Abstract
A new natural IND-type metallo-β-lactamase variant, IND-5, was identified in a clinical isolate of Chryseobacterium indologenes. IND-5 shared 92.8% and 92.4% amino acid homology with IND-1 and IND-3, respectively. Purified enzyme (pI = 8.8, Mr = 25,000) was able to hydrolyze penicillins, some narrow- and expanded-spectrum cephalosporins, and carbapenems but not monobactams.
Metallo-β-lactamases (MBLs) are a group of enzymes that exhibit broad-spectrum substrate specificity and include carbapenems (5, 8, 12). MBL genes are found as resident chromosomal genes in some species of the family Flavobacteriaceae, including Chryseobacterium indologenes, which is the most common flavobacterial species encountered in clinical specimens (10). The resident MBL of C. indologenes, named IND, belongs to subclass B1 and exhibits notable allelic diversity: five IND-type enzymes that may diverge up to 20% from each other at the sequence level have been identified (1). Knowledge of the biochemical properties of these enzymes is still very limited, and kinetic data are available only for IND-1 and IND-2 (1).
The aim of this study was to characterize a novel IND-type MBL variant, IND-5, identified in a clinical isolate of C. indologenes from an Italian hospital.
(Part of this work was presented at the 16th European Congress of Clinical Microbiology and Infectious Diseases, abstr. 934, 2006.)
C. indologenes NF16 (CDC group Iib) was isolated from the sputum of a patient at the Hematology Unit of Ferrarotto Hospital (Catania, southern Italy). Genomic and plasmidic DNAs were purified as previously described (9). In order to amplify the blaIND gene and its flanking regions from genomic DNA, the following primers were used: INDA_for, 5′GGGCATATGAAAAAAAGAATTCAGTTCTTTA; INDB_for, 5′GGGGATATCATGAAAAAAAGAATTCAGTTCTTTA; and IND_rev, 5′GGGGGATCCTTATTTTTTGTTAAGAAGTTCAAGA (boldface type indicates the NdeI, EcoRV, and BamHI restriction sites, respectively, in the three primers). Amplicons obtained by PCR using INDA_for and IND_rev were cloned into pET-9a (Novagen, Inc., Madison, WI) vectors, whereas amplicons obtained using INDB_for and IND_rev were cloned into pBC-SK (Stratagene, Inc., La Jolla, CA) vectors. The recombinant plasmids, named pET-IND-5 and pBC-IND-5, were introduced by transformation into Escherichia coli BL21(DE3) and HB101 competent cells, respectively. The fidelity of the cloned fragments was confirmed by sequencing both strands. Conjugation experiments were carried out using Escherichia coli K-12 as a recipient and at an initial donor/recipient ratio of 0.1. Transconjugants were selected on Mueller-Hinton agar containing streptomycin (1,000 μg/ml) and imipenem (2 μg/ml).
In vitro susceptibility testing was performed by agar dilution using a bacterial inoculum of 5 × 105 CFU per spot according to CLSI guidelines (2). E. coli ATCC 25922 was used as a quality control strain for antimicrobial susceptibility testing.
IND-5 was purified from 2 liters of E. coli BL21(DE3)/pET-IND-5 grown at 37°C for about 6 h (A600 = 0.8) and for a further 2 h at 20°C after induction with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). The bacterial suspension was pelleted, resuspended in 40 ml of 50 mM HEPES (pH 7.5) plus 50 μM ZnSO4, sonicated (five cycles at 30 W for 30 s each time), and centrifuged for 30 min at 105,000 × g at 4°C. The cleared extract was loaded onto a Mono Q Sepharose column equilibrated with 20 mM ethanolamine buffer (pH 8.6) plus 50 μM ZnSO4. The active fractions were eluted during the washing step, whereas the remaining proteins (about 77%) bound to the column. The active fractions were dialyzed against 0.025 M diethanolamine buffer (pH 9.0) plus 50 μM ZnSO4 and loaded onto a Mono P HR 5/20 column. The enzyme was eluted with 1:10 Polybuffer 96 (pH range, 9.0 to 6.0). At the end of each purification step, the β-lactamase activity was tested by measuring the hydrolysis of 200 μM imipenem using 50 mM HEPES (pH 7.5) plus 50 μM ZnSO4. The Mr and pI values of IND-5 were determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (4) and isoelectric focusing analysis (6), respectively. Steady-state kinetic parameters (Km and kcat) were determined for purified enzyme by measuring substrate hydrolysis under initial rate conditions and by using the Hanes linearization of the Michaelis-Menten equation as described previously (11) with 30 mM sodium cacodylate buffer (pH 6.5) plus 50 μM ZnSO4. The thermal stability of the IND-5 enzyme was determined by measuring the hydrolysis of 200 μM imipenem after incubation of the enzyme at 40°C for a maximum of 120 min, at 45°C for a maximum of 90 min, and at 55°C for a maximum of 60 min. The interaction with the chelating agents EDTA, 1,10-o-phenanthroline, and pyridine-2,6-dicarboxylic (dipicolinic) acid was determined by incubating 0.1 μM enzyme at 30°C for 15 min with increasing inhibitor concentrations and using 100 μM nitrocefin as a reporter substrate (3).
The C. indologenes NF16 clinical isolate was resistant to carbapenems and intermediate to cefotaxime but appeared to be susceptible to piperacillin, piperacillin-tazobactam, ceftazidime, cefepime, and aztreonam (Table 1).
TABLE 1.
Pattern of resistance mediated by the IND-5 metallo-β-lactamase gene in E. coli and C. indologenes strains
| Antibiotica | Concn (μg/ml) at which indicated organism showed resistance
|
||
|---|---|---|---|
| C. indologenes NF16 | E. coli HB101/pBC-IND-5 | E. coli HB101/pBC-SK | |
| PIP | 16 | 32 | 1 |
| TZP | 8 | 16 | 1 |
| AMX | >64 | >64 | 2 |
| AMC | 32 | 32 | 2 |
| CFZ | 64 | 64 | 1 |
| CTX | 32 | 16 | <0.125 |
| CAZ | 1 | 0.5 | <0.5 |
| FEP | 0.5 | 0.5 | <0.125 |
| ATM | 32 | <0.125 | <0.125 |
| IPM | 32 | 1 | 0.5 |
| MEM | 16 | 1 | <0.125 |
PIP, piperacillin; TZP, piperacillin-tazobactam; AMX, amoxicillin; AMC, amoxicillin-clavulanate; CFZ, cefazolin; CTX, cefotaxime; CAZ, ceftazidime; FEP, cefepime; ATM, aztreonam; IPM, imipenem; MEM, meropenem. Tazobactam was used at fixed concentration of 4 μg/ml, and clavulanate was used at a 1:2 ratio with amoxicillin.
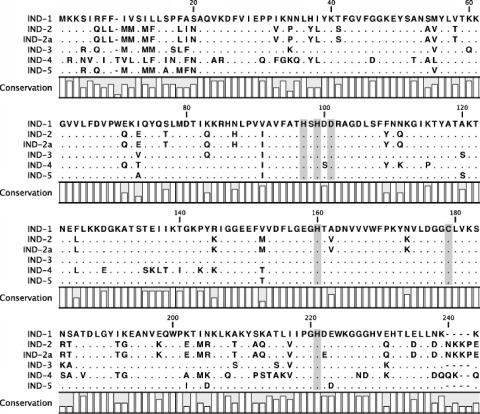
A PCR for blaIND alleles, performed with genomic DNA, yielded an amplicon of 720 bp. Direct amplicon sequencing revealed a blaIND allele encoding a putative 240-amino-acid preprotein named IND-5. When compared to IND-1 (1), IND-5 showed 92.8% amino acid identity and carried the following substitutions: S4R, R6Q, I9M, I12M, L13M, S15A, F17M, A18F, and S19N in the signal peptide region and M54V, I72A, V90I, A119S, V151T, K201I, N204D, and E222D in the mature protein. When compared to IND-3 (1), IND-5 showed 92.4% amino acid identity (Fig. 1).
FIG. 1.
Amino acid sequence of the IND-5 (corresponding to GenBank accession number AY504627) metallo-β-lactamase compared to those of IND-1 (AF099139), IND-2 (AF219129), IND-2a (AF219130), IND-3 (AF219133), and IND-4 (AF219135). Dots indicate identical amino acids. Shading indicates conserved residues in carbapenem-hydrolyzing β-lactamases.
No plasmids were detected in C. indologenes NF16, and direct conjugation experiments failed to transfer any β-lactam resistance marker from the wild-type organism to E. coli K-12. Preliminary experiments using Southern blotting on genomic DNA confirmed the chromosomal localization of the gene (data not shown).
IND-5 was purified from E. coli BL21(DE3)/pET-IND-5 by two chromatographic steps. The protein preparation was estimated to be >95% pure, as estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (data not shown). The Mr and pI values of IND-5 were estimated to be 25,000 and 8.8, respectively, in accordance with theoretical calculations (Mr 24,535 and pI 8.89).
As shown in Table 2, the IND-5 enzyme was able to hydrolyze amino-, carboxy-, and ureidopenicillins, cefazolin, cefotaxime, and carbapenems. Benzylpenicillin was the best substrate, showing a kcat/Km value about 57-fold and 35-fold higher than those of meropenem and imipenem, respectively. Hydrolysis of ceftazidime, cefepime, aztreonam, and moxalactam was not detectable. Two classical inactivators of active-site serine β-lactamases that are in current clinical use have been investigated. Sulbactam was hydrolyzed by the IND-5 enzyme with a non-negligible hydrolysis (kcat/Km = 2.5 × 10−3 μM−1s−1), as previously observed with other zinc enzymes (7). Conversely, tazobactam was stable in the presence of this enzyme. Overall, kinetic data were consistent with the results of susceptibility testing when the IND-5 enzyme was expressed in E. coli (Table 1). Among IND-like enzymes, a comparison of the kinetic parameters of IND-5 is possible only with IND-2 and IND-1 (in the case of IND-1, it is possible to examine only the Km values) (1). It is interesting that the catalytic efficiencies of IND-2 toward imipenem and meropenem are about 25-fold and 13-fold higher than those of IND-5, respectively. We suggest that among the amino acid substitutions found in the IND-5 metallo-β-lactamase, the residue at position 222 could be involved in the drastic reduction in catalytic efficiency of this enzyme relative to those of other IND-2 enzymes (1). The glutamic acid at position 222, strictly conserved in all IND-type β-lactamases, is close to the histidine 220 residue (located at the Cys site) that is involved in the binding of the Zn(II).
TABLE 2.
Kinetic parameters of purified IND-5 β-lactamasea
| Substrate | Km (μM) | kcat (s−1) | kcat/Km (μM−1s−1) | Relative kcat/Km (%) |
|---|---|---|---|---|
| Benzylpenicillin | 13 ± 3 | 10.92 | 0.84 | 100 |
| Ampicillin | 150 ± 15 | 10.30 | 0.069 | 8.2 |
| Piperacillin | 375 ± 30 | 14.61 | 0.039 | 4.6 |
| Carbenicillin | 320 ± 20 | 16.96 | 0.053 | 6.3 |
| Meropenem | 280 ± 30 | 4.13 | 0.015 | 1.8 |
| Imipenem | 300 ± 25 | 7.28 | 0.024 | 2.8 |
| Cefazolin | 25 ± 3 | 0.46 | 0.018 | 2.1 |
| Cefotaxime | 49 ± 8 | 1.53 | 0.031 | 3.7 |
| Ceftazidime | NHb | NH | NH | NDc |
| Cefepime | NH | NH | NH | ND |
| Moxalactam | NH | NH | NH | ND |
| Tazobactam | NH | NH | NH | ND |
| Sulbactam | 1,300 ± 50 | 3.30 | 2.5 × 10−3 | 0.3 |
| Aztreonam | NH | NH | NH | ND |
| Nitrocefin | 8 ± 2 | 4.4 | 0.55 | 65 |
Km and kcat values are the means of three measurements. The standard deviations were always lower than 10%.
NH, no hydrolysis.
ND, not determined.
Thermal stability assays showed that the enzyme exhibited a half-life of 90 min at 40°C, 25 min at 45°C, and less than 1 min at 55°C. Concerning the interaction with zinc ion chelators, inhibition appeared to follow competitive behavior, with Ki values of 3.0, 7.3, and 815 μM for dipicolinic acid, 1,10-o-phenanthroline, and EDTA, respectively.
The importance of chromosomal MBLs from the Flavobacteriaceae has been established in the last decade because of their involvement in resistance in pathogens like C. indologenes and Elizabethkingia meningoseptica. Further biochemical and molecular characterizations of different variants of these enzymes could be of interest to provide insight into their structure-function relationships.
Nucleotide sequence accession number.
The nucleotide sequence of the blaIND-5 gene will appear in the GenBank database under accession number AY504627.
Acknowledgments
This work was supported by grants to G.A. and M.P. from MURST (60% from the MIUR [Ministero dell'Istruzione, dell'Universita e della Ricerca]).
Footnotes
Published ahead of print on 30 April 2007.
REFERENCES
- 1.Bellais, S., L. Poirel, S. Leotard, T. Naas, and P. Nordmann. 2000. Genetic diversity of carbapenem-hydrolyzing metallo-β-lactamases from Chryseobacterium (Flavobacterium) indologenes. Antimicrob. Agents Chemother. 44:3028-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clinical and Laboratory Standards Institute. 2005. Performance standards for antimicrobial susceptibility testing. CLSI document M100-S15. Clinical and Laboratory Standards Institute, Wayne, PA.
- 3.De Meester, F., B. Joris, G. Reckinger, C. Bellefroid-Bourguignon, J. M. Frère, and S. G. Waley. 1987. Automated analysis of enzyme inactivation phenomena. Application to β-lactamases and DD-peptidases. Biochem. Pharmacol. 36:2393-2403. [DOI] [PubMed] [Google Scholar]
- 4.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 5.Nordmann, P., and L. Poirel. 2002. Emerging carbapenemases in Gram-negative aerobes. Clin. Microbiol. Infect. 8:321-331. [DOI] [PubMed] [Google Scholar]
- 6.Perilli, M., A. Felici, N. Franceschini, A. De Santis, L. Pagani, F. Luzzaro, A. Oratore, G. M. Rossolini, J. R. Knox, and G. Amicosante. 1997. Characterization of a new TEM-derived β-lactamase produced in a Serratia marcescens strain. Antimicrob. Agents Chemother. 41:2374-2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prosperi-Meys, C., G. Llabres, D. de Seny, R. P. Soto, M. H. Valladares, N. Laraki, J. M. Frère, and M. Galleni. 1999. Interaction between class B β-lactamases and suicide substrates of active-site serine β-lactamases. FEBS Lett. 443:109-111. [DOI] [PubMed] [Google Scholar]
- 8.Rasmussen, B. A., and K. Bush. 1997. Carbapenem-hydrolyzing β-lactamases. Antimicrob. Agents Chemother. 41:223-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 10.Schreckenberger, P. C., M. I. Daneshvar, R. S. Weyant, and D. G. Hollis. 2003. Acinetobacter, Achromobacter, Cryseobacterium, Moraxella, and other nonfermentative gram-negative rods, p. 749-779. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed. American Society for Microbiology, Washington, DC.
- 11.Segel, I. H. 1975. Enzyme kinetics, behavior and analysis of rapid equilibrium and steady-state enzyme systems, p. 210-212. John Wiley & Sons, Inc., New York, NY.
- 12.Walsh, T. R., M. A. Toleman, L. Poirel, and P. Nordmann. 2005. Metallo-β-lactamases: the quiet before the storm? Clin. Microbiol. Rev. 18:306-325. [DOI] [PMC free article] [PubMed] [Google Scholar]