Abstract
The ATP-dependent phosphorolytic excision of nucleoside analogue reverse transcriptase inhibitors can diminish their inhibitory effects on human immunodeficiency virus replication. Previous studies have shown that excision can occur only when the reverse transcriptase complex exists in its pretranslocational state. Binding of the next complementary nucleotide causes the formation of a stable dead-end complex in the posttranslocational state, which blocks the excision reaction. To provide mechanistic insight into the excision of the acyclic phosphonate nucleotide analog tenofovir, we compared the efficiencies of the reaction in response to changes in the translocation status of the enzyme. We found that rates of excision of tenofovir with wild-type reverse transcriptase can be as high as those seen with 3′-azido-3′-deoxythymidine monophosphate (AZT-MP). Thymidine-associated mutations, which confer >100-fold and 3-fold decreased susceptibility to AZT and tenofovir, respectively, caused substantial increases in the efficiency of excision of both inhibitors. However, in contrast to the case for AZT-MP, the removal of tenofovir was highly sensitive to dead-end complex formation. Site-specific footprinting experiments revealed that complexes with AZT-terminated primers exist predominantly pretranslocation. In contrast, complexes with tenofovir-terminated primers are seen in both configurations. Low concentrations of the next nucleotide are sufficient to trap the complex posttranslocation despite the flexible, acyclic character of the compound. Thus, the relatively high rate of excision of tenofovir is partially neutralized by the facile switch to the posttranslocational state and by dead-end complex formation, which provides a degree of protection from excision in the cellular environment.
Nucleoside analogue reverse transcriptase (RT) inhibitors (NRTI) remain important components in drug regimens used to treat infection with human immunodeficiency virus type 1 (HIV-1). The following eight different NRTI are currently in clinical use: 3′-azido-3′-deoxythymidine (zidovudine [AZT]), 2′,3′-didehydro-3′-dideoxythymidine (stavudine [d4T]), 2′,3′-dideoxyinosine (didanosine [ddI]), 2′,3′-dideoxycytidine (zalcitabine), (−)-β-2′,3′-dideoxy-3′-thiacytidine (lamivudine), its fluorinated derivative (−)-β-2′,3′-dideoxy-5-fluoro-3′-thiacytidine (emtricitabine), (1S,4R)-4-[2-amino-6-(cyclopropyl-amino)-9H-purin-9-yl]-2-cyclopentene-1-methanol succinate (abacavir), and the acyclic 9-[2-(R)-(phosphonomethoxy)propyl]adenine (tenofovir [TFV]). All NRTI require phosphorylation to their triphosphate (TP) forms, while metabolic activation of TFV requires phosphorylation to its diphosphate form (TFV-DP) (2, 34). The TFV acyclic linker between the adenine base and the phosphonate moiety is another unique feature that is not shared by either the other NRTI or natural deoxynucleoside triphosphates (dNTPs). Despite such substantial structural differences, TFV is efficiently incorporated by HIV-1 RT and was also shown to act as a chain-terminating nucleotide as a consequence of a missing hydroxyl group in the linker region (3). However, as demonstrated for AZT and other chain terminators (1, 26, 27), the incorporated TFV can be excised in the presence of pyrophosphate (PPi) or a PPi-donor, such as ATP (5, 43, 44). The balance between nucleotide excision and excision protection is regulated in a complex manner and depends critically on the chemical nature of the inhibitor (14, 23, 26). The factors and mechanisms that determine the efficiency of excision of TFV and its blockage are poorly understood.
The ATP-dependent excision reaction provides an important mechanism for resistance to NRTI. Rates of excision of AZT-monophosphate (AZT-MP) are significantly increased with mutant enzymes that contain classical AZT resistance mutations, including M41L, D67N, K70R, L210W, T215F/Y, and K219Q/E/N (6, 26). Differences between wild-type (WT) and mutant enzymes are most pronounced in the presence of ATP compared to the reaction with PPi, which suggests that some of these mutations facilitate binding of ATP in an orientation that allows excision to occur more efficiently (6, 33). The aforementioned mutations emerge under the selective pressure of AZT or d4T and are therefore referred to as thymidine-associated mutations (TAM). However, clinical data have shown that the accumulation of three or more TAM can also reduce susceptibility to nonthymidine analogues, including TFV (31). Biochemical data have shown that HIV-1 RT enzymes containing various combinations of TAM facilitate the ATP-dependent removal of literally all clinically available chain terminators, albeit at significantly different rates. For instance, high rates of excision have been reported for AZT-MP, while excision of ddAMP, i.e., the active metabolite of ddI, is sometimes difficult to detect (26).
Progress has been made in relating specific structural features of AZT-MP and deoxynucleoside monophosphates to differences seen in the rates of excision. Excision can occur only in the pretranslocation complex, when the 3′ end of the primer occupies the nucleotide binding site (N site) (6, 22, 35, 36). In contrast, binding of the next complementary nucleotide can occur only when the N site is freely accessible in the posttranslocation complex. Translocation of RT relative to its bound primer/template substrate clears the N site and brings the 3′ end of the primer to the product or priming site (P site). This configuration allows binding of the next nucleotide, resulting in a dead-end complex (DEC) that blocks excision.
Polymerase translocation is not kinetically defined (16). However, the development of site-specific footprinting techniques has allowed us to distinguish between pre- and posttranslocational states (N complex and P complex, respectively). It appears that the RT enzyme can rapidly shuttle between both configurations, which defines a translocational equilibrium (9, 10, 23, 24). An increasing concentration of the next nucleotide substrate can trap the complex in the posttranslocational state. Both band shift and site-specific footprinting experiments revealed that the stability of such DEC and, in turn, blockage of the excision reaction depend crucially on the chemical nature of the chain terminator (23, 39). Relatively high concentrations of the next nucleotide are required to form or stabilize a ternary complex with AZT-terminated primer strands, while d4T- or ddA-terminated primers do not appear to diminish the stability of a ternary complex (26, 35, 44).
The factors that govern the ATP-dependent excision of TFV in the context of both WT and TAM-containing enzymes remain to be characterized. Biochemical data point to relatively high levels of excision of TFV (5, 43, 44); however, it is unclear how the acyclic structure and the phosphonate moiety affect the translocational equilibrium and the formation of a DEC. Based on the structure of the RT-DNA complex with a TFV-terminated primer (P complex), it has been suggested that the flexibility of the acyclic nucleotide may provide a certain degree of protection from excision (41). The stability or catalytic efficiency of the pretranslocation complex could be diminished by multiple conformations of TFV, which may disfavor excision. Alternatively, the formation or stability of a ternary DEC in the posttranslocational state could likewise be diminished, which may favor excision.
To provide mechanistic insight, we studied the three A analogues ddAMP, TFV, and AZA-MP (the adenosine analogue of AZT-MP) and the thymidine analogue AZT-MP with respect to their influence on the translocational equilibrium in the absence or presence of nucleotides. Together, our results suggest that TFV-terminated primers have sufficient access to the N site in the pretranslocation complex. In the absence of natural nucleotides, excision of TFV occurs efficiently, and the rates are significantly increased with TAM-containing mutant enzymes. However, slight increases in the concentration of the next templated dNTP shift the translocational equilibrium towards posttranslocation, which provides a degree of protection from excision.
MATERIALS AND METHODS
Recombinant HIV-1 production and antiviral susceptibility assays.
PCR fragments corresponding to the first 1,000 bp of the HIV-1 RT gene were amplified from plasmids containing the RT gene of HIV-1 and cotransfected with the RT-deleted HIV-1 proviral molecular clone pHXB2Δ2-261RT (a gift from C. Boucher, Utrecht University, The Netherlands) as previously described (4, 30). The virus 4Y (with M41L, D67N, L210W, and T215Y RT mutations) was constructed by oligonucleotide-based site-directed mutagenesis of HXB2D RT and homologous recombination. Replication-competent viruses generated by homologous recombination were harvested after 8 to 18 days, when the cultures contained notable syncytia. The genotypes of the recombinant viruses were determined by RT-PCR of viral supernatants followed by sequencing using an ABI PRISM 3100 genetic analyzer (Applied Biosystems, Foster City, CA). The susceptibilities of the recombinant 4Y virus and the WT HIV-1 molecular clone HXB2D to TFV, AZT, d4T, ddI (Sigma, St. Louis, MO), abacavir (GlaxoSmithKline, Research Triangle Park, NC), and lamivudine were evaluated using an XTT-based viability assay (Sigma, St. Louis, MO) with MT-2 cells as previously described (7). All infections were performed with 1.2 × 106 cells at a multiplicity of infection of approximately 0.001, which resulted in equal levels of cell killing in the absence of drug over the 5-day assay period. Fifty percent effective concentration (EC50) values were calculated as averages for 8 to 20 experiments. Statistical significance (P < 0.05) was determined using two-tailed Student's t tests. t tests were performed on the array of EC50 values from all experimental data for each drug for 4Y compared to the WT data sets.
Enzymes, nucleic acids, and nucleotides.
Heterodimeric p66/p51 WT RT and the M41L/D67N/L210W/T215Y (4Y) mutant enzyme were purified as recently described (20, 44). Mutant enzymes were generated through site-directed mutagenesis using a Stratagene QuikChange kit according to the manufacturer's protocol. Custom DNA oligonucleotides with the following sequences were purchased from Invitrogen (Carlsbad, CA): PPT-57T, 5′-CGTTGGGAGTGAATTAGCCCTTCCAGTCCCCCCTTTTCTTTTAAAAAGTGGCTAAGA-3′; and PPT-57A, 5′-CGTTGGGAGTGAATTAGCCCTTCCAGACCCCCCTTTTCTTTTAAAAAGTGGCTAAGA-3′. Nucleotides in bold highlight the difference between the two oligonucleotides used as template strands. PPT-17 (5′-TTAAAAGAAAAGGGGGG-3′) was used as a primer that is complementary to the underlined region. The same sequences have previously been utilized to study the relationship between nucleotide excision and the translocation status of RT (23). Chemically synthesized oligonucleotides were purified in 12% polyacrylamide-7 M urea gels containing 50 mM Tris-borate (pH 8.0) and 1 mM EDTA. Purified nucleic acids were eluted from gel slices in a buffer containing 500 mM ammonium acetate and 0.1% sodium dodecyl sulfate. 5′-End labeling of oligonucleotides was conducted with [γ-32P]ATP and T4 polynucleotide kinase. Labeled oligonucleotides were gel purified as described previously (23). ATP, dNTPs, and ddNTPs were obtained from Fermentas Life Sciences (Hanover, MD), Roche Diagnostics (Indianapolis, IN), or Sigma-Aldrich Inc. (St. Louis, MO). AZT-TP and AZA-TP were purchased from TriLink Biotechnologies (Sorrento Mesa, CA). TFV-DP was synthesized at Gilead Sciences, Inc. (Foster City, CA). All dNTPs, dNTP analogues, and ATP were treated with inorganic pyrophosphatase in order to eliminate contaminating pyrophosphate. dNTPs were incubated in a buffer containing 1 mM dithiothreitol (DTT), 50 mM Tris-HCl (pH 8), 50 mM KCl, and 6 mM MgCl2 with 0.5 unit of inorganic pyrophosphatase (Roche Diagnostics) in a final volume of 100 μl at 37°C for 1 h. The nucleotides were purified by filtration through a 10-kDa-cutoff Microcon filter (Millipore, Billerica, MA).
Nucleotide incorporation.
Steady-state kinetic constants were determined using activated calf thymus DNA (Amersham Pharmacia Biotech, Piscataway, NJ) and rate determination by linear regression of [3H]dNTP incorporation over a time course, using GraphPad Prism, version 3.03 (GraphPad Software, San Diego, CA), followed by Km and Ki determinations using the competitive inhibition equation with simple weighting, using the Marquardt-Levenberg algorithm (EnzFitter, version 2.0.16; Biosoft, Stapleford, Cambridge, United Kingdom) as described previously (44). In these experiments, the template was present in excess, and competitive inhibition was observed based on a constant Vmax when increasing concentrations of NRTI were assayed. Statistical significance (P < 0.05) was determined using two-tailed Student's t tests.
Excision and rescue of chain-terminated DNA synthesis.
Excision and the ensuing rescue of DNA synthesis were studied at a single template position, using an assay similar to that described earlier (11). The primer-template duplex was preformed in a buffer containing 50 mM Tris-HCl (pH 7.8) and 50 mM NaCl. For this purpose, the 5′-end-labeled DNA primer (8.5 pmol) was hybridized to the cDNA template (25.5 pmol) by denaturing at 95°C for 2 min, followed by annealing at 72°C for 15 min and slow cooling to room temperature. The duplex was incubated with 25.5 pmol HIV-1 RT in a buffer containing 50 mM Tris-HCl (pH 7.8), 50 mM NaCl, and 6 mM MgCl2, followed by the addition of 10 μM ddATP, TFV-DP, AZA-TP, or AZT-TP. Reactions were allowed to proceed for 20 min to ensure complete incorporation of the nucleotide analogue. Excision and the ensuing rescue of chain-terminated DNA synthesis were monitored in time course experiments after the addition of ATP (3.5 mM) and a dNTP cocktail consisting of 100 μM dATP, 10 μM dCTP, and 100 μM ddTTP for adenosine analogs and 100 μM dTTP, 10 μM dCTP, and 100 μM ddGTP for AZT. Reactions were performed at 37°C and stopped at different times by adding 1-μl aliquots of reaction mixture to 9 μl of 95% formamide containing 40 mM EDTA. Samples were heat denatured for 5 min at 95°C and finally resolved in 8% polyacrylamide-7 M urea gels. Kinetic parameters for the combined excision and rescue reaction were determined at different concentrations of the next complementary nucleotide. Gels were scanned and analyzed with a phosphorimager (Molecular Imager FX; Bio-Rad). The results were quantified using ImageQuant 5.2 (Amersham) and Prism 4.0 (GraphPad).
Site-specific footprinting.
Potassium peroxynitrite (KOONO) was prepared by stirring 10 ml of 1.2 M KNO2 with 1.4 ml of 30% H2O2 on ice. Ten milliliters of 1.4 M HCl was added to the stirring solution and immediately quenched with 10 ml of 2 M NaOH. Aliquots were stored at −80°C. For preparation of the footprinting reaction, 2.7 pmol of PPT-57 in combination with a trace amount of 3′-32P-labeled PPT-57 was heat annealed with 8 pmol of PPT-17 (23). The DNA-DNA hybrid was incubated with 16 pmol of RT and the chain terminator (10 μM) in a buffer containing cacodylate (pH 7; 120 mM), NaCl (20 mM), DTT (0.5 mM), and MgCl2 (6 mM) in a final volume of 20 μl for 20 min at 37°C in order to form the chain-terminated complex. Different concentrations of the next templated nucleotide were then added to the reaction mix, followed by incubation for 10 min at 37°C. The complex was finally treated with 1.5 μl of KOONO (100 mM). The samples were analyzed as described above. The experiments were repeated at least three times, and representative images are shown in Fig. 3 and Fig. 5.
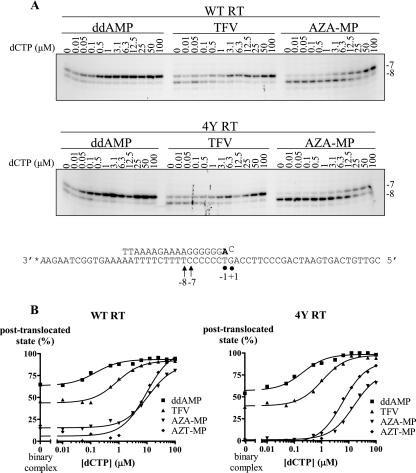
FIG. 3.
Site-specific footprints of chain-terminated complexes. (A) RT complexes with radiolabeled template and chain-terminated primers were treated with KOONO as described in Materials and Methods. The next templated nucleotide (dCTP) was added at increasing concentrations prior to KOONO cleavage. The sequence underneath the gels shows the positions of site-specific cleavage with KOONO treatment. Cleavage at position −8 is indicative of the pretranslocation complex, and cleavage at position −7 is indicative of the posttranslocation complex. (B) Graphical representation of data shown in panel A.
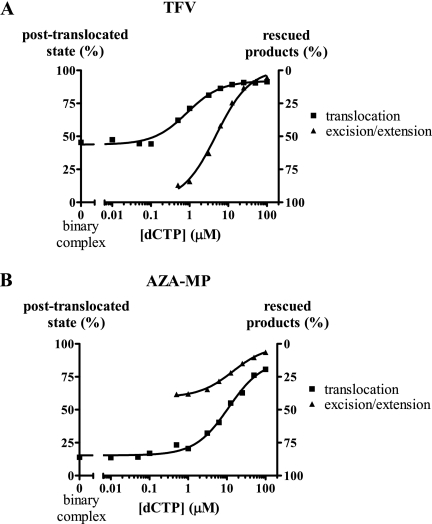
FIG. 5.
Correlation between excision and the translocation status of HIV-1 RT. Combined excision/rescue reactions with (A) TFV- and (B) AZA-MP-terminated primers were performed in the presence of increasing concentrations of the next templated nucleotide (dCTP). The results were graphed together with the corresponding footprinting experiments shown in Fig. 2. The graphs point to an inverse correlation between rates of excision (▴) and the shift toward the posttranslocation state (▪).
Gel mobility shift assay.
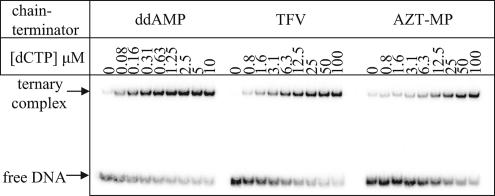
One picomole of the 3′-32P-labeled template PPT-31T (5′-CCTTCCAGTCCCCCCTTTTCTTTTAAAAAGT-3′) or PPT-31A (5′-CCTTCCAGACCCCCCTTTTCTTTTAAAAAGT-3′) was heat annealed to 3 pmol of PPT-17 primer. The DNA-DNA hybrid (1 pmol) was incubated with 5 pmol of HIV-1 RT in a buffer containing 50 mM Tris-HCl (pH 7.8), 80 mM NaCl, and 6 mM MgCl2 in a final volume of 20 μl. The DNA primer was first chain terminated with ddATP, TFV-DP, or AZT-TP. The next complementary nucleotide was then added at different concentrations prior to the addition of heparin (1.5 μg/μl). Complexes were incubated for 60 min at room temperature before the addition of 20 μl of 50% sucrose and trace amounts of bromophenol blue. The products were resolved in native 6% polyacrylamide-Tris-borate gels and visualized as described above. The results were quantified using ImageQuant software (Amersham) and fitted to a “one site binding” hyperbola [y = Bmax × x/(Kd + x), where Bmax is maximum binding], using GraphPad Prism. Kd values were determined from three independent experiments, and a representative image is shown in Fig. 4.
FIG. 4.
Formation of ternary complexes. Preformed complexes of HIV-1 RT and a DNA-DNA substrate were incubated with increasing concentrations of the templated nucleotide before the addition of a trap. The primers were chain terminated with ddAMP (left), TFV (middle), or AZT-MP (right).
RESULTS
Cell-based drug susceptibility measurements.
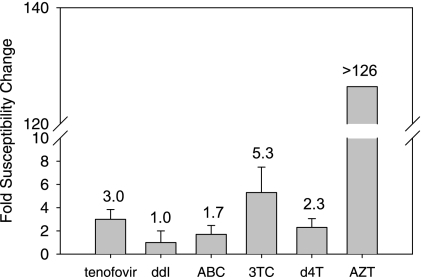
TAM are not selected under pressure of TFV; however, the accumulation of three or more of these mutations that include M41L or L210W was shown to reduce susceptibility to the drug in vitro and clinically (31, 42). We have generated an HIV-1 variant, designated 4Y, that contains the M41L-plus-D67N-plus-L210W-plus-T215Y cluster of TAM and determined its phenotypic susceptibility to different NRTI, including TFV. The 4Y TAM cluster conferred a threefold increase in EC50 values of TFV compared to those for the WT virus (Fig. 1). In accordance with published data, susceptibility to AZT was markedly reduced (>100-fold) in this mutational context, while susceptibility to ddI remained largely unaffected (19, 21). Cell toxicity prevented accurate susceptibility measurements with AZA.
FIG. 1.
Phenotypic susceptibility of the 4Y variant to different NRTI compared to that of WT HIV-1. Drug susceptibility measurements were conducted with MT-2 cells as described in Materials and Methods. Susceptibility changes (x-fold) for the 4Y mutant compared to the WT are indicated above the bars. ABC, abacavir; 3TC, lamivudine.
Efficiency of nucleotide incorporation and excision.
Steady-state kinetic measurements showed no significant differences between WT HIV-1 RT and mutant 4Y RT in analyzing the efficiency of binding or incorporation of different NRTI. We found that the ratio of Ki to Km was only marginally reduced for TFV-DP, which is not surprising considering that TAM are not associated with resistance mechanisms that involve substrate discrimination (17, 18, 33, 37) (Table 1). Hence, we looked at the efficiency of ATP-dependent excision reactions and the ensuing rescue of DNA synthesis of ddAMP-, AZA-MP-, and TFV-terminated primers (Fig. 2). In this setup, ATP was added together with a mixture of dNTP substrates required to produce a longer product upon NRTI excision that is distinguishable from the chain-terminated DNA. The concentration of dCTP, i.e., the next complementary nucleotide, was 10 μM, which reasonably mimics physiologically relevant conditions (8, 12, 40).
TABLE 1.
Ki and Ki/Km values for wild-type and mutant HIV-1 RT enzymes
| NRTI | WT RT
|
4Y RT
|
||
|---|---|---|---|---|
| Ki (μM)a (fold change)b | Ki/Km (μM) (fold change)c | Ki (μM)a (fold change)b | Ki/Km (μM) (fold change)c | |
| TFV-DP | 0.21 ± 0.07 (1.0) | 0.47 (1.0) | 0.11 ± 0.01d (0.5) | 0.19 (0.4) |
| ddATP | 0.09 ± 0.03 (1.0) | 0.20 (1.0) | 0.05 ± 0.01 (0.6) | 0.09 (0.5) |
| AZA-TP | 0.23 ± 0.07 (1.0) | 0.51 (1.0) | 0.15 ± 0.04 (0.65) | 0.26 (0.5) |
| AZT-TP | 0.041 ± 0.010 (1.0) | 0.09 (1.0) | 0.047 ± 0.009 (1.2) | 0.09 (1.0) |
Ki values are averages for at least three experiments ± standard deviations.
Relative to that for the wild type.
Relative to that for the wild type; however, no statistics were generated for this ratio.
P < 0.01 compared to the Km for the wild type by two-tailed Student's t test.
FIG. 2.
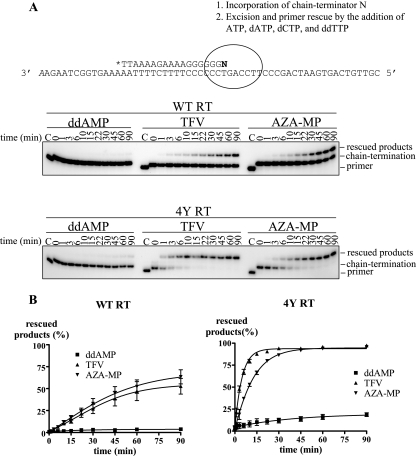
Excision of chain-terminating nucleotides with WT RT and the 4Y mutant. (A) Combined excision/rescue reactions were monitored in time course experiments with a radiolabeled primer (lanes C). The primer/template sequences and nucleotide mixtures used in this setup are shown above the gels. The primer was initially chain terminated with 10 μM ddAMP (left), 10 μM TFV-DP (middle), or 10 μM AZA-MP (right). The reaction was then initiated by the addition of ATP (3.5 mM) and a mixture of dATP (100 μM), dCTP (10 μM), and ddTTP (100 μM) to generate a defined reaction product, referred to as the rescued product, which can be distinguished from the chain-terminated primer. Reactions were stopped at the indicated time points, and samples were analyzed in denaturing 12% polyacrylamide gels. (B) Graphical representation of three data sets. Representative images are shown in panel A.
The rescue of ddAMP-terminated DNA synthesis with WT HIV-1 RT was almost undetectable under these conditions, which is in good agreement with previous studies (25, 44). The 4Y mutant enzyme increased the efficiency of the reaction. In contrast, WT RT was able to rescue TFV- and AZA-MP-terminated DNA synthesis, and the efficiencies of the reactions were substantially increased with the 4Y mutant RT. These reactions involve the same template position and the same nucleotide base; thus, any differences in rates of the reactions can be assigned to structural differences related to the sugar-phosphate moieties of ddAMP and AZA-MP or to the acyclic linker and phosphonate moiety of TFV. Nucleotide excision can be highly efficient, despite the lack of a 3′-azido group or any other equivalent bulky substituents that could diminish the stability of the posttranslocation state. These data suggest that TFV-terminated primers have relatively good access to the N site in the pretranslocation state.
Site-specific footprinting of binary and ternary complexes.
To study the influence of the chemical nature of incorporated chain terminators on the translocational equilibrium of HIV-1 RT, we determined the relative proportions of complexes that existed either pre- or posttranslocation (Fig. 3). We utilized site-specific footprinting techniques in the presence of increasing concentrations of the next complementary nucleotide (dCTP) to characterize conditions that promote a shift toward posttranslocation (Table 2). These quantitative footprinting experiments were conducted with KOONO, which is a metal-free source of hydroxyl radicals that mediates hyperreactive cleavage at template positions −7 and −8, depending on the translocational state (23). A cut at position −7 is indicative of the posttranslocation state, while a cut at position −8 is indicative of the pretranslocation state. Cleavage is mediated through residue C280, which in the large subunit p66 is located in the vicinity of the template. The relative distribution of the two bands at positions −7 and −8 reflects the relative distribution of pre- and posttranslocational stages within the time interval of a few seconds.
TABLE 2.
Effects of inhibitor and enzyme on the translocation status of RT
| Chain terminator | Posttranslocated state | % or concna
|
|
|---|---|---|---|
| WT RT | 4Y RT | ||
| ddAMP | No dCTP | 65% | 54% |
| IC50(dCTP) | ND | ND | |
| IC80(dCTP) | 0.2 μM | 0.3 μM | |
| TFV | No dCTP | 45% | 38% |
| IC50(dCTP) | 0.1 μM | 0.3 μM | |
| IC80(dCTP) | 3 μM | 2 μM | |
| AZA-MP | No dCTP | 14% | 1% |
| IC50(dCTP) | 10 μM | 27 μM | |
| IC80(dCTP) | 100 μM | >100 μM | |
| AZT-MP | No dCTP | 6% | 0% |
| IC50(dCTP) | 8 μM | 8 μM | |
| IC80(dCTP) | 31 μM | 50 μM | |
Percentage of complexes that existed posttranslocationally in the absence of nucleotide substrate or nucleotide concentration required to form 50% or 80% of the complex population in its posttranslocation state. Percentages are means for at least three independent experiments. The standard deviation was below 10%. ND, not determined because the posttranslocated complex was >50% in the absence of dCTP.
We found that the binary complex of WT RT with a ddAMP-terminated primer exists predominantly in its posttranslocation state (65%) (Fig. 3 and Table 2). Submicromolar concentrations of the next nucleotide were sufficient to obtain 80% of the complex population in the posttranslocational state. This value is referred to as the IC80(dNTP). The IC50(dNTP) value (concentration of dCTP required to obtain 50% of the complex population in the posttranslocation state) cannot be determined for ddAMP, because the posttranslocational complex is already favored in the absence of dNTP substrates.
Translocation appears to be diminished with TFV-terminated primers compared to that with ddAMP. For TFV, 45% of the complex population preexisted in the posttranslocational state in the absence of nucleotides, and IC50(dNTP) and IC80(dNTP) values of 0.1 μM and 3 μM, respectively, were obtained in the presence of the next nucleotide. For comparative purposes, another template was evaluated that allowed us to monitor RT translocation and excision of the T analogue AZT-MP at the same position as the A analogues. In contrast to TFV and ddAMP, both AZT-MP and AZA-MP promoted formation of the pretranslocational state. In the absence of nucleotides, the complex population existed almost exclusively in its pretranslocational state, and high concentrations of the next templated nucleotide were required to obtain the majority of complexes in the posttranslocational state. We measured IC80(dNTP) values of >100 μM (AZA-MP) and 31 μM (AZT-MP), respectively. The difference between the two 3′-azido-containing nucleotides may not be significant. Taken together, the footprints show that the concentration of the next nucleotide required to trap the complex posttranslocation follows the order AZA-MP ≈ AZT-MP > TFV > ddAMP.
We next analyzed the influence of the chain terminator on the stability of the ternary complex directly. Previous band shift studies have shown that binary complexes of HIV-1 RT and primer/template are unstable when challenged with a trap, e.g., heparin. However, the presence of the dNTP substrate can stabilize the complex. The concentration of the next nucleotide required to maintain a band shift was significantly higher with AZT-terminated primers than with other chain terminators, such as ddATP (39). Here we found that TFV-terminated primers behave between the two extremes (Fig. 4). The apparent Kds follow the order AZT-MP > TFV > ddAMP (24.9 ± 1.4 μM, 3.2 ± 0.12 μM, and 0.1 ± 0.007 μM, respectively). Relatively large amounts of stable binary complex (≈40%) prevented accurate measurements with AZA-terminated primers, while <10% binary complex was seen with the other chain terminators under these conditions. Overall, the data are in good agreement with the results of our footprinting experiments.
The four TAM did not appear to largely affect the translocational equilibrium of HIV-1 RT (Fig. 3 and Table 2). Footprints in the absence of dNTP substrates point to slight increases in the stability of the pretranslocation complex; however, these differences are within a margin of error of approximately 10% translocation. In the presence of the next dNTP, no significant differences between WT RT and the 4Y mutant RT were observed with ddAMP-terminated primers, which is consistent with the observation that this combination of TAM does not greatly affect susceptibility to the drug or rates of excision. In contrast, the 4Y cluster facilitated the efficient removal of TFV and AZA/T-MP; however, our footprint results show that such increased rates of excision are not due to increased stability of the pretranslocation complex in the context of TAM.
It is conceivable that differences between WT and mutant enzymes may become evident when footprint experiments are conducted in the presence of the pyrophosphate donor ATP, although it should be noted that these conditions promote excision, which in turn affects the positioning of the RT enzyme. However, ATP binding and the ensuing removal of the nucleotide analogue can be assessed through enzyme kinetic measurements. The rate of the reaction will crucially depend on the concentration of the next templated nucleotide. Thus, to establish meaningful reaction conditions for more detailed kinetic measurements, we determined the amount of rescued product in the presence of multiple concentrations of the next complementary nucleotide.
Influence of the next dNTP on RT translocation and the efficiency of excision.
The next experiments were limited to excision reactions with WT RT in the context of TFV- and AZA-MP-terminated primer strands. The lowest nucleotide concentration was 0.5 μM in these experiments. Lower concentrations compromised the forward reaction following excision of the nucleotide analogue (data not shown). Moreover, following the removal of A analogues, the absence of the next nucleotide promoted incorporation of ATP and prevented accurate quantification of excision (data not shown). The efficiency of rescue of AZA-MP- and TFV-terminated DNA synthesis correlated inversely with the formation of the posttranslocation complex, as expected (Fig. 5A and B). However, the sensitivity to the dNTP substrate depended crucially on the chemical nature of the chain terminator. TFV was almost completely removed at submicromolar concentrations of the next templated nucleotide, while the removal of AZA-MP was less efficient under the same conditions. This was surprising at first glance, because the amount of complex that existed pretranslocation was reduced compared to that of AZA-MP. Thus, the data suggest that the N site is used more efficiently for excision when the primer is terminated with TFV under these conditions. However, the presence of the next templated nucleotide, which at low micromolar concentrations mimics the cellular milieu, caused substantial declines in the removal of TFV. In contrast, reactions with AZA-MP were less sensitive to changes with respect to the concentration of the next nucleotide. The same trend was seen with respect to dNTP dependence of the translocational equilibrium.
Kinetic parameters for ATP-dependent excision of TFV.
To better understand how the four TAM in 4Y RT can facilitate the removal of incorporated TFV, we measured and compared kinetic parameters for reactions conducted with WT RT and the 4Y mutant in the context of TFV, AZA-MP, and AZT-MP. Km and Vmax values for ATP were determined in the presence of 0.5 μM and 10 μM dCTP to assess the sensitivity to inhibition of excision by the next templated nucleotide. Ten micromolar dCTP may represent a concentration that is well within a physiologically relevant window (8, 12, 40). Excision of TFV was increased 15- to 20-fold with the 4Y mutant, while values for the excision of AZA-MP and AZT-MP showed 3-fold and 7-fold changes, respectively (Table 3). Of note, almost identical values were obtained under steady-state and pre-steady-state conditions when looking at the excision of these two 3′-azido-containing nucleotides (26, 37).
TABLE 3.
Efficiency of combined ATP-dependent excision/extension reaction
| Chain terminator | Enzyme | dCTP concn (μM) | Km (mM)a | kcat (min−1 [10−3])a | kcat/Km (min−1 M−1) | Catalytic efficiencyb (fold change) | Sensitivity to dNTP inhibitionc |
|---|---|---|---|---|---|---|---|
| TFV | WT RT | 0.5 | 1.0 ± 0.11 | 1.1 ± 0.04 | 1.1 | ||
| 10 | 1.9 ± 0.3 | 0.28 ± 0.02 | 0.15 | 7.3 | |||
| 4Y RT | 0.5 | 0.59 ± 0.05 | 9.7 ± 0.24 | 16.4 | 14.9 | ||
| 10 | 0.96 ± 0.24 | 2.8 ± 0.24 | 2.9 | 19.3 | 5.7 | ||
| AZA-MP | WT RT | 0.5 | 1.7 ± 0.21 | 0.84 ± 0.044 | 0.49 | ||
| 10 | 1.2 ± 0.21 | 0.44 ± 0.03 | 0.37 | 1.3 | |||
| 4Y RT | 0.5 | 2.0 ± 0.49 | 3.2 ± 0.37 | 1.6 | 3.3 | ||
| 10 | 2.0 ± 0.36 | 2.4 ± 0.22 | 1.2 | 3.2 | 1.3 | ||
| AZT-MP | WT RT | 0.5 | 1.3 ± 0.34 | 0.25 ± 0.026 | 0.19 | ||
| 10 | 1.0 ± 0.25 | 0.16 ± 0.014 | 0.16 | 1.2 | |||
| 4Y RT | 0.5 | 1.4 ± 0.27 | 1.8 ± 0.14 | 1.3 | 6.8 | ||
| 10 | 1.2 ± 0.19 | 1.3 ± 0.076 | 1.08 | 6.8 | 1.2 |
Values are means for at least three independent experiments ± standard deviations.
Catalytic efficiency is given as the ratio of [kcat/Km(4Y RT)] to [kcat/Km(WT RT)].
Sensitivity to dNTP-dependent inhibition is given as the ratio of [kcat/Km(0.5 μM dCTP)] to [kcat/Km(10 μM dCTP)].
The kinetic measurements indicate that the removal of TFV is highly sensitive to inhibition in the presence of the next nucleotide. We calculated a sensitivity index as the ratio of the efficiencies of excision measured at 0.5 μM and 10 μM of the next templated nucleotide, i.e., dCTP. WT RT and the 4Y mutant showed relatively high values for TFV, of 7.7 and 5.6, respectively, while the corresponding values with AZA-MP and AZT-MP were just slightly above 1.
DISCUSSION
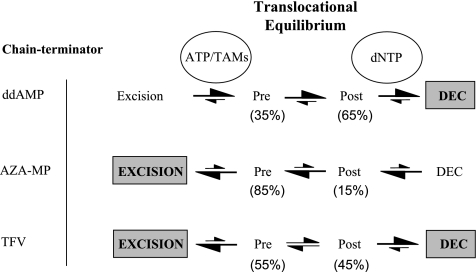
The translocational equilibrium of HIV-1 RT may be seen as a distinct parameter that can influence the excision reaction. We have previously shown that the ratio of complexes that exist either pre- or posttranslocation can depend on the primer/template sequence, the temperature, and the chemical nature of the chain terminator (9, 23, 24). Moreover, different classes of RT inhibitors can specifically bind to the pre- or posttranslocated state, which validates this equilibrium as a distinct viral target (15, 24). Here we characterized and quantitatively assessed the impact of the translocational equilibrium on the overall efficiency of excision of TFV in comparison to that of ddAMP, AZA-MP, and AZT-MP. Our results are summarized in the model shown in Fig. 6.
FIG. 6.
Effects of translocation status of HIV-1 RT on the ATP-dependent excision of NRTI. Site-specific footprinting experiments revealed that the RT enzyme can oscillate between pre- and posttranslocational states. The ratio of both complexes is given as a percentage for WT RT and is dependent upon the nature of the chain terminator present at the 3′ end of the primer. Differences in the sizes of the arrows under “translocational equilibrium” represent differences in the translocational equilibrium measured in the absence of dNTP. The formation of the DEC depends on the dNTP concentration. Differences in the sizes of the arrows under “dNTP” reflect differences seen in footprints with increasing concentrations of the next templated nucleotide. The efficiency of the excision seen under steady-state conditions depends upon the chemical nature of the inhibitor, the concentration of ATP, and the presence of resistance-conferring mutations (TAM) in the RT enzyme. Dominant pathways are boxed. The excision of TFV is efficient at low concentrations of dNTP, while increases to the low micromolar range favor DEC formation and chain terminator stability.
Effects of TFV-terminated primers on translocational equilibrium.
The relatively high rate of excision of TFV, despite its more limited access to the pretranslocational state compared to that of AZA/T-MP, suggests that the phosphonate and/or the flexibility of the acyclic inhibitor is a factor that can affect the efficiency of the excision reaction. Comparative experiments with nucleotides containing a phosphonate or a phosphate with identical sugar and base moieties are required to elucidate the contribution of each of the two structural features individually.
Effects of TFV on DEC formation.
The ratio of complexes that exist pre- and posttranslocation in the absence of the next templated nucleotide appears to be indicative of the efficiency with which a ternary DEC is formed in the presence of nucleotides (Fig. 6). The formation of a DEC depends on the posttranslocation position of the 3′ end of the primer and follows the order ddAMP > TFV > AZA/T-MP. The diminished ability of RT to form ternary complexes with AZA/T-terminated primer strands is consistent with previous modeling studies and crystallographic data pointing to steric problems between the bulky 3′-azido group and the incoming dNTP substrate (6, 35). In contrast, despite the lack of a sugar moiety, the DEC with a TFV-terminated primer strand is remarkably stable, which suggests that base stacking between the bound nucleotide, the incorporated TFV, and the penultimate primer base is relatively stable. This was unexpected, considering that crystallographic models in the absence of the next nucleotide show multiple conformations of the incorporated acyclic nucleotide (41). Thus, the combined biochemical and crystallographic data suggest that the incoming nucleotide may freeze a conformation that allows base stacking with the incoming nucleotide. The formation of a stable ternary complex is consistent with the diminished rates of excision of TFV in the presence of 10 μM nucleotide substrate.
Effects of TAM on the excision of TFV.
TAM-containing mutant enzymes can cause significant increases in the rate of excision of TFV. Like the case for the WT enzyme, the presence of 10 μM of the next templated nucleotide caused severe reductions in the combined excision and rescue reaction with TFV-terminated primer strands. However, the efficiency of the reaction was still remarkably high, and the mutant conferred a clear advantage over the WT enzyme. In contrast, cell-based susceptibility measurements point to 3-fold and >100-fold increases in EC50 values for TFV and AZT, respectively, when comparing the TAM-containing virus with WT HIV-1. A possible explanation is that the concentration of 10 μM of the next templated nucleotide used in our biochemical experiments may not be high enough to adequately mimic conditions under which drug susceptibility measurements are performed in vitro. Values in the range of 10 μM may well represent average concentrations of intracellular dNTP pools; however, dNTP concentrations can fluctuate, depending on the cell cycle, the intracellular compartment, the cell type, and the transformation status of the cell (8, 12, 40).
The threshold for dNTP concentrations required to completely block excision was also shown to depend critically on the primer/template sequence (23, 29). In addition, both site-specific footprinting experiments and binding studies have shown that the translocational equilibrium depends on the sequence (13, 23, 24). Our experiments were performed with a sequence that facilitates the study of RT translocation close to equilibrium. We detected very similar amounts of pre- and posttranslocated states in the absence of a chain terminator or with ddAMP-terminated primers. Thus, this sequence permits the analysis of changes with respect to the ratio of complexes that exist either pre- or posttranslocation. Sequences that promote translocation may have a greater impact on excision protection of TFV, because this inhibitor facilitates translocation and the formation of a DEC per se. In contrast, AZT-MP, which compromises translocation and disfavors the formation of a DEC, may still be excised in a particular sequence context that diminishes or even prevents excision of TFV.
Several studies have suggested that TAM facilitate binding of ATP in a catalytically competent mode, which helps to explain the increased rates of excision of multiple chain terminators in the presence of these mutations (28, 32, 33). This appears to be the dominant role played by the T215F/Y substitution. The effects of TAM on the translocational equilibrium are subtle. The presence of the T69S-SS finger insertion against a background of TAM shows a stronger bias towards pretranslocation, which probably reflects the diminished ability of the enzyme to form a DEC (23, 25, 38, 43).
Taken together, the data shown in this study suggest that the ATP-dependent excision of TFV is driven by efficient excision at the N site in the pretranslocation complex. High rates of excision can be partially neutralized by a facile switch to the posttranslocational state and the subsequent formation of a DEC in the presence of relatively low concentrations of the next templated nucleotide. These results are in agreement with clinical data which have shown decreased virological responses in TFV-disoproxil fumarate-treated patients who have multiple TAM in their HIV-1 strains (31). In addition, the results of this study warrant further investigation with regard to the sequence dependence of the translocational equilibrium and its possible impact on nucleotide excision in the context of both WT and mutant RT enzymes. It will be important to screen for and to identify sequences that promote enzyme dissociation and/or formation of the posttranslocation state in order to determine the lower limits of the concentration of the next templated nucleotide required to render chain termination irreversible.
Acknowledgments
We thank Suzanne McCormick, Meoïn Hagège, and Adrian Ray for excellent technical assistance, Joy Feng for a review of the manuscript, and Margaret Benton for administrative assistance.
The work performed by B.M. was in partial fulfillment of the requirements for a Ph.D. degree by the Faculty of Graduate Studies, McGill University. B.M. is the recipient of a doctoral research award from the Canadian Institutes of Health Research (CIHR). M.G. is the recipient of a national career award from the CIHR. This research was funded by the CIHR and Gilead Sciences, Inc.
Footnotes
Published ahead of print on 21 May 2007.
REFERENCES
- 1.Arion, D., N. Kaushik, S. McCormick, G. Borkow, and M. A. Parniak. 1998. Phenotypic mechanism of HIV-1 resistance to 3′-azido-3′-deoxythymidine (AZT): increased polymerization processivity and enhanced sensitivity to pyrophosphate of the mutant viral reverse transcriptase. Biochemistry 37:15908-15917. [DOI] [PubMed] [Google Scholar]
- 2.Balzarini, J., and E. De Clercq. 1991. 5-Phosphoribosyl 1-pyrophosphate synthetase converts the acyclic nucleoside phosphonates 9-(3-hydroxy-2-phosphonylmethoxypropyl)adenine and 9-(2-phosphonylmethoxyethyl)adenine directly to their antivirally active diphosphate derivatives. J. Biol. Chem. 266:8686-8689. [PubMed] [Google Scholar]
- 3.Balzarini, J., Z. Hao, P. Herdewijn, D. G. Johns, and E. De Clercq. 1991. Intracellular metabolism and mechanism of anti-retrovirus action of 9-(2-phosphonylmethoxyethyl)adenine, a potent anti-human immunodeficiency virus compound. Proc. Natl. Acad. Sci. USA 88:1499-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boucher, C. A., W. Keulen, T. van Bommel, M. Nijhuis, D. de Jong, M. D. de Jong, P. Schipper, and N. K. Back. 1996. Human immunodeficiency virus type 1 drug susceptibility determination by using recombinant viruses generated from patient sera tested in a cell-killing assay. Antimicrob. Agents Chemother. 40:2404-2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyer, P. L., T. Imamichi, S. G. Sarafianos, E. Arnold, and S. H. Hughes. 2004. Effects of the Delta67 complex of mutations in human immunodeficiency virus type 1 reverse transcriptase on nucleoside analog excision. J. Virol. 78:9987-9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyer, P. L., S. G. Sarafianos, E. Arnold, and S. H. Hughes. 2001. Selective excision of AZTMP by drug-resistant human immunodeficiency virus reverse transcriptase. J. Virol. 75:4832-4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cherrington, J. M., M. D. Fuller, A. S. Mulato, S. J. Allen, S. C. Kunder, M. A. Ussery, Z. Lesnikowski, R. F. Schinazi, J. P. Sommadossi, and M. S. Chen. 1996. Comparative kinetic analyses of interaction of inhibitors with Rauscher murine leukemia virus and human immunodeficiency virus reverse transcriptases. Antimicrob. Agents Chemother. 40:1270-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao, W.-Y., A. Cara, R. C. Gallo, and F. Lori. 1993. Low levels of deoxynucleotides in peripheral blood lymphocytes: a strategy to inhibit human immunodeficiency virus type 1 replication. Proc. Natl. Acad. Sci. USA 90:8925-8928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gotte, M. 2006. Effects of nucleotides and nucleotide analogue inhibitors of HIV-1 reverse transcriptase in a ratchet model of polymerase translocation. Curr. Pharm. Des. 12:1867-1877. [DOI] [PubMed] [Google Scholar]
- 10.Gotte, M. 2004. Inhibition of HIV-1 reverse transcription: basic principles of drug action and resistance. Expert Rev. Anti Infect. Ther. 2:707-716. [DOI] [PubMed] [Google Scholar]
- 11.Gotte, M., D. Arion, M. A. Parniak, and M. A. Wainberg. 2000. The M184V mutation in the reverse transcriptase of human immunodeficiency virus type 1 impairs rescue of chain-terminated DNA synthesis. J. Virol. 74:3579-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hauschka, P. V. 1973. Analysis of nucleotide pools in animal cells. Methods Cell Biol. 7:361-462. [DOI] [PubMed] [Google Scholar]
- 13.Ignatov, M. E., A. J. Berdis, S. F. Le Grice, and M. D. Barkley. 2005. Attenuation of DNA replication by HIV-1 reverse transcriptase near the central termination sequence. Biochemistry 44:5346-5356. [DOI] [PubMed] [Google Scholar]
- 14.Isel, C., C. Ehresmann, P. Walter, B. Ehresmann, and R. Marquet. 2001. The emergence of different resistance mechanisms toward nucleoside inhibitors is explained by the properties of the wild type HIV-1 reverse transcriptase. J. Biol. Chem. 276:48725-48732. [DOI] [PubMed] [Google Scholar]
- 15.Jochmans, D., J. Deval, B. Kesteleyn, H. Van Marck, E. Bettens, I. De Baere, P. Dehertogh, T. Ivens, M. Van Ginderen, B. Van Schoubroeck, M. Ehteshami, P. Wigerinck, M. Gotte, and K. Hertogs. 2006. Indolopyridones inhibit human immunodeficiency virus reverse transcriptase with a novel mechanism of action. J. Virol. 80:12283-12292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kati, W. M., K. A. Johnson, L. F. Jerva, and K. S. Anderson. 1992. Mechanism and fidelity of HIV reverse transcriptase. J. Biol. Chem. 267:25988-25997. [PubMed] [Google Scholar]
- 17.Kerr, S. G., and K. S. Anderson. 1997. Pre-steady-state kinetic characterization of wild type and 3′-azido-3′-deoxythymidine (AZT) resistant human immunodeficiency virus type 1 reverse transcriptase: implication of RNA directed DNA polymerization in the mechanism of AZT resistance. Biochemistry 36:14064-14070. [DOI] [PubMed] [Google Scholar]
- 18.Krebs, R., U. Immendorfer, S. H. Thrall, B. M. Wohrl, and R. S. Goody. 1997. Single-step kinetics of HIV-1 reverse transcriptase mutants responsible for virus resistance to nucleoside inhibitors zidovudine and 3-TC. Biochemistry 36:10292-10300. [DOI] [PubMed] [Google Scholar]
- 19.Larder, B. A., and S. D. Kemp. 1989. Multiple mutations in HIV-1 reverse transcriptase confer high-level resistance to zidovudine (AZT). Science 246:1155-1158. [DOI] [PubMed] [Google Scholar]
- 20.Le Grice, S. F., and F. Gruninger-Leitch. 1990. Rapid purification of homodimer and heterodimer HIV-1 reverse transcriptase by metal chelate affinity chromatography. Eur. J. Biochem. 187:307-314. [DOI] [PubMed] [Google Scholar]
- 21.Lin, P. F., H. Samanta, R. E. Rose, A. K. Patick, J. Trimble, C. M. Bechtold, D. R. Revie, N. C. Khan, M. E. Federici, H. Li, et al. 1994. Genotypic and phenotypic analysis of human immunodeficiency virus type 1 isolates from patients on prolonged stavudine therapy. J. Infect. Dis. 170:1157-1164. [DOI] [PubMed] [Google Scholar]
- 22.Marchand, B., and M. Gotte. 2004. Impact of the translocational equilibrium of HIV-1 reverse transcriptase on the efficiency of mismatch extensions and the excision of mispaired nucleotides. Int. J. Biochem. Cell. Biol. 36:1823-1835. [DOI] [PubMed] [Google Scholar]
- 23.Marchand, B., and M. Gotte. 2003. Site-specific footprinting reveals differences in the translocation status of HIV-1 reverse transcriptase. J. Biol. Chem. 278:35362-35372. [DOI] [PubMed] [Google Scholar]
- 24.Marchand, B., E. P. Tchesnokov, and M. Gotte. 2007. The pyrophosphate analogue foscarnet traps the pre-translocational state of HIV-1 reverse transcriptase in a Brownian ratchet model of polymerase translocation. J. Biol. Chem. 282:3337-3346. [DOI] [PubMed] [Google Scholar]
- 25.Meyer, P. R., J. Lennerstrand, S. E. Matsuura, B. A. Larder, and W. A. Scott. 2003. Effects of dipeptide insertions between codons 69 and 70 of human immunodeficiency virus type 1 reverse transcriptase on primer unblocking, deoxynucleoside triphosphate inhibition, and DNA chain elongation. J. Virol. 77:3871-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer, P. R., S. E. Matsuura, A. M. Mian, A. G. So, and W. A. Scott. 1999. A mechanism of AZT resistance: an increase in nucleotide-dependent primer unblocking by mutant HIV-1 reverse transcriptase. Mol. Cell 4:35-43. [DOI] [PubMed] [Google Scholar]
- 27.Meyer, P. R., S. E. Matsuura, A. G. So, and W. A. Scott. 1998. Unblocking of chain-terminated primer by HIV-1 reverse transcriptase through a nucleotide-dependent mechanism. Proc. Natl. Acad. Sci. USA 95:13471-13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer, P. R., S. E. Matsuura, A. A. Tolun, I. Pfeifer, A. G. So, J. W. Mellors, and W. A. Scott. 2002. Effects of specific zidovudine resistance mutations and substrate structure on nucleotide-dependent primer unblocking by human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 46:1540-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyer, P. R., A. J. Smith, S. E. Matsuura, and W. A. Scott. 2004. Effects of primer-template sequence on ATP-dependent removal of chain-terminating nucleotide analogues by HIV-1 reverse transcriptase. J. Biol. Chem. 279:45389-45398. [DOI] [PubMed] [Google Scholar]
- 30.Miller, M. D., K. E. Anton, A. S. Mulato, P. D. Lamy, and J. M. Cherrington. 1999. Human immunodeficiency virus type 1 expressing the lamivudine-associated M184V mutation in reverse transcriptase shows increased susceptibility to adefovir and decreased replication capability in vitro. J. Infect. Dis. 179:92-100. [DOI] [PubMed] [Google Scholar]
- 31.Miller, M. D., N. Margot, B. Lu, L. Zhong, S.-S. Chen, A. Cheng, and M. Wulfsohn. 2004. Genotypic and phenotypic predictors of the magnitude of response to tenofovir disoproxil fumarate in antiretroviral-experienced patients. J. Infect. Dis. 189:837-846. [DOI] [PubMed] [Google Scholar]
- 32.Naeger, L. K., N. A. Margot, and M. D. Miller. 2002. ATP-dependent removal of nucleoside reverse transcriptase inhibitors by human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 46:2179-2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ray, A. S., E. Murakami, A. Basavapathruni, J. A. Vaccaro, D. Ulrich, C. K. Chu, R. F. Schinazi, and K. S. Anderson. 2003. Probing the molecular mechanisms of AZT drug resistance mediated by HIV-1 reverse transcriptase using a transient kinetic analysis. Biochemistry 42:8831-8841. [DOI] [PubMed] [Google Scholar]
- 34.Robbins, B. L., J. Greenhaw, M. C. Connelly, and A. Fridland. 1995. Metabolic pathways for activation of the antiviral agent 9-(2-phosphonylmethoxyethyl)adenine in human lymphoid cells. Antimicrob. Agents Chemother. 39:2304-2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sarafianos, S. G., A. D. Clark, Jr., K. Das, S. Tuske, J. J. Birktoft, P. Ilankumaran, A. R. Ramesha, J. M. Sayer, D. M. Jerina, P. L. Boyer, S. H. Hughes, and E. Arnold. 2002. Structures of HIV-1 reverse transcriptase with pre- and post-translocation AZTMP-terminated DNA. EMBO J. 21:6614-6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarafianos, S. G., A. D. Clark, Jr., S. Tuske, C. J. Squire, K. Das, D. Sheng, P. Ilankumaran, A. R. Ramesha, H. Kroth, J. M. Sayer, D. M. Jerina, P. L. Boyer, S. H. Hughes, and E. Arnold. 2003. Trapping HIV-1 reverse transcriptase before and after translocation on DNA. J. Biol. Chem. 278:16280-16288. [DOI] [PubMed] [Google Scholar]
- 37.Sluis-Cremer, N., D. Arion, U. Parikh, D. Koontz, R. F. Schinazi, J. W. Mellors, and M. A. Parniak. 2005. The 3′-azido group is not the primary determinant of 3′-azido-3′-deoxythymidine (AZT) responsible for the excision phenotype of AZT-resistant HIV-1. J. Biol. Chem. 280:29047-29052. [DOI] [PubMed] [Google Scholar]
- 38.Terada, N., T. Hamazaki, M. Oka, M. Hoki, D. M. Mastalerz, Y. Nakano, E. M. Meyer, L. Morel, B. E. Petersen, and E. W. Scott. 2002. Bone marrow cells adopt the phenotype of other cells by spontaneous cell fusion. Nature 416:542-545. [DOI] [PubMed] [Google Scholar]
- 39.Tong, W., C.-D. Lu, S. K. Sharma, S. Matsuura, A. G. So, and W. A. Scott. 1997. Nucleotide-induced stable complex formation by HIV-1 reverse transcriptase. Biochemistry 36:5749-5757. [DOI] [PubMed] [Google Scholar]
- 40.Traut, T. 1994. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 140:1-22. [DOI] [PubMed] [Google Scholar]
- 41.Tuske, S., S. G. Sarafianos, A. D. Clark, Jr., J. Ding, L. K. Naeger, K. L. White, M. D. Miller, C. S. Gibbs, P. L. Boyer, P. Clark, G. Wang, B. L. Gaffney, R. A. Jones, D. M. Jerina, S. H. Hughes, and A. Eddy. 2004. Structures of HIV-1 RT-DNA complexes before and after incorporation of the anti-AIDS drug tenofovir. Nat. Struct. Mol. Biol. 11:469-474. [DOI] [PubMed] [Google Scholar]
- 42.White, K. L., J. M. Chen, J. Y. Feng, N. A. Margot, J. K. Ly, A. S. Ray, H. L. MacArthur, M. J. McDermott, S. Swaminathan, and M. D. Miller. 2006. The K65R reverse transcriptase mutation in HIV-1 reverses the excision phenotype of zidovudine resistance mutations. Antivir. Ther. 11:155-163. [DOI] [PubMed] [Google Scholar]
- 43.White, K. L., J. M. Chen, N. A. Margot, T. Wrin, C. J. Petropoulos, L. K. Naeger, S. Swaminathan, and M. D. Miller. 2004. Molecular mechanisms of tenofovir resistance conferred by human immunodeficiency virus type 1 reverse transcriptase containing a diserine insertion after residue 69 and multiple thymidine analog-associated mutations. Antimicrob. Agents Chemother. 48:992-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.White, K. L., N. A. Margot, J. K. Ly, J. M. Chen, A. S. Ray, M. Pavelko, R. Wang, M. McDermott, S. Swaminathan, and M. D. Miller. 2005. A combination of decreased NRTI incorporation and decreased excision determines the resistance profile of HIV-1 K65R RT. AIDS 19:1751-1760. [DOI] [PubMed] [Google Scholar]