Abstract
Clinical isolates of Neisseria meningitidis with reduced susceptibility to penicillin G (intermediate isolates, PenI) harbor alterations in the penA gene encoding the penicillin binding protein 2 (PBP2). A 402-bp DNA fragment in the 3′ half of penA was sequenced from a collection of 1,670 meningococcal clinical isolates from 22 countries that spanned 60 years. Phenotyping, genotyping, and the determination of MICs of penicillin G were also performed. A total of 139 different penA alleles were detected with 38 alleles that were highly related, clustered together in maximum-likelihood analysis and corresponded to the penicillin G-susceptible isolates. The remaining 101 penA alleles were highly diverse, corresponded to different genotypes or phenotypes, and accounted for 38% of isolates, but no clonal expansion was detected. Analysis of the altered alleles that were represented by at least five isolates showed high correlation with the PenI phenotype. The deduced amino acid sequence of the corresponding PBP2 comprised five amino acid residues that were always altered. This correlation was not complete for rare alleles, suggesting that other mechanisms may also be involved in conferring reduced susceptibility to penicillin. Evidence of mosaic structures through events of interspecies recombination was also detected in altered alleles. A new website was created based on the data from this work (http://neisseria.org/nm/typing/penA). These data argue for the use of penA sequencing to identify isolates with reduced susceptibility to penicillin G and as a tool to improve typing of meningococcal isolates, as well as to analyze DNA exchange among Neisseria species.
The natural habitat of Neisseria meningitidis is the nasopharynx where it is encountered in ca. 10% of the general population (asymptomatic carriers) (10), but it can also cause serious invasive infections, mainly septicemia and meningitis, provoking public health concern. Prompt treatment is critical to the management of invasive meningococcal diseases. Penicillin G remains, in several countries, the antibiotic of first choice in the treatment of invasive meningococcal diseases, particularly when the bacteriological diagnosis has been established (20, 23). However, isolates with reduced susceptibility to penicillin G (PenI) are increasingly being reported worldwide (29) and have led clinicians to use third generation cephalosporins such as ceftriaxone for initial treatment (20). The PenI isolates are defined phenotypically by showing a MIC of penicillin G ranging between 0.094 and 1 mg/liter (8). A recent questionnaire among European laboratory members of the European Monitoring Group on Meningococci (EMGM) revealed heterogeneous definitions of penicillin susceptibility among these laboratories (9). Consequently, the percentage of agreement on susceptibility to penicillin using Etest (AB Biodisk, Solna, Sweden) was not optimal among these laboratories and varied between 24 and 100% (33).
N. meningitidis is a transformable bacterium that undergoes frequent horizontal DNA transfer. The alteration of the penA gene encoding the penicillin binding protein 2 (PBP2) through horizontal DNA transfer was suggested as the major mechanism for the emergence of PenI isolates (5, 24, 26, 31). The modifications of PBP2 result in a decrease in the affinity of PBP2 to penicillin G, as well as in modifications in the structure of peptidoglycan in the bacterial cell wall that are responsible for the PenI phenotype (5). We have previously reported penicillin-binding experiments using membrane extracts or purified PBP2 proteins to show that modification of PBP2 are correlated with reduction in binding affinity of PBPs for [3H]benzylpenicillin (5). The modifications of PBP2 that confer the PenI phenotype are located in the C-terminal half of the protein that binds penicillin and harbors the transpeptidase region (5). Indeed, we have previously reported that transformation with DNA (both genomic DNA or penA PCR products) from PenI isolates from several countries conferred the PenI phenotype on a PenS strain, indicating that this phenotype is directly related to changes in penA (5, 7). Several polymorphic positions were observed in this part of penA with alterations in the corresponding amino acid residues. Transformation of a susceptible isolate by a 3′ fragment of penA (encoding the C-terminal half of the protein) harboring these polymorphisms was sufficient to confer the PenI phenotype (5).
The impact of these alterations on the spread of PenI meningococcal isolates and the structure of the bacterial population is not clear. The aims of the present study were to analyze the penA sequences, the corresponding deduced amino acid sequences of the encoded PBP2, as well as the phenotypic susceptibility to penicillin of a large collection of meningococcal isolates to investigate the spread of PenI isolates. Such approaches may also allow the establishment of a general molecular scheme to define bacterial susceptibility and/or resistance to different antibiotics by strategies of modifications of key (target) genes involved in this process.
MATERIALS AND METHODS
Meningococcal isolates and conventional bacteriological methods.
Meningococcal isolates were received by the participating laboratories and National Reference Centers and had been identified by using standard culture and identification methods. The MIC of penicillin G was determined as previously described (33). Several participating laboratories performed the following analysis themselves, and some laboratories sent isolates for analysis in one of the other participating laboratories. However, all laboratories followed the methods described in the references or in the text below.
Serogroup was determined by agglutination with serogroup-specific antisera according to the standard procedure of each laboratory. Further phenotyping (serotyping and serosubtyping) was performed as previously described (1). Genotyping, using multilocus sequence typing (MLST), porA typing, and fetA typing were performed as previously described (12, 14, 18, 27, 28, 30, 32). Sequence types (STs) and FetA and PorA types were determined through MLST websites (http://pubmlst.org and http://neisseria.org). Data on geographical location, year, and anatomical site of isolation were obtained.
DNA sequencing and analysis of penA.
Two primers were designed to amplify the penA gene between the positions 4948 and 5459 (according to EMBL/GenBank accession number AE002397). These primers are penA1F (the upstream oligonucleotide; 5′-gttttcccagtcacgacgttgtaATCGAACAGGCGACGATGTC-3′) and penA1R (the downstream oligonucleotide; 5′-ttgtgagcggataacaatttcGATTAAGACGGTGTTTTGACGG-3′). The universal forward and reverse sequences were added as adapters to the 5′ end upstream and downstream from the oligonucleotides (shown in lowercase letters). The universal forward and reverse sequences were then used for sequencing. A DNA fragment of 402 bp of the penA gene, which corresponds to the residues 441 to 574 of the PBP2, was extracted from the DNA sequence. Alignments were made by using the MacMolly program (Mologen, Berlin, Germany). Sequences differing by at least one nucleotide were assigned a unique penA allele sequence number. Some laboratories used other protocols and primers to obtain the same 402-bp fragment of penA described above (8, 31).
A new database on penA typing was especially created based on the data obtained in the present study, and information regarding all included isolates and DNA sequences are available (http://neisseria.org/nm/typing/penABlast) (3); clustering analyses were made by the PHYML program of maximum-likelihood phylogenies using the penA sequences (16) through the website of the Institut Pasteur, Paris, France (http://www.pasteur.fr). The penA sequences from N. perflava (accession number X76423), N. mucosa (accession number X59635), N. cinerea (accession number Z17310), and N. flavescens (accession number M26645) were also obtained through the website (http://www.pasteur.fr). The identification of potential recombination events between two penA sequences was performed by using the maximum chi-squared test in the START package available through (http://pubmlst.org) using an input of two sequences and 1,000 resamplings (17, 19).
Statistical analysis.
Qualitative data were analyzed by using the chi-squared test. A P value of ≤0.05 was considered to be statistically significant. Geometric means, as well as lower and upper 95% confidence limits, were calculated using GraphPad InStat version 3.06 (GraphPad Software, San Diego, CA). For reliable calculation of the geometric means and 95% confidence intervals, only alleles that were represented by at least five isolates were included. Geometric mean was used since it better evaluates the MIC of different isolates.
RESULTS
General characteristics of meningococcal isolates.
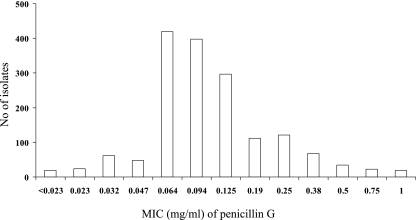
A large number (n = 1,670) of clinical isolates of N. meningitidis were included from 22 different countries worldwide spanning 60 years (1945 to 2006) (Table 1). Most isolates were from reference laboratories of European countries that are members of the EMGM and represented mostly invasive isolates (n = 1393, 83%). Isolates from the cerebrospinal fluid were the most frequent (n = 808 [48%]), followed by isolates from blood (585 [35%]). Some of the isolates were from United States and several African countries, as well as French overseas territories. Representative isolates for which the penicillin G MIC was ≥0.06 mg/liter were tested. A few isolates with lower penicillin G MICs (n = 153) were also analyzed. For some countries (e.g., Denmark and France), all of the invasive isolates from or since 2005 were included, in addition to several isolates from other years. The distribution of isolates with known MIC (n = 1,644 [98%]) is shown in Fig. 1. According to the phenotypic definition of PenI isolates (MICs ranging between 0.094 and 1 mg/liter), 1,072 of 1,644 isolates (65%) showed reduced susceptibility to penicillin G. The number of isolates per country and their MICs are available online (http://neisseria.org/nm/typing/penA).
TABLE 1.
Distribution of the major penAps and altered penA alleles according to countrya
| Country | No. of alleles
|
|||||
|---|---|---|---|---|---|---|
| penAps | penA14 | penA9 | penA12 | Other altered alleles | Total | |
| Austria | 61 | 0 | 0 | 3 | 8 | 72 |
| Belgium | 1 | 4 | 1 | 1 | 1 | 8 |
| Congo | 1 | 0 | 0 | 0 | 0 | 1 |
| Croatia | 7 | 1 | 0 | 0 | 2 | 10 |
| Czech Republic | 113 | 0 | 0 | 0 | 24 | 137 |
| Denmark | 62 | 0 | 0 | 5 | 0 | 67 |
| France (domestic) | 463 | 38 | 49 | 20 | 124 | 694 |
| France (overseas territories) | 19 | 0 | 1 | 0 | 3 | 23 |
| Germany | 63 | 2 | 2 | 4 | 9 | 80 |
| Greece | 19 | 19 | 1 | 0 | 19 | 58 |
| Ireland | 32 | 10 | 2 | 0 | 13 | 57 |
| Italy | 14 | 8 | 11 | 75 | 15 | 123 |
| Madagascar | 7 | 0 | 0 | 0 | 0 | 7 |
| Niger | 18 | 0 | 0 | 0 | 0 | 18 |
| Norway | 1 | 1 | 0 | 0 | 0 | 2 |
| Poland | 20 | 0 | 1 | 0 | 7 | 28 |
| Romania | 2 | 0 | 0 | 0 | 8 | 10 |
| Scotland | 34 | 15 | 0 | 2 | 12 | 63 |
| Slovenia | 2 | 3 | 0 | 0 | 9 | 14 |
| Spain | 21 | 3 | 10 | 30 | 30 | 94 |
| Sweden | 45 | 0 | 0 | 1 | 2 | 48 |
| Tunisia | 15 | 0 | 3 | 0 | 3 | 21 |
| United States | 24 | 2 | 1 | 0 | 8 | 35 |
| Total | 1,044 | 106 | 82 | 141 | 297 | 1,670 |
penAps, penicillin-susceptible alleles.
FIG. 1.
Phenotype distribution among the tested meningococcal isolates for which the penicillin G MICs are known (n = 1,644, 98% of total isolates).
Serogroups were determined for 1,625 isolates (97%). The serogroup distribution for the five major serogroups was as follows: A, 1.4%; B, 55%;, C, 29%; Y, 4.5%; and W-135, 6.1%. This distribution was similar to the general distribution observed in Europe in 2004 (0.4, 70, 18.5, 2.6, and 2.8%, respectively) (see http://www.euibis.org). MLST was performed on 636 isolates representing 193 different STs. Most of these isolates (56%) represented STs that belong to the five major hypervirulent clonal complexes in Europe (11, 13, 35): ST-8 complex/Cluster A4 (16%), ST-11 complex/ET-37 complex (14%), ST-32 complex/ET-5 complex (10%), ST-41/44 complex/Lineage 3 (12%), and ST-269 complex (4%). Isolates belonging to the ST-22 complex represented 3% and were most frequently isolated from elderly individuals (data not shown).
Characteristics of penA.
The sequence of a 402-bp DNA fragment of the 3′ part of penA gene was obtained for all of the 1,670 isolates of the present study and allowed the identification of 139 different alleles of penA that were named penA1 to penA139. The frequencies of theses alleles varied from 1 up to 428 isolates per allele and are available online (http://neisseria.org/nm/typing/penA). The most frequent allele among the tested isolates was penA1 (n = 428 isolates [26%]), and it was distributed globally. When all alleles were aligned with the penA1 allele, they showed homologies of between 99.75% (1 polymorphic site) and 80.60% (78 polymorphic sites). The maximum-likelihood analysis clustered together a group of 38 alleles that shared at least 98.5% homology (no more than six polymorphic sites). These 38 highly related alleles corresponded to 1,043 isolates (62%). This cluster harbored the two most frequent alleles, penA1 and penA3, together representing 46% of isolates (428 and 337 isolates, respectively). Most of these alleles (23 of 38) had an identical deduced amino acid sequence (only silent, synonymous DNA polymorphisms) for the corresponding 402-bp fragment and accounted for 1,018 isolates. The other 15 alleles belonging to this particular cluster differed by no more than two amino acids and were rare (only 25 isolates). Consequently, this cluster of 38 alleles corresponded to isolates with highly related penA sequences with low levels of alterations (if any) of the PBP2 protein. The remaining 101 alleles, corresponding to 627 isolates (38% of the total isolates) were distinctly separated from the first cluster into several other clusters and showed up to 21 amino acid changes from the most frequent sequence described above encoded by the penA1 allele (data not shown). Forty different altered penA alleles were observed among all of the invasive meningococcal strains isolated in France since 2005 (http://neisseria.org/nm/typing/penA). These 101 alleles corresponded to the group of altered alleles with an altered PBP2 protein as deduced from the penA allele sequences.
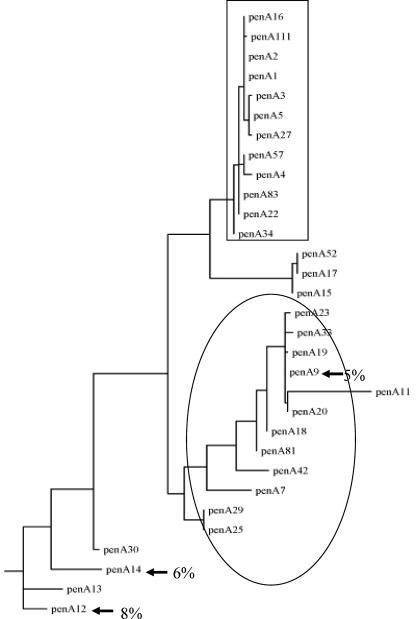
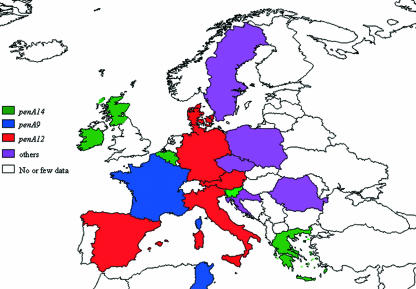
Three alleles (penA12, penA14, and penA9) were the most frequent among these altered alleles and accounted for 8% (n = 141), 6% (n = 106), and 5% (n = 82) of the total isolates, respectively. The maximum-likelihood clustering of penA alleles is shown in Fig. 2 for the 31 penA alleles that were represented by at least five isolates. The three frequent altered alleles showed disparity in their geographical distribution, penA9 and penA14 being frequent in France and Denmark, respectively, while penA12 was frequent in Spain and Italy. These three alleles were not observed in, for example, the Czech Republic (Fig. 3 and Table 1). The penA9 allele was observed in isolates of several different phenotypes and genotypes. Moreover, the maximum-likelihood analysis clustered several other altered penA alleles together with the allele penA9 with identical deduced amino acid sequences for some of them (penA9, penA19, penA20, penA23, and penA33) (Fig. 2).
FIG. 2.
Schematic illustration of phylogenic analysis by maximum likelihood of the 31 penA alleles that were identified among the tested isolates and were represented by at least five isolates. The group of penAps alleles that were highly related and corresponded to susceptible isolates is indicated in a box. The most frequent altered alleles (penA12, penA14, and penA9) are indicated by arrows, and their percentages are indicated. The “penA9 cluster” is shown in an ellipse.
FIG. 3.
Geographic distribution of the most frequent altered penA alleles (penA12, penA14, and penA9). The colors correspond to the most frequent allele in each country. “Other” indicates countries where other alleles were observed, such as penA132 (Czech Republic), penA32 (Romania), and penA52 (Sweden).
Correlation between MIC and alterations of PBP2.
Transformation using PCR products of altered penA alleles (including the most frequent alleles penA12, penA14, and penA9) confers PenI phenotype and reduction of [3H]benzylpenicillin binding to PBP2 (5, 7; data not shown). We therefore analyzed the associations between penA alleles and susceptibility to penicillin G; only penA alleles that were represented by at least five isolates were considered (see Materials and Methods). The selected alleles (n = 31) corresponded to 1,520 isolates (91% of the total isolates). Among these alleles, 12 were from the cluster of the highly related penA that encoded one identical PBP2, except for penA111, which encoded a PBP2 with one amino acid change (P551 to S), while the remaining 19 penA alleles encoded 15 distinct altered PBP2 (Fig. 2 and Table 2). The geometric mean MICs (MICgm) of penicillin G were calculated for the corresponding isolates of each selected allele. The MICgm for isolates harboring the alleles from the cluster of highly related penA varied between 0.055 and 0.094 mg/liter (Table 2). This range was 0.055 to 0.076 mg/liter if only alleles with more than 10 isolates were considered. The narrow 95% confidence intervals had an upper limit of 0.090 mg/liter (Table 2). This correlation suggests that the cluster of highly related penA is linked to meningococcal susceptibility to penicillin G. The corresponding penA alleles are designated henceforth “penicillin-susceptible alleles penAps,” and the corresponding PBP2 are designated “PBP2ps.” In contrast, the isolates of other clusters (with altered PBP2) showed higher MICgm ranging from 0.112 to 0.511 mg/liter; an MIC of amoxicillin of >0.25 mg/liter was also correlated with altered PBP2 (data not shown). When these altered PBP2 were aligned to PBP2ps, five positions corresponding to residues F504, A510, I515, H541, and I566 in the wild-type PBP2 were always modified in all altered PBP2 (Fig. 4). These positions were also always modified in the PBP2 encoded by altered penA alleles that were represented by three and four isolates (data not shown). These residues are located in the transpeptidase domain of PBP2. However, the KTG motif was always conserved in all PBP2 encoded by the 139 penA alleles (Fig. 4).
TABLE 2.
Characteristics of penicillin G MIC and PBP2 alterations among the tested isolates (n = 1,520)a
| penA allele | No. of isolates | MIC range (mg/liter) | Geometric mean MIC (mg/liter) | 95% Confidence interval | No. of alterations in PBP2b |
|---|---|---|---|---|---|
| 83 | 17 | 0.023-0.380 | 0.055 | 0.040-0.076 | 0 |
| 57 | 19 | 0.008-0.125 | 0.058 | 0.041-0.084 | 0 |
| 5 | 38 | 0.015-0.190 | 0.066 | 0.056-0.077 | 0 |
| 2 | 39 | 0.015-0.125 | 0.066 | 0.056-0.077 | 0 |
| 34 | 20 | 0.032-0.125 | 0.067 | 0.057-0.079 | 0 |
| 3 | 333 | 0.004-0.250 | 0.070 | 0.067-0.074 | 0 |
| 27 | 34 | 0.032-0.125 | 0.070 | 0.060-0.081 | 0 |
| 4 | 16 | 0.032-0.125 | 0.072 | 0.060-0.087 | 0 |
| 16 | 8 | 0.047-0.125 | 0.074 | 0.055-1.004 | 0 |
| 1 | 423 | 0.008-0.250 | 0.076 | 0.073-0.079 | 0 |
| 22 | 43 | 0.023-0.250 | 0.077 | 0.064-0.090 | 0 |
| 111c | 7 | 0.094-0.094 | 0.094 | 0.094-0.094 | 0 |
| 25 | 5 | 0.064-0.190 | 0.112 | 0.073-0.193 | 5 |
| 29 | 12 | 0.094-0.125 | 0.119 | 0.111-0.128 | 5 |
| 52 | 20 | 0.064-0.500 | 0.123 | 0.096-0.157 | 5 |
| 15 | 24 | 0.094-0.250 | 0.145 | 0.126-0.167 | 5 |
| 12 | 141 | 0.064-1.000 | 0.162 | 0.148-0.176 | 5 |
| 30 | 6 | 0.125-0.380 | 0.170 | 0.102-0.281 | 5 |
| 23 | 5 | 0.064-0.380 | 0.171 | 0.073-0.340 | 5 |
| 42 | 10 | 0.094-0.250 | 0.172 | 0.137-0.216 | 5 |
| 14 | 102 | 0.064-1.000 | 0.173 | 0.155-0.194 | 5 |
| 19 | 11 | 0.064-0.500 | 0.174 | 0.115-0.262 | 5 |
| 7 | 17 | 0.064-0.750 | 0.193 | 0.146-0.254 | 5 |
| 17 | 5 | 0.125-0.380 | 0.201 | 0.122-0.330 | 5 |
| 20 | 7 | 0.094-0.380 | 0.227 | 0.124-0.414 | 5 |
| 81 | 5 | 0.190-0.750 | 0.321 | 0.149-0.691 | 5 |
| 9 | 81 | 0.064-1.000 | 0.328 | 0.282-0.381 | 5 |
| 33 | 14 | 0.125-1.000 | 0.340 | 0.242-0.477 | 5 |
| 13 | 14 | 0.125-0.750 | 0.351 | 0.250-0.493 | 5 |
| 18 | 12 | 0.094-1.000 | 0.388 | 0.250-0.603 | 5 |
| 11 | 12 | 0.094-1.000 | 0.511 | 0.331-0.787 | 5 |
Only data for alleles that were represented by at least five isolates are shown.
There were no alterations in the five amino acid positions F504, A510, I515, H54,1 and I566.
The PBP2 encoded by penA111 differs from susceptible PBP2 by one amino acid residue (P551).
FIG. 4.
Partial sequences of the C-terminal part of PBP2 (amino acids 441 to 574) deduced from the sequences of penA genes from isolates of N. meningitidis tested in the present study. Only the PBP2 encoded by the alleles that were represented by at least five isolates are shown. Ditto symbols (″) indicate identical residues. Polymorphic residues are indicated in one-letter code. The five positions that were always modified in PenI isolates are indicated by asterisks (F504, A510, I515, H541, and I566). The KTG motif is underlined. The corresponding alleles are indicated on the left. PBP2 from susceptible penA alleles were identical except for PBP2 encoded by penA111 that differed by one residue (P551). Both of these wild-type PBP2 sequences are shown.
Distribution of altered penA alleles.
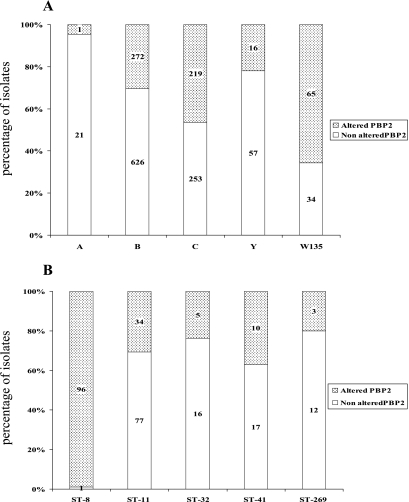
Association between serogroups and alterations of PBP2 was analyzed for the 1,625 isolates with determined serogroups. A significant association was observed between isolates of serogroup W135 and alterations of PBP2 at the five amino acid positions (P < 0.00001) (Fig. 5A). Indeed, 65 of the 99 isolates belonging to the serogroup W135 (66%) harbored altered PBP2 proteins. Moreover, 59 of these isolates shared the same altered allele (penA14), which was also one of the most frequent altered penA alleles in the present study and was detected in 106 isolates. Isolates of serogroup A were mostly from Africa and harbored penAps alleles (particularly penA83). Only one serogroup A isolate (from Romania) harbored an altered penA allele (penA32) that was the most frequent altered penA allele in Romania.
FIG. 5.
Distribution of isolates with altered or nonaltered PBP2 according to serogroup (A) or according to ST (B). The number of isolates is also indicated for each corresponding category of isolates.
The association between an altered PBP2 and the major invasive STs (ST-8, ST-11, ST-22, ST-32, ST-41, and ST-269) was analyzed for the 636 isolates with determined STs. A significant association with alterations of PBP2 was observed in the case of ST-8 isolates (P < 0.0001) (Fig. 5B). These isolates were mostly of the phenotype C:2b:P1.5,2. Moreover, all of these isolates shared the same altered penA allele (penA12). Most of these isolates were from Spain and Italy.
A higher percentage of altered PBP2 was observed in ST-22 isolates, but a statistical test could not be performed due to the low number of isolates. Several of the ST-22 isolates were of the phenotype W135:NT:NST. No other obvious association was observed among a particular ST and altered penA alleles.
FetA types were obtained for 208 isolates. No obvious correlation was observed between FetA types and penA alleles. For instance, 16 different FetA types were with penA1 allele. The same FetA type was observed with several penA alleles. For instance, FetA type 1-5 was observed in 19 isolates showing penAps alleles (penA1, penA3, penA5, penA22, and penA57) or altered alleles (penA7, penA19, penA131, and penA135).
Mosaic structure of altered penA alleles.
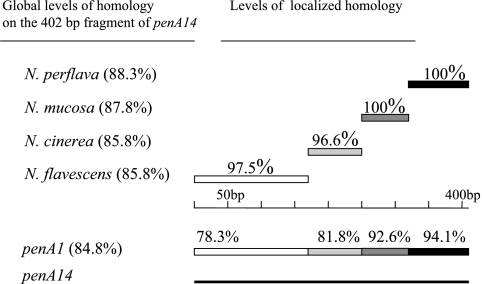
BLAST analysis was performed on the altered penA alleles and detected substantial homologies with penA genes from other Neisseria species, in particular with commensal species (data not shown). For instance, one of the most frequent altered allele (penA14) showed identity scores of 88.3, 87.8, 85.8, and 85.8% with the corresponding 402 bp of N. perflava, N. mucosa, N. cinerea, and N. flavescens, respectively (Fig. 6). However, this 402 bp corresponding to penA14 shared an identity of 84.8% with the most frequent allele (penA1). Analysis for identity and recombination sites between penA14 and the four penA genes from N. perflava, N. mucosa, N. cinerea, and N. flavescens allowed the construction of a combined illustration demonstrating regions of high homology and even complete identity between penA14 and the other four penA genes from the commensal Neisseria species (Fig. 6). Analysis using the START package revealed significant putative recombination sites after nucleotides 87 and 147 on penA14 with penA genes of N. cinerea and N. perflava, respectively. These results strongly suggest a mosaic structure of penA14. Similar results were obtained when other altered penA alleles were analyzed (data not shown).
FIG. 6.
Schematic and combined representation of DNA homology between penA14 and the corresponding penA genes from other Neisseria species: N. perflava (accession number X76423), N. mucosa (accession number X59635), N. cinerea (accession number Z17310), and N. flavescens (accession number M26645). The global homology on the 402-bp fragment between penA14 and each of these penA genes, as well as with meningococcal penA1, are indicated in parentheses on the left. The levels of localized homology with different segments of penA are indicated on the right.
DISCUSSION
The prevalence of resistance due to beta-lactamase production is very low among isolates of N. meningitidis, and the modifications of PBP2 appear to be the most common mechanism of resistance to penicillin G (25). This mechanism of resistance is clinically highly relevant since it is being detected in an increasing number of invasive isolates (6). Moreover, the collection of isolates tested in the present study represents isolates with similar phenotypic and genotypic characteristics to invasive strains currently circulating worldwide, particularly in Europe (21). Amoxicillin and cefotaxime (a broad-spectrum cephalosporin) MICs were higher among PenI isolates, suggesting that cross-resistance to other beta-lactams may be of clinical concern (6). This cross-resistance was also reported in the closely related species, N. gonorrhoeae, where resistance to penicillin G and cefixime has been described to be associated with alterations in penA (4). The alterations of penA that seem to be directly linked to the PenI phenotype are located in the transpeptidase-encoding domain (5). Five polymorphic sites that changed their corresponding amino acid residues were detected in all isolates with MICgm ranging between 0.112 and 0.511 mg/liter (Table 2). Additional polymorphic sites were also observed, but not in all altered penA alleles, and may correspond to allelic association with some genotypes. A significant finding of the present study is a molecular structure-based definition of PenI isolates. Indeed, for alleles represented by at least five isolates, an MICgm of >0.094 mg/liter always correlated with altered penA alleles and five modified amino acid positions in PBP2 (F504, A510, I515, H541, and I566). The isolates showing lower MICgm correlated well with the presence of penAps alleles (Table 2). One definition of PenI isolates could be a combination of an MIC of >0.094 mg/liter with alterations of penA encoding the five modified positions in PBP2. The amoxicillin MIC, when available, also correlated well with this definition. The MICs of penicillin G and amoxicillin can thus be used together to better define PenI isolates (see http://neisseria.org/nm/typing/penA). Alleles that were represented by three or four isolates also fit well with this breakpoint (data not shown). However, discrepancies were observed for alleles that were represented by less than three isolates. Some rare intermediate isolates may also lack some of the alterations as was previously described (31). The low number of isolates may be responsible for disagreement in the correlation between wild-type/altered penA and a susceptible or intermediate susceptibility to penicillin G. Moreover, disagreement in MIC determination between laboratories may also account for this discrepancy (33). Alternatively, other mechanisms may be responsible for reduced susceptibility to penicillin G among rare isolates that showed an MIC of >0.094 mg/liter with no alteration in penA (22).
The most common alleles (penA1 and penA3) were found in isolates belonging to the major hypervirulent genotypes (in particular ST-11 isolates). However, several susceptible isolates of ST-11 showed different susceptible penAps alleles, suggesting a higher diversity of penA, most likely through horizontal DNA exchanges, compared to the housekeeping genes used in MLST analysis. The three most frequent altered penA alleles (penA12, penA14, and penA9) were found in several different genotypes. The preferential association between isolates of ST-8 and penA12 may reflect a particular geographic spread of these isolates. Indeed, several of these isolates were from Spain and were responsible for an epidemic wave of meningococcal disease during 1996 and 1997 (2). The association of penA14 and ST-22 isolates needs to be confirmed. ST-22 isolates are frequently encountered in carriage state (http://pubmlst.org), suggesting that the distribution of penA alleles among carriage isolates may differ from that observed among invasive isolates. Altered alleles were genetically diverse and harbored fewer isolates in each allele than susceptible alleles. The majority of these altered alleles (61 of 101) were represented by only one isolate. The distribution of these altered alleles is unlikely to be due to inclusion bias as was observed for France, where all invasive isolates were included since the year 2005. These results suggest frequent emergence of altered alleles without clonal expansion of a particular altered penA allele among invasive isolates. This may be due to a biological cost of the modification of penA that may decrease meningococcal survival by, for example, enhancing bacterial clearance in blood and hence reducing meningococcal virulence. This would diminish clonal expansion. Supporting this hypothesis are the heterogeneous phenotypes and genotypes of the isolates that harbored altered penA alleles of the cluster of penA9 allele (Fig. 2). Indeed, modifications of PBP2 in meningococci have been shown to provoke changes in peptidoglycan structure, with increasing amounts of pentapeptide-containing muropeptides indicating a defect in peptidoglycan biosynthesis (5). Meningococcal muropeptides showed variable activation of the transcription factor NF-κB pathway (15).
Our data suggest that altered penA alleles are most likely to appear through interspecies recombination with other Neisseria species, as also suggested by previous studies (26, 34). Interestingly, the allele penA14 showed higher levels of homology with penA genes of N. perflava, N. mucosa, N. cinerea, and N. flavescens than with the most common meningococcal penA susceptible allele (penA1). Evidence of a mosaic-like structure in the penA14 allele was detected (Fig. 6), suggesting that the penA14 allele evolved by recombination with penA from other Neisseria species. In N. gonorrhoeae, some regions in the transpeptidase-encoding domain in this penA gene were also similar to those in the penA genes of N. meningitidis, N. perflava, N. cinerea, and N. flavescens (4). These latter three commensal species are intrinsically less susceptible to penicillin G than meningococcal isolates. The detection of several points of crossovers on penA14 may indicate independent events of recombination and random crossovers points. The data presented here are valuable in establishing the molecular basis for the phenotypic and/or genotypic surveillance of meningococcal resistance to beta-lactam antibiotics. These findings also provide a database that may contribute to the analysis of genetic exchange among the Neisseria species.
Acknowledgments
We are indebted to all laboratory staff who have worked on these isolates, as well as to the various laboratories for submitting isolates to their reference laboratories throughout Europe. We thank the following individuals for excellent assistance and support in this project: Pavla Urbaskova (Czech Republic), Lene Berthelsen (Denmark), Jean-Michel Alonso and Corinne Ruckly (France), and Per Olcén (Sweden).
A.S. is supported by a fellowship from the Community's Sixth Framework Program (Marie Curie Action). The penA alleles of Polish meningococci were obtained during FEMS Research Fellowship (number 2005-1) for M.K. in the Spanish laboratory. R.E. is the recipient of predoctoral fellowships from the Institute of Health Carlos III (ISCIII 04/0021). MLST characterization of Czech isolates was supported by grant IGA MZ C̆R NR/1A8688-3. This study was partially supported in the Spanish laboratory by a grant FIS PI060297. J.A.V. was partially supported by a grant from the Sociedad Española de Enfermedades Infecciosas y Microbiología Clínica.
Footnotes
Published ahead of print on 21 May 2007.
REFERENCES
- 1.Abdillahi, H., and J. T. Poolman. 1988. Neisseria meningitidis group B serosubtyping using monoclonal antibodies in whole-cell ELISA. Microb. Pathog. 4:27-32. [DOI] [PubMed] [Google Scholar]
- 2.Alcala, B., C. Salcedo, L. Arreaza, S. Berron, L. De La Fuente, and J. A. Vázquez. 2002. The epidemic wave of meningococcal disease in Spain in 1996-1997: probably a consequence of strain displacement. J. Med. Microbiol. 51:1102-1106. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ameyama, S., S. Onodera, M. Takahata, S. Minami, N. Maki, K. Endo, H. Goto, H. Suzuki, and Y. Oishi. 2002. Mosaic-like structure of penicillin-binding protein 2 Gene (penA) in clinical isolates of Neisseria gonorrhoeae with reduced susceptibility to cefixime. Antimicrob. Agents Chemother. 46:3744-3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antignac, A., I. G. Boneca, J. C. Rousselle, A. Namane, J. P. Carlier, J. A. Vázquez, A. Fox, J. M. Alonso, and M. K. Taha. 2003. Correlation between alterations of the penicillin-binding protein 2 and modifications of the peptidoglycan structure in Neisseria meningitidis with reduced susceptibility to penicillin G. J. Biol. Chem. 278:31529-31535. [DOI] [PubMed] [Google Scholar]
- 6.Antignac, A., M. Ducos-Galand, A. Guiyoule, R. Pires, J. M. Alonso, and M. K. Taha. 2003. Neisseria meningitidis strains isolated from invasive infections in France (1999-2002): phenotypes and antibiotic susceptibility patterns. Clin. Infect. Dis. 37:912-920. [DOI] [PubMed] [Google Scholar]
- 7.Antignac, A., P. Kriz, G. Tzanakaki, J. M. Alonso, and M. K. Taha. 2001. Polymorphism of Neisseria meningitidis penA gene associated with reduced susceptibility to penicillin. J. Antimicrob. Chemother. 47:285-296. [DOI] [PubMed] [Google Scholar]
- 8.Arreaza, L., C. Salcedo, B. Alcala, M. J. Uria, R. Abad, R. Enríquez, and J. A. Vázquez. 2004. Sequencing of Neisseria meningitidis penA gene: the key to success in defining penicillin G breakpoints. Antimicrob. Agents Chemother. 48:358-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Block, C. 2001. Antibiotic susceptibility testing, p. 89-106. In A. J. Pollard and M. C. J. Maiden (ed.), Meningococcal disease: methods and protocols. Humana Press, Inc., Totowa, NJ.
- 10.Cartwright, K. A., J. M. Stuart, D. M. Jones, and N. D. Noah. 1987. The Stonehouse survey: nasopharyngeal carriage of meningococci and Neisseria lactamica. Epidemiol. Infect. 99:591-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caugant, D. A. 1998. Population genetics and molecular epidemiology of Neisseria meningitidis. APMIS 106:505-525. [PubMed] [Google Scholar]
- 12.Clarke, S. C., M. A. Diggle, and G. F. Edwards. 2001. Semiautomation of multilocus sequence typing for the characterization of clinical isolates of Neisseria meningitidis. J. Clin. Microbiol. 39:3066-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diggle, M. A., and S. C. Clarke. 2005. Increased genetic diversity of Neisseria meningitidis isolates after the introduction of meningococcal serogroup C polysaccharide conjugate vaccines. J. Clin. Microbiol. 43:4649-4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diggle, M. A., and S. C. Clarke. 2002. Rapid assignment of nucleotide sequence data to allele types for multi-locus sequence analysis (MLSA) of bacteria using an adapted database and modified alignment program. J. Mol. Microbiol. Biotechnol. 4:515-517. [PubMed] [Google Scholar]
- 15.Girardin, S. E., I. G. Boneca, L. A. Carneiro, A. Antignac, M. Jehanno, J. Viala, K. Tedin, M. K. Taha, A. Labigne, U. Zahringer, A. J. Coyle, P. S. DiStefano, J. Bertin, P. J. Sansonetti, and D. J. Philpott. 2003. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science 300:1584-1587. [DOI] [PubMed] [Google Scholar]
- 16.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 17.Jolley, K. A., E. J. Feil, M. S. Chan, and M. C. Maiden. 2001. Sequence type analysis and recombinational tests (START). Bioinformatics 17:1230-1231. [DOI] [PubMed] [Google Scholar]
- 18.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maynard-Smith, J. M. 1992. Analyzing the mosaic structure of genes. J. Mol. Evol. 34:126-129. [DOI] [PubMed] [Google Scholar]
- 20.Nadel, S., and J. S. Kroll. 2007. Diagnosis and management of meningococcal disease: the need for centralized care. FEMS Microbiol. Rev. 31:71-83. [DOI] [PubMed] [Google Scholar]
- 21.Noah, N., and B. Henderson. 2001. Surveillance of bacterial meningitis in Europe 1999-2000. PHLS, London, United Kingdom.
- 22.Orus, P., and M. Vinas. 2001. Mechanisms other than penicillin-binding protein-2 alterations may contribute to moderate penicillin resistance in Neisseria meningitidis. Int. J. Antimicrob. Agents 18:113-119. [DOI] [PubMed] [Google Scholar]
- 23.Quagliarello, V. J., and W. M. Scheld. 1997. Treatment of bacterial meningitis. N. Engl. J. Med. 336:708-716. [DOI] [PubMed] [Google Scholar]
- 24.Saez Nieto, J. A., J. A. Vázquez, and C. Marcos. 1990. Meningococci moderately resistant to penicillin. Lancet 336:54. [DOI] [PubMed] [Google Scholar]
- 25.Spratt, B. G. 1994. Resistance to antibiotics mediated by target alterations. Science 264:388-393. [DOI] [PubMed] [Google Scholar]
- 26.Spratt, B. G., L. D. Bowler, Q. Y. Zhang, J. Zhou, and J. M. Smith. 1992. Role of interspecies transfer of chromosomal genes in the evolution of penicillin resistance in pathogenic and commensal Neisseria species. J. Mol. Evol. 34:115-125. [DOI] [PubMed] [Google Scholar]
- 27.Sullivan, C. B., J. M. Jefferies, M. A. Diggle, and S. C. Clarke. 2006. Automation of MLST using third-generation liquid-handling technology. Mol. Biotechnol. 32:219-226. [DOI] [PubMed] [Google Scholar]
- 28.Taha, M. K., D. Giorgini, M. Ducos-Galand, and J. M. Alonso. 2004. Continuing diversification of Neisseria meningitidis W135 as a primary cause of meningococcal disease after emergence of the serogroup in 2000. J. Clin. Microbiol. 42:4158-4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taha, M. K., M. L. Zarantonelli, A. Neri, R. Enríquez, J. A. Vázquez, and P. Stefanelli. 2006. Interlaboratory comparison of PCR-based methods for detection of penicillin G susceptibility in Neisseria meningitidis. Antimicrob. Agents Chemother. 50:887-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson, E. A., I. M. Feavers, and M. C. Maiden. 2003. Antigenic diversity of meningococcal enterobactin receptor FetA, a vaccine component. Microbiology 149:1849-1858. [DOI] [PubMed] [Google Scholar]
- 31.Thulin, S., P. Olcén, H. Fredlund, and M. Unemo. 2006. Total variation in the penA gene of Neisseria meningitidis: correlation between susceptibility to beta-lactam antibiotics and penA gene heterogeneity. Antimicrob. Agents Chemother. 50:3317-3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Urwin, R., J. E. Russell, E. A. Thompson, E. C. Holmes, I. M. Feavers, and M. C. Maiden. 2004. Distribution of surface protein variants among hyperinvasive meningococci: implications for vaccine design. Infect. Immun. 72:5955-5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vázquez, J. A., L. Arreaza, C. Block, I. Ehrhard, S. J. Gray, S. Heuberger, S. Hoffmann, P. Kriz, P. Nicolas, P. Olcen, A. Skoczynska, L. Spanjaard, P. Stefanelli, M. K. Taha, and G. Tzanakaki. 2003. Interlaboratory comparison of agar dilution and Etest methods for determining the MICs of antibiotics used in management of Neisseria meningitidis infections. Antimicrob. Agents Chemother. 47:3430-3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vázquez, J. A., S. Berron, M. O'Rourke, G. Carpenter, E. Feil, N. H. Smith, and B. G. Spratt. 1995. Interspecies recombination in nature: a meningococcus that has acquired a gonococcal PIB porin. Mol. Microbiol. 15:1001-1007. [DOI] [PubMed] [Google Scholar]
- 35.Yazdankhah, S. P., P. Kriz, G. Tzanakaki, J. Kremastinou, J. Kalmusova, M. Musilek, T. Alvestad, K. A. Jolley, D. J. Wilson, N. D. McCarthy, D. A. Caugant, and M. C. Maiden. 2004. Distribution of serogroups and genotypes among disease-associated and carried isolates of Neisseria meningitidis from the Czech Republic, Greece, and Norway. J. Clin. Microbiol. 42:5146-5153. [DOI] [PMC free article] [PubMed] [Google Scholar]