of LigAMt has been described previously (33). Note that this protein contains a His10 tag within an extra 21 amino acids (2.5 kDa) at the N terminus. LigAMt was purified using nickel affinity chromatography (His-Bind column; Novagen) and was used to raise a primary rabbit polyclonal antibody (Davids Biotechnologie).
Total protein isolation and Western blotting.
Preparation of mycobacterial cell lysates by bead beating (using 0.1-mm zirconia beads) and detection of LigA by Western blotting using polyclonal antibodies were done essentially as described previously (6). The total protein concentration was determined by the Bradford method (22). For quantitative immunoblotting, the same concentration of total protein extracts was loaded onto sodium dodecyl sulfate-polyacrylamide gels. Western blots were processed using the Amersham Pharmacia ECL chemiluminescence kit and protocol. The LigA bands were visualized on Hyperfilm ECL (Amersham) and scanned for densitometry with FluorChem (Alpha Innotech Corp.).
RESULTS
NAD+-dependent DNA ligase is essential for viability of mycobacteria.
NAD+-dependent DNA ligases are highly conserved across all bacterial genomes (32), allowing ready identification of homologous genes by bioinformatics analysis (Fig. 1). The genomes of fast- and slow-growing mycobacteria carry a single gene that is homologous to ligA, with the activity of their expressed NAD+-dependent DNA ligases confirmed by in vitro studies (10, 27, 28, 33). Since NAD+-dependent DNA ligases are not present in the genomes of cells targeted by mycobacterial pathogens, they have been postulated to be a potential target for new antibiotics (5, 24, 29, 32). The potential usefulness of this approach is dependent on the extent to which these enzymes are essential for the viability of mycobacteria.
To allow a careful evaluation of the essentiality of bacterial ligA, we used M. smegmatis as a model experimental system. A two-step recombination protocol (17) was used to construct SCO strains carrying both wt ligA and a mutated copy of ligA (ΔligA). Further processing of SCO strains for a second step of recombination should generate strains carrying either wt ligA or only the ΔligA gene (mut-DCO) (Fig. 2A). The identification of mut-DCO strains would be possible only if ligA is not essential for the viability of mycobacteria. PCR analysis of more than 50 individual DCO colonies identified wt DCO exclusively, confirming that knockout mutations in ligA are lethal for mycobacteria.
Even though the wt gene is essential, we wondered if it would be possible to delete it from the bacterial chromosome if another copy of the gene at a different location supported the expression of required protein. Moreover, by controlling expression from the additional gene by an inducible promoter, a conditional mutant might be obtained. To prepare such mutant strains, M. smegmatis ligA (ligAMs) and M. tuberculosis ligA (ligAMt) were cloned under the control of the tetracycline (Ptet) or acetamidase (Pami) promoter. These recombinant genes were introduced into SCO strains by integration into the host chromosome (genes induced from Pami) or as a self-replicating vector (genes induced from Ptet). The resultant mutant strains (SCO-Ptet/Pami ligAMs/ligAMt) carrying wt ligA, ΔligA, and Ptet/Pami ligAMs/ligAMt were processed in the presence of the appropriate inducer to obtain DCO mutant strains. mut-DCO strains carrying ligAMs/ligAMt controlled by the Pami or Ptet promoter as the sole intact copy of ligA were identified by PCR and Southern hybridization (Fig. 2B). The successful engineering of such strains confirmed the essentiality of ligA in mycobacteria and showed that ligAMt can replace its M. smegmatis ortholog.
NAD+-dependent DNA ligase cannot be replaced by overproduction of ATP-dependent DNA ligases of mycobacteria.
In addition to an NAD+-dependent DNA ligase, mycobacteria encode three different ATP-dependent ligases (9, 10, 31). In fact, M. smegmatis has the potential to express four ATP-dependent DNA ligases since it has two closely related copies of ligC (Fig. 1). BLAST analysis identified that each of these ATP-dependent DNA ligases contains a domain that is well conserved in other DNA ligases, as indicated by their Pfam nomenclature (8): Pfam01068 relates to the conserved catalytic (adenylation) domain of DNA ligases. Notably, this domain is present in the ATP-dependent DNA ligase of enterobacteriophage T4 (Fig. 1), which is able to replace the function of ligAEc (13). Although the essential nature of ligA shows that the ATP-dependent DNA ligases cannot normally replace LigA, we wondered if these proteins may be able to replace the function of LigA if they were overexpressed. Such a phenomenon could lead to the selection of resistant strains if LigA were to be inactivated with an antibiotic drug. Therefore, by following the procedures outlined above to obtain conditional mutants, we assessed the potential redundancy of function between the mycobacterial DNA ligases by testing whether ligA is still essential in genetic backgrounds that overexpress ATP-dependent ligases.
To simplify the DCO screening and to allow the examination of a larger number of colonies, the SCO strain was modified by the introduction of a hygromycin resistance cassette into ΔligA (ΔligA::Hygr). This allowed the resultant mut-DCO and wt DCO to be distinguished by supplementation of hygromycin B into the screening media, with only the mut-DCO strains being able to grow under such conditions. The ATP-dependent ligases of M. smegmatis (ligB, ligC1, ligC2, and ligD) were cloned under the control of the Pami promoter and introduced into the attB site of an SCO strain. Although we did not have antibodies to test of the level of expression for each of these proteins, the expression system is known to be efficient (6), and it clearly worked for the ligA homologs (Fig. 2A). The screening for DCO strains was performed in the presence of the Pami inducer and in the presence or absence of hygromycin B. No colonies with the expected DCO phenotype (white and Kms) grew in the presence of hygromycin B in the media. For SCO strains carrying a gene for each of the ATP-dependent ligases, all DCO strains obtained without hygromycin revealed the wt genotype (wt DCO) when verified by PCR (data not shown). These results demonstrate that ATP-dependent DNA ligases cannot substitute for NAD+-dependent enzymes, again confirming the essentiality of ligA in M. smegmatis.
E. coli NAD+-dependent DNA ligase and ATP-dependent ligase of bacteriophage T4 can substitute for LigA in mycobacteria.
The data described above show that the NAD+-dependent DNA LigA of M. smegmatis can be substituted with its ortholog from M. tuberculosis. This result is expected since there is very close homology across mycobacterial gene sequences. Since mycobacterial genomes are G+C rich, we wondered if they may have evolved specialized versions of ligA. We tested whether complementation of LigAMs might extend to gene sequences that are less well conserved. In these experiments, we used genes that are much less G+C rich, namely, those that are the paradigms for DNA ligases that participate in replication: the NAD+-dependent DNA LigA from the gram-negative bacterium E. coli and the ATP-dependent ligase from enterobacteriophage T4. Both genes were amplified by PCR, cloned under the control of the Pami promoter, and introduced into the attB site of an SCO strain already carrying ΔligA. The resultant strains carrying wt ligA, ΔligA, and either ligAEc or the ATP-dependent ligase of bacteriophage T4 (ligT4) expressed from the Pami promoter were processed to obtain DCO strains. PCR and Southern hybridization analysis revealed mut-DCO strains without an intact copy of ligAMs (Fig. 3). Thus, ligAMs could be complemented with either ligAEc or ligT4. Significantly, in the latter case, the resultant strains of M. smegmatis carry genes for ATP-dependent DNA ligases exclusively.
The replacement of LigAMs with its counterpart from M. tuberculosis or E. coli did not produce a significant effect on the growth of M. smegmatis (Fig. 4A and B). Mutant strains carrying ligT4 to complement ligA from M. smegmatis grew more slowly, taking an extra 24 h to reach stationary phase, especially if its expression was not induced to a high level with acetamide (Pami inducer) (Fig. 4C). The growth dynamics of the strain carrying ligT4 were most affected by a delay in achieving exponential growth. Since each culture was inoculated with the same number of actively growing cells, such a delay in growth is likely to result from an increase in doubling time compared to the wt strain. Although the specific reasons behind this delay are not yet known, it is clear that M. smegmatis is sensitive to the type of replicative DNA ligase that is present within the cell.
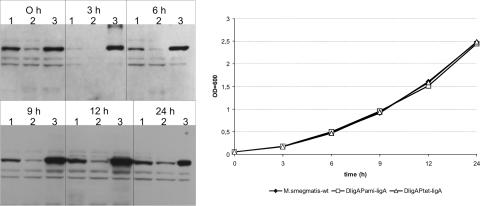
FIG. 4.
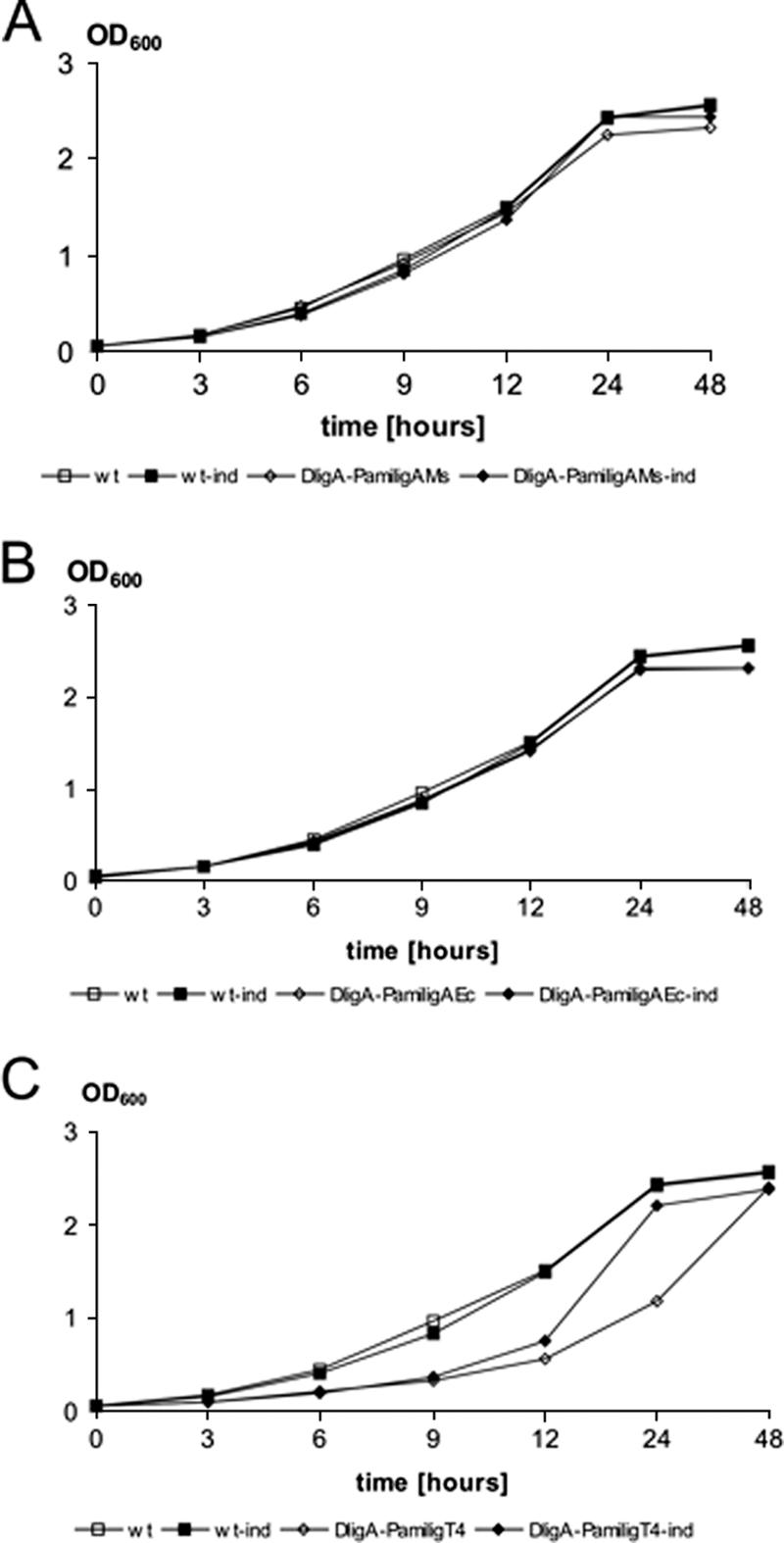
M. smegmatis carrying only ATP-dependent ligases is defective in growth. The M. smegmatis wt control strain (wt) and conditional mutant strains carrying ligase genes driven by Pami promoter were grown in Middlebrook 7H9/oleic acid-albumin-dextrose catalase medium with and without the induction of Pami. All conditional mutant strains carried a single intact copy of the named ligA substituting for a deletion in M. smegmatis ligA as follows: (A) ΔligA-Pami ligAMs, ΔligA mutant complemented with M. smegmatis ligA; (B) ΔligA-Pami ligAEc, ΔligA mutant complemented with E. coli ligA; (C) ΔligA-Pami ligT4, ΔligA mutant complemented with the gene for T4 DNA ligase. The growth of each culture was monitored by optical density analysis. Growth experiments were repeated three times, with the representative result being presented in the figure. OD600, optical density at 600 nm.
The growth of M. smegmatis is insensitive to the level of LigA.
A reliable antibiotic target should be essential for bacterial viability, and a significant decrease in its activity should have a similarly significant effect on bacterial growth. To assess this property of LigAMs, the constructed conditional mutant strains carrying a single ligA gene under the control of inducible promoters (Ptet/Pami) were used to analyze the influence of the LigA amount on bacterial growth.
Strains were grown as required, and the level of LigA was estimated using polyclonal antibodies raised against a recombinant form of LigAMs. Initial experiments confirmed that this antibody had cross-reactivity with native LigA expressed from the chromosome of M. smegmatis (data not shown). The mutant strains were able to grow without inducer supplementing the media, indicating that both promoters are leaky and allow low levels of constitutive expression. The protein analysis showed that the Ptet promoter leaks more than Pami, but its fully induced level of expression was much higher (Fig. 5). These observations are likely to be due to the fact that Ptet expressed genes from plasmid DNA (five to six copies) but that Pami expressed genes from a single copy in the chromosome. The strains expressing different levels of LigA protein were cultured: the wt M. smegmatis strain, the ΔligA-Pami ligA strain growing without acetamide in the medium, and the ΔligA-Ptet ligA strain growing in medium supplemented with anhydrotetracycline. The growth of these three strains was determined by optical density measurement, and the level of LigA was determined by Western blot analysis (Fig. 5). The Western blot analysis revealed a large overproduction of LigA in the ΔligA-Ptet ligA strain growing in the presence of inducer, up to 11 times more than that of the wt strain as detected by densitometry (data not shown). By contrast, very low amounts of LigA were present in the ΔligA-Pami ligA mutant strain (about three times less than the wt strain). However, the growth dynamics of the strains were not affected by the different levels of LigA and were comparable to the growth of the wt strain (Fig. 5).
FIG. 5.
Growth of M. smegmatis is insensitive to the amount of expressed LigA. As shown in the panel on the right, optical density (OD) analysis determined the extent of growth of M. smegmatis strains carrying a natural level of LigA (lane 1, wt strain), a low level of LigA (lane 2, ΔligA-Pami ligAMs, without inducer), and an overproduced level of LigA (lane 3, ΔligA-Ptet ligAMs, with inducer). In the left panel, the level of LigA was determined by Western blot analysis with antibodies raised against LigA of M. tuberculosis. For each lane, 12 μg of total proteins was loaded.
It is likely that drug treatments decrease the level of functional target protein, which inhibits or prevents bacterial growth over long-term exposure. To determine if the depletion of LigA over long periods may affect the viability of M. smegmatis, the wt strain and ΔligA-Pami ligA mutant strains were incubated for 3 weeks. No inducer was added to the medium in this experiment, so there was limited expression of LigA in the ΔligA-Pami ligA strains. The number of viable cells was counted, and the level of LigA was determined by Western blot analysis. During these experiments, lasting for 21 days, no significant differences in viability were observed between a wt strain carrying normal levels of LigA and mutant strains carrying as little as 30% of LigA (data not shown). Thus, the results presented here suggest that any compound targeting LigA would need to reduce the activity of this essential enzyme by more than 70% if is to be able to act effectively as an antibiotic.
DISCUSSION
Mycobacteria contain genes for several ATP-dependent DNA ligases and a single NAD+-dependent DNA ligase encoded by ligA. Previous studies are consistent with ligA being essential in M. smegmatis (9) and M. tuberculosis (23). The data presented here support these suggestions and provide the most direct genetic evidence yet obtained that NAD+-dependent DNA ligases are essential in bacteria. Thus, these experiments in mycobacteria support proposals from experiments with other bacteria, including E. coli (14), Salmonella enterica serovar Typhimurium (18), Bacillus subtilis (19), and Staphylococcus aureus (11). These findings confirm that NAD+-dependent DNA ligases may provide useful targets for broad-spectrum antibacterial compounds (5, 24, 29, 32) and suggest that it is worthwhile to focus effort on identifying specific inhibitors of them (2, 26-28).
Complementation of M. smegmatis ΔligA strains by expression of LigA from E. coli confirms that the function of this protein is conserved across organisms that are widely divergent in evolutionary terms. By contrast, the mycobacterial ATP-dependent DNA ligases are unable to replace the function of LigA. This is not due simply to the use of ATP as the cofactor since the M. smegmatis ΔligA strains are complemented by the ATP-dependent DNA ligase of bacteriophage T4. In terms of the protein sequences of different DNA ligases, the catalytic region is well conserved among all ATP-dependent DNA ligases used in this study (Fig. 1). Since the M. smegmatis ATP-dependent DNA ligases were unable to produce the same complementation as the T4 DNA ligase, this shows that some other parts of their polypeptide sequences influence their activity and prevent them from functioning in DNA replication. This observation is consistent with findings that the ATP-dependent DNA ligases of bacteria participate in DNA repair pathways rather than DNA replication (1, 24, 29).
The observation that the M. smegmatis ΔligA strains can be complemented by the ATP-dependent DNA ligase of bacteriophage T4 is consistent with findings for other bacteria (13, 18, 21). Although bacteriophage T4 does not naturally infect mycobacteria, this observation raises the possibility that if genes encoding ATP-dependent ligases were transferred to the mycobacterial chromosome (e.g., by horizontal gene transfer), they may be able to substitute for LigA and therefore impact the targeting of DNA ligases by antibiotics. On the other hand, horizontal transmission of resistance factors is not generally seen with tubercle bacilli.
Evaluation of the expression of LigA shows that M. smegmatis grows similarly across levels of expression of the protein that varied by approximately 10-fold. This is reminiscent of findings for E. coli, where it has been estimated that 1 to 3% of LigA is sufficient to support growth under laboratory conditions (14, 16). It is not fully understood why bacterial cells have evolved to express such an additional “capacity” of DNA ligase as a standard, but it may relate to the fact that these enzymes participate in many aspects of DNA metabolism. The activity of ligase required for standard laboratory conditions may be relatively low, since the enzyme will be involved primarily with DNA replication. However, at certain times, such as after extensive DNA damage, the increased flux through DNA repair/recombination pathways will mean that the cells require a much larger DNA ligase activity. The cells may therefore find it an advantage to have a built-in extra capacity so that they can respond quickly to times of increased DNA stress. Future experiments will test the effects of DNA damage on the strains produced in this study.
The high level of identity between M. smegmatis and M. tuberculosis DNA ligases and the complementation of the M. smegmatis ΔligA mutant strain with intact ligAMt without a detectable effect on viability or growth rate suggest that our findings are not limited to nonpathogenic mycobacteria. Our experiments with complemented strains of M. smegmatis raise at least two important issues that should be evaluated in the context of targeting NAD+-dependent DNA ligases with antibiotics. First, complementation with DNA ligase from bacteriophage T4 raises the possibility that proteins other than mycobacterial LigA may influence the efficacy of an inhibitor. Second, the overcapacity in terms of the amount of LigA available to M. smegmatis suggests that an irreversible inhibitor will need to shut down LigA activity extremely efficiently. Presently, it is unclear if inhibitors of NAD+-dependent DNA ligases would be able to act so specifically and efficiently. Although good progress is being made in the development of potential compounds to target NAD+-dependent DNA ligases (2, 26-28), the concerns raised in this study make it clear that these compounds will need to be tested in good experimental models. The mycobacterial strains described here will provide useful tools for such a detailed evaluation.
Acknowledgments
We thank Laura Bowater for comments on the manuscript, T. Parish for the Pami expression vector and the p2NIL/pGOAL17 recombination system, and Sabine Ehrt for the Ptet expression system.
We acknowledge financial support from travel grants awarded by the British Council (YSP) and the Wellcome Trust.
Footnotes
Published ahead of print on 4 June 2007.
REFERENCES
- 1.Bowater, R. P., and A. J. Doherty. 2006. Making ends meet: repairing breaks in bacterial DNA by non-homologous end-joining. PLoS Genet. 2:93-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brotz-Oesterhelt, H., I. Knezevic, S. Bartel, T. Lampe, U. Warnecke-Eberz, K. Ziegelbauer, D. Habich, and H. Labischinski. 2003. Specific and potent inhibition of NAD+-dependent DNA ligase by pyridochromanones. J. Biol. Chem. 278:39435-39442. [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 2006. Emergence of Mycobacterium tuberculosis with extensive resistance to second-line drugs—worldwide, 2000-2004. Morb. Mortal. Wkly. Rep. 55:301-305. [PubMed] [Google Scholar]
- 4.Dahle, U. R., P. Sandven, E. Heldal, and D. A. Caugant. 2003. Continued low rates of transmission of Mycobacterium tuberculosis in Norway. J. Clin. Microbiol. 41:2968-2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doherty, A. J., and S. W. Suh. 2000. Structural and mechanistic conservation in DNA ligases. Nucleic Acids Res. 28:4051-4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dziadek, J., M. V. Madiraju, S. A. Rutherford, M. A. Atkinson, and M. Rajagopalan. 2002. Physiological consequences associated with overproduction of Mycobacterium tuberculosis FtsZ in mycobacterial hosts. Microbiology 148:961-971. [DOI] [PubMed] [Google Scholar]
- 7.Ehrt, S., X. V. Guo, C. M. Hickey, M. Ryou, M. Monteleone, L. W. Riley, and D. Schnappinger. 2005. Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucleic Acids Res. 33:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finn, R. D., J. Mistry, B. Schuster-Bockler, S. Griffiths-Jones, V. Hollich, T. Lassmann, S. Moxon, M. Marshall, A. Khanna, R. Durbin, S. R. Eddy, E. L. L. Sonnhammer, and A. Bateman. 2006. Pfam: clans, web tools and services. Nucleic Acids Res. 34:D247-D251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gong, C., P. Bongiorno, A. Martins, N. C. Stephanou, H. Zhu, S. Shuman, and M. S. Glickman. 2005. Mechanism of nonhomologous end-joining in mycobacteria: a low-fidelity repair system driven by Ku, ligase D and ligase C. Nat. Struct. Mol. Biol. 12:304-312. [DOI] [PubMed] [Google Scholar]
- 10.Gong, C., A. Martins, P. Bongiorno, M. Glickman, and S. Shuman. 2004. Biochemical and genetic analysis of the four DNA ligases of mycobacteria. J. Biol. Chem. 279:20594-20606. [DOI] [PubMed] [Google Scholar]
- 11.Kaczmarek, F. S., R. P. Zaniewski, T. D. Gootz, D. E. Danley, M. N. Mansour, M. Griffor, A. V. Kamath, M. Cronan, J. Mueller, D. Sun, P. K. Martin, B. Benton, L. McDowell, D. Biek, and M. B. Schmid. 2001. Cloning and functional characterization of an NAD+-dependent DNA ligase from Staphylococcus aureus. J. Bacteriol. 183:3016-3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korycka-Machala, M., A. Brzostek, S. Rozalska, A. Rumijowska-Galewicz, R. Dziedzic, R. Bowater, and J. Dziadek. 2006. Distinct DNA repair pathways involving RecA and nonhomologous end joining in Mycobacterium smegmatis. FEMS Microbiol. Lett. 258:83-91. [DOI] [PubMed] [Google Scholar]
- 13.Lavesa-Curto, M., H. Sayer, D. Bullard, A. MacDonald, A. Wilkinson, A. Smith, L. Bowater, A. Hemmings, and R. Bowater. 2004. Characterisation of a temperature-sensitive DNA ligase from Escherichia coli. Microbiology 150:4171-4180. [DOI] [PubMed] [Google Scholar]
- 14.Lehman, I. R. 1974. DNA ligase: structure, mechanism, and function. Science 186:790-797. [DOI] [PubMed] [Google Scholar]
- 15.Letunic, I., R. R. Copley, B. Pils, S. Pinkert, J. Schultz, and P. Bork. 2006. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 34:D257-D260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Modrich, P., and I. R. Lehman. 1973. Deoxyribonucleic acid ligase. A steady state kinetic analysis of the reaction catalyzed by the enzyme from Escherichia coli. J. Biol. Chem. 248:7502-7511. [PubMed] [Google Scholar]
- 17.Parish, T., and N. G. Stoker. 2000. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146:1969-1975. [DOI] [PubMed] [Google Scholar]
- 18.Park, U. E., B. M. Olivera, K. T. Hughes, J. R. Roth, and D. R. Hillyard. 1989. DNA ligase and the pyridine nucleotide cycle in Salmonella typhimurium. J. Bacteriol. 171:2173-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petit, M. A., and S. D. Ehrlich. 2000. The NAD-dependent ligase encoded by yerG is an essential gene of Bacillus subtilis. Nucleic Acids Res. 28:4642-4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raviglione, M. 2006. XDR-TB: entering the post-antibiotic era? Int. J. Tuberc. Lung Dis. 10:1185-1187. [PubMed] [Google Scholar]
- 21.Ren, Z. J., R. G. Baumann, and L. W. Black. 1997. Cloning of linear DNAs in vivo by overexpressed T4 DNA ligase: construction of a T4 phage hoc gene display vector. Gene 195:303-311. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 23.Sassetti, C. M., D. H. Boyd, and E. J. Rubin. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48:77-84. [DOI] [PubMed] [Google Scholar]
- 24.Shuman, S., and C. D. Lima. 2004. The polynucleotide ligase and RNA capping enzyme superfamily of covalent nucleotidyltransferases. Curr. Opinion. Struct. Biol. 14:757-764. [DOI] [PubMed] [Google Scholar]
- 25.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911-1919. [DOI] [PubMed] [Google Scholar]
- 26.Srivastava, S. K., D. Dube, V. Kukshal, A. K. Jha, K. Hajela, and R. Ramachandran. 7 June 2007, posting date. NAD+-dependent DNA ligase (Rv3014c) from Mycobacterium tuberculosis: novel structure-function relationship and identification of a specific inhibitor. Proteins doi: 10.1002/prot.21457. [DOI] [PubMed]
- 27.Srivastava, S. K., D. Dube, N. Tewari, N. Dwivedi, R. P. Tripathi, and R. Ramachandran. 2005. Mycobacterium tuberculosis NAD+-dependent DNA ligase is selectively inhibited by glycosylamines compared with human DNA ligase I. Nucleic Acids Res. 33:7090-7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Srivastava, S. K., R. P. Tripathi, and R. Ramachandran. 2005. NAD+-dependent DNA ligase (Rv3014c) from Mycobacterium tuberculosis. Crystal structure of the adenylation domain and identification of novel inhibitors. J. Biol. Chem. 280:30273-30281. [DOI] [PubMed] [Google Scholar]
- 29.Tomkinson, A. E., S. Vijayakumar, J. M. Pascal, and T. Ellenberger. 2006. DNA ligases: structure, reaction mechanism, and function. Chem. Rev. 106:687-699. [DOI] [PubMed] [Google Scholar]
- 30.Triccas, J. A., T. Parish, W. J. Britton, and B. Gicquel. 1998. An inducible expression system permitting the efficient purification of a recombinant antigen from Mycobacterium smegmatis. FEMS Microbiol. Lett. 167:151-156. [DOI] [PubMed] [Google Scholar]
- 31.Weller, G. R., B. Kysela, R. Roy, L. Tonkin, E. Scanlan, M. Della, S. K. Devine, J. P. Day, A. Wilkinson, F. d'Adda di Fagagna, K. Devine, R. P. Bowater, P. Jeggo, S. P. Jackson, and A. J. Doherty. 2002. Identification of a DNA non-homologous end-joining complex in bacteria. Science 297:1686-1689. [DOI] [PubMed] [Google Scholar]
- 32.Wilkinson, A., J. Day, and R. Bowater. 2001. Bacterial DNA ligases. Mol. Microbiol. 40:1241-1248. [DOI] [PubMed] [Google Scholar]
- 33.Wilkinson, A., H. Sayer, D. Bullard, A. Smith, J. Day, T. Kieser, and R. Bowater. 2003. NAD+-dependent DNA ligases of Mycobacterium tuberculosis and Streptomyces coelicolor. Proteins 51:321-326. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, Y., C. Vilcheze, and W. R. Jacobs, Jr. 2005. Mechanisms of drug resistance in Mycobacterium tuberculosis, p. 115-142. In S. T. Cole, K. D. Eisenach, D. N. McMurray, and W. R. Jacobs, Jr. (ed.), Tuberculosis and the tubercle bacillus. ASM Press, Washington, DC.