Abstract
Nonobligate chain terminators, such as 2′-C-methylated nucleotides, block RNA synthesis by the RNA-dependent RNA polymerase (RdRp) of hepatitis C virus (HCV). Previous studies with related viral polymerases have shown that classical chain terminators lacking the 3′-hydroxyl group can be excised in the presence of pyrophosphate (PPi), which is detrimental to the inhibitory activity of these compounds. Here we demonstrate that the HCV RdRp enzyme is capable of removing both obligate and clinically relevant nonobligate chain terminators. Pyrimidines are more efficiently excised than are purines. The presence of the next complementary templated nucleotide literally blocks the excision of obligate chain terminators through the formation of a dead-end complex (DEC). However, 2′-C-methylated CMP is still cleaved efficiently under these conditions. These findings show that a 2′-methylated primer terminus impedes nucleotide binding. The S282T mutation, associated with resistance to 2′-C-methylated nucleotides, does not affect the excision patterns. Thus, the decreased susceptibility to 2′-C-methylated nucleotides appears to be based solely on improved discrimination between the inhibitor and its natural counterpart. In conclusion, our data suggest that the phosphorolytic excision of nonobligate, pyrimidine-based chain terminators can diminish their potency. The templated nucleotide does not appear to provide protection from excision through DEC formation.
Hepatitis C virus (HCV) infection is a serious public health concern that affects 170 million people worldwide (33, 42). Among those infected, approximately 20 to 30% develop severe liver disease, such as chronic hepatitis, liver cirrhosis, or hepatocellular carcinoma (2). The combined use of the nucleoside analogue ribavirin and pegylated alpha interferon is the current treatment standard; however, success in treatment depends largely on the viral genotype, and this drug combination has also been associated with severe side effects (30, 43). Thus, the development of novel, more potent, specific drugs is urgent.
HCV belongs to the Flaviviridae family. The HCV RNA genome consists of approximately 10 kb, encoding a polyprotein which is processed into several smaller polypeptides, including the capsid protein (C), the envelope proteins (E1 and E2), and the nonstructural proteins (NS2, NS3, NS4A, NS4B, NS5A, and NS5B). Initial cis cleavage through NS2-NS3 releases the NS3 protein, which in turn continues to process the precursor. Promising compounds with the ability to inhibit the viral protease (NS3) and the polymerase (NS5B) have been identified.
NS5B is a 65-kDa RNA-dependent RNA polymerase capable of initiating RNA synthesis de novo in the absence of a primer (16, 17, 25, 27, 45). Three classes of inhibitors of HCV NS5B have been developed, namely, nucleoside analogue inhibitors, nonnucleoside analogue inhibitors, and pyrophosphate (PPi) analogues. Nonnucleoside inhibitors and PPi analogues are still under preclinical evaluation, while a 2′-modified nucleoside analogue has advanced to clinical trials (for a recent review, see reference 7). Nonnucleoside inhibitors of NS5B bind reversibly to distinct allosteric sites of the enzyme (4, 9, 15, 41).
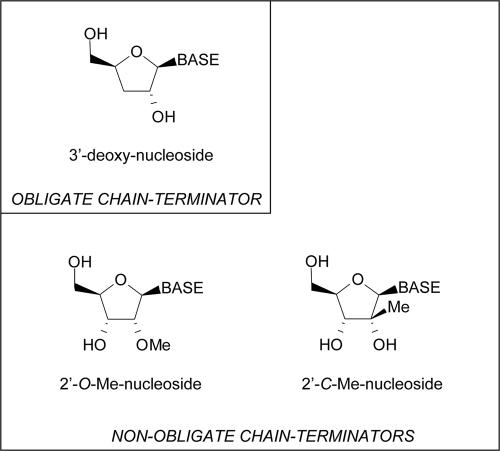
The prodrug of 2′-C-methyl-cytidine, i.e., NM283, has advanced to clinical trials (7). It appears that the methylation at the sugar moiety may be sufficient to cause chain termination, although the 3′-hydroxyl group is not removed from the sugar moiety (5). Compounds with these features are referred to as nonobligate chain terminators (Fig. 1). Once incorporated, they diminish binding and/or incorporation of the next nucleotide. Earlier work with structurally different nonobligate chain terminators has shown that these compounds may allow the incorporation of the next templated nucleotide and compromise subsequent elongation events. 9-(1,3-Dihydroxy-2-propoxymethyl)guanine (DHPG), which inhibits DNA polymerases of members of the Herpesviridae family, is a prominent example in this regard (29). Obligate chain terminators that inhibit HCV replication have not yet been developed, presumably because the presence of the 3′-hydroxyl group is a crucial structural determinant for the intracellular phosphorylation of ribonucleosides (34, 40).
FIG. 1.
Nucleoside analogue inhibitors of HCV NS5B. Chemical structures are given for an obligate chain terminator that lacks the 3′-hydroxyl group and three distinct nonobligate chain terminators that are modified at the 2′ or 4′ position of the sugar moiety.
Chain termination is not necessarily an irreversible process. For instance, the 3′-5′ exonuclease activity of viral DNA polymerases of the Herpesviridae family can remove incorporated nucleotide analogues. Moreover, amino acid changes in the exonuclease domain of these enzymes have been linked to drug resistance (38). HCV NS5B does not possess intrinsic exonuclease activity. However, the pyrophosphorolytic excision of incorporated chain terminators is a possible alternative mechanism that can diminish the efficiency of these compounds. We recently reported that the related bovine viral diarrhea virus (BVDV) NS5B enzyme is capable of using PPi to excise nucleotide analogues that lack the 3′-hydroxyl group (6). The excision of nucleotide analogue inhibitors of the reverse transcriptase (RT) of human immunodeficiency virus type 1 (HIV-1) was also shown to play an important role in the development of resistance to this class of compounds (3, 23).
Here we asked whether obligate and nonobligate chain terminators can be excised by HCV NS5B through pyrophosphorolysis. We compared 2′-C-methyl-cytidine (2′-C-Me-CTP) and 2′-O-methyl-cytidine (2′-O-Me-CTP) with the corresponding classical chain terminator 3′-dCTP and found that all inhibitors are excised in the presence of physiologically relevant concentrations of PPi.
MATERIALS AND METHODS
Chemicals and nucleic acids.
The following RNA template sequences were used in this study: 5′-AACCGUAUCCAAAACAGUCC-3′ (T20), 5′-AACAGUUUCCUUUUCUCUCC-3′ (T20-C16), and 5′-AACCUGAGAAGGAGAAAGCC-3′ (T20-A16). The underlined bases show the unique sites for incorporation of C and A analogues, respectively. The dinucleotide 5′-GG-3′ served as a primer (GG). All RNA oligonucleotides were chemically synthesized and purified in 12% polyacrylamide-7 M urea gels containing 50 mM Tris-borate, pH 8, and 1 mM EDTA, followed by elution from gel slices in a buffer containing 500 mM ammonium acetate and 0.1% sodium dodecyl sulfate. 5′-End labeling of the GG primer was conducted with [γ-32P]ATP and T4 polynucleotide kinase according to the manufacturer's recommendations (Invitrogen). Ribonucleoside triphosphates were purchased from Roche, and 3′-deoxynucleoside triphosphates (3′-dNTPs) and 2′-O-Me nucleotides were purchased from TriLink Biotechnologies. 2′-C-Me nucleotides were generously provided by Merck.
Expression and purification of HCV NS5B.
The HCV NS5B sequence, inserted into the expression vector pET-22 (Novagen), was expressed as a C-terminally truncated enzyme (Δ21) in Escherichia coli BL21(DE3) and purified utilizing a combination of metal ion affinity and ion-exchange chromatography (44). An S282T mutant enzyme was generated through site-directed mutagenesis, using a Stratagene QuikChange kit according to the manufacturer's protocol. Sequences were confirmed by sequencing at the McGill University Genome Quebec Innovation Centre.
RNA synthesis and chain termination.
Unless otherwise specified, standard reaction mixtures consisted of 1 μM RNA template (T20, T20-C16, or T20-A16), 1.5 μM HCV NS5B, and 0.25 μM radiolabeled GG primer in a buffer containing 40 mM HEPES, pH 8, 10 mM NaCl, 1 mM dithiothreitol, and 0.2 mM MnCl2. Reactions that involved multiple sites of chain termination (T20) were started by adding a 10 μM concentration of each NTP in the presence of a 3.7 to 100 μM concentration of each 3′-dNTP for 30 min at room temperature. Chain termination at a single template position was conducted with 10 μM GTP-ATP and 3 μM cytidine analogue (T20-C16) or with 10 μM CTP-UTP and 10 μM adenosine analogue (T20-A16). Reaction products were precipitated with isopropanol, heat denatured for 5 minutes at 95°C, and resolved in 12% polyacrylamide-7 M urea gels. The experiments were repeated, and representative images are shown in Fig. 2, 3, and 4. The concentration of chain terminator required to inhibit 50% of full-length product formation (IC50) was determined for a single site of nucleotide analogue incorporation with templates T20-C16 and T20-A16. Reactions were allowed to proceed for 30 min in the presence of a 3 μM concentration of each NTP, with the exception of the competing nucleotide (300 nM). Gels were scanned and analyzed with a phosphorimager (Molecular Imager FX; Bio-Rad). IC50 values were calculated with Prism software (GraphPad, Inc.), using the four-parameter logistic equation y = min(max − min)/{1 + 10̂[(log IC50 − x) × hill slope]}, where x is the logarithm of the concentration, “min” and “max” are the minimum and maximum response plateaus, and “hill slope” is the slope of the curve at its midpoint.
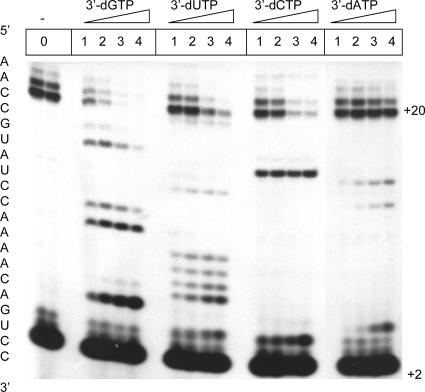
FIG. 2.
Inhibition of RNA synthesis with obligate chain terminators. The extension of a 5′-end-radiolabeled dinucleotide (GG) primer to a 20-mer full-length RNA product was monitored in the presence of a 10 μM concentration of each NTP (lane 0). Inhibition of RNA synthesis was studied in the presence of increasing concentrations of 3′-dGTP, 3′-dUTP, 3′-dCTP, or 3′-dATP. Lanes 1 to 4, 3.7, 11, 33, and 100 μM 3′-dNTP, respectively. Reactions were allowed to proceed for 30 min.
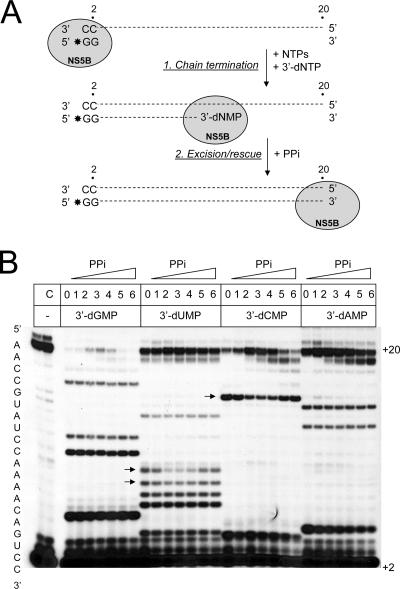
FIG. 3.
PPi-dependent rescue of chain-terminated RNA synthesis. (A) Principle of the reaction. The inclusion of obligate chain terminators in the GG-primed reaction yields various abortive products. The subsequent addition of PPi provides conditions that allow excision of incorporated 3′-dNMPs and, in turn, rescue of RNA synthesis. (B) RNA synthesis was allowed to proceed for 30 min in the presence of a 3 μM concentration of each NTP and 10 μM 3′-dGTP, 3′-dUTP, 3′-dCTP, or 30 μM 3′-dATP (lanes 0). Rescue of RNA synthesis was then monitored over another 30 min through the addition of increasing concentrations of PPi. Lanes 1 to 6, 31, 62, 125, 250, 500, and 1,000 μM PPi. Arrows point to rescued primers. Controls in lane C show reactions in the absence of obligate chain terminators.
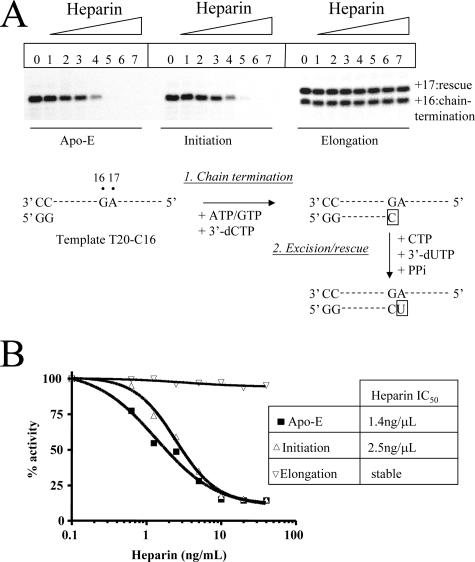
FIG. 4.
Stability of NS5B-RNA complexes. (A) RNA synthesis was monitored with the T20-C16 template, which allows the incorporation and excision of chain-terminating 3′-dCMP at a single position (+16), as outlined in the experimental design under the image. Heparin was used as a protein trap to analyze enzyme dissociation under different conditions, including preincubation of heparin and NS5B (Apo-E) prior to the addition of RNA and the start of the reaction, preincubation of heparin with NS5B bound to the template and the GG primer, and the addition of heparin after elongation and chain termination. In the last case, the activity of NS5B was measured in the presence of 125 μM PPi and a mixture of 10 μM CTP and 3′-dUTP that is required to rescue RNA synthesis for a single-nucleotide incorporation event. The loss of activity is a measure of enzyme dissociation, which was monitored in the presence of increasing concentrations of heparin. Lanes 0 to 7, 0, 0.037, 0.073, 0.146, 0.293, 0.586, 1.17, and 2.34 μM heparin. (B) The results were quantified and expressed as concentrations of heparin required to inhibit 50% of the enzymatic activity (IC50s).
Excision and DEC formation.
Excision and the ensuing rescue of RNA synthesis at multiple template positions (T20) were initiated by the addition of 125 μM PPi and a 100 μM concentration of each NTP. Reactions were allowed to proceed for 30 min at room temperature. For the combined excision and rescue reaction at a single template position, we used 125 μM PPi, 100 μM CTP, and 5 μM 3′-dUTP (T20-C16) or 125 μM PPi, 100 μM ATP, and 5 μM 3′-dGTP (T20-A16). Aliquots of the reaction mixes were taken at 1, 2, 5, 10, 20, 30, and 45 min. The formation of rescued products was fitted with the following burst equation as previously described (28): y = ymax × [1 − exp(−krescue × t)], where ymax is the amplitude of the burst, krescue is the velocity of the reaction, and t is the time, in minutes. Formation of a dead-end complex (DEC) was assayed under similar conditions, except that 3′-dUTP was replaced with increasing concentrations of UTP. We also included 6 mM MgCl2 to avoid nonspecific inhibition of the reaction at high NTP concentrations. The concentration of the next correct nucleotide required to inhibit 50% of primer rescue was also determined with the four-parameter logistic equation described above. The experiments were repeated, and representative images are shown in Fig. 5, 6, and 7.
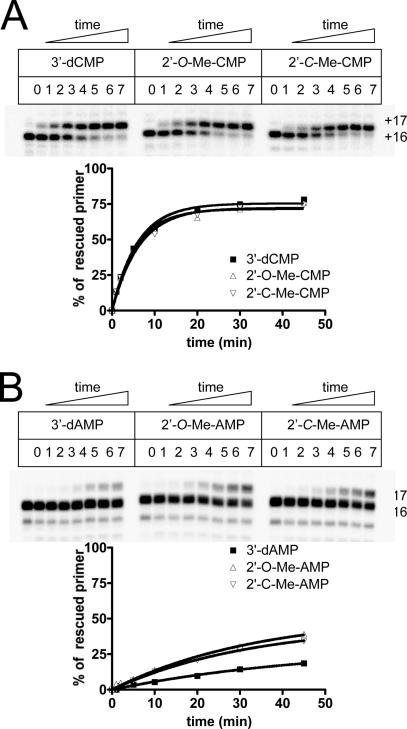
FIG. 5.
Kinetics of excision/rescue of purine and pyrimidine analogues. (A) The velocity of the combined excision/rescue reaction was analyzed with template T20-C16 and 3′-dCMP-, 2′-O-Me-CMP-, and 2′-C-Me-CMP-terminated primers (lanes 0). The reaction was monitored in a time course experiment for 1, 2, 5, 10, 20, 30, and 45 min (lanes 1 to 7, respectively). The percentage of rescued primer (position +17) was quantified relative to the amount of total RNA synthesis products. (B) The same experiment was performed with 3′-dAMP, 2′-O-Me-AMP, and 2′-C-Me-AMP, using the T20-A16 RNA template. Rates of the reactions are reported in Table 1.
FIG. 6.
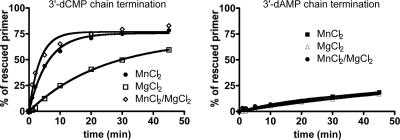
Effects of metal ions on the efficiency of rescue of RNA synthesis. RNA synthesis and chain termination with either 3′-dCMP or 3′-dAMP were performed under the same conditions as those in the legend to Fig. 5, in the presence of 0.2 mM Mn2+, 6 mM Mg2+, or a mixture of both divalent metal ions. The excision reaction was monitored in a time course experiment for 1, 2, 5, 10, 20, 30, and 45 min, and the percentage of rescued primer was quantified relative to the amount of total RNA synthesis products.
FIG. 7.
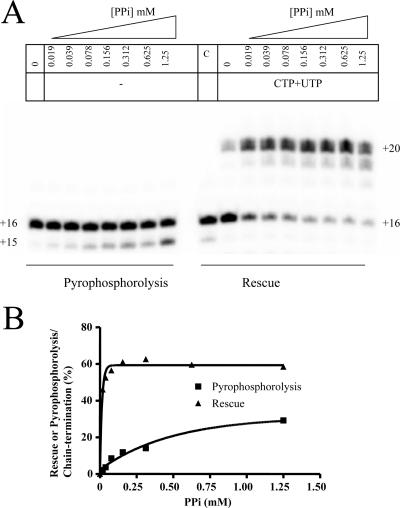
Comparison of efficiencies of pyrophosphorolysis and rescue of RNA synthesis. (A) RNA synthesis and chain termination were performed with a mixture of a 10 μM concentration (each) of GTP, ATP, and 2′-C-Me-CMP, using the template T20-C16. Excision of the nucleotide analogue was monitored in the presence of increasing concentrations of PPi (left). The rescue reaction was initiated by adding a mixture of 50 μM CTP, 10 μM UTP, and different concentrations of PPi (right). Lane C is a control reaction in the absence of nucleotides and in the presence of 0.125 mM PPi. (B) Quantification of the data shown in panel A. All reactions in this experiment were performed in the presence of 0.2 mM Mn2+ and 6 mM Mg2+.
RESULTS
Chain termination of RNA synthesis with 3′-dNTPs.
HCV NS5B is capable of initiating RNA synthesis either de novo or in the presence of short RNA primers. Throughout this study, we measured enzymatic activity from the extension of a radiolabeled dinucleotide (GG) primer complementary to the 3′ end of a short, heteropolymeric RNA model template. Chain termination and inhibition of full-length RNA synthesis were initially analyzed in the presence of the four 3′-dNTPs lacking the 3′-hydroxyl group (Fig. 2). Increasing concentrations of 3′-dNTPs gave rise to chain termination in accordance with the template sequence. In this setup, the efficiency of inhibition of full-length RNA synthesis followed the order 3′-dGTP > 3′-dCTP ≥ 3′-dUTP > 3′-dATP. The total number of incorporation events as well as the efficiency of incorporation of a given chain terminator plays a role in this regard.
Phosphorolytic excision of incorporated 3′-dNMPs.
We next studied the effects of adding increasing concentrations of PPi to reaction mixtures that contained the chain-terminated products described in the previous experiment (Fig. 3A). The disappearance of specific abortive reaction products, concomitant with the accumulation of full-length products, is indicative of excision and the ensuing rescue of RNA synthesis. Such rescue events were seen predominantly with pyrimidine-terminated reactions. Our data suggest that specific 3′-dUMP- and 3′-dCMP-terminated products are susceptible to PPi-mediated excision. This was most pronounced for the 3′-dCMP-terminated product at position +16, which was ultimately converted into full-length RNA at PPi concentrations between 62 and 250 μM (Fig. 3B, 3′-dCMP panel, lane 3). In contrast, the disappearance of 3′-dGMP- or 3′-dAMP-terminated products was not evident, and there were only small increases in the amounts of the full-length RNA products at the optimal PPi concentration of 125 μM (Fig. 3B, 3′-dGMP panel, lane 3). These findings point to inefficient rescue events at positions +11 and/or +6, which showed the largest amounts of chain-terminated products. Together, our data suggest that incorporated pyrimidines may be more susceptible to pyrophosphorolysis than are purines. Reactions with concentrations of PPi that exceeded 500 μΜ showed decreases in primer rescue, presumably by depleting concentrations of free divalent metal ions required for catalysis. Small RNA products were also resistant to PPi-mediated excision, which probably reflects the frequent dissociation of the NS5B-RNA complex during early stages of RNA synthesis (10, 15, 35).
Stability of the elongation complex.
To translate the data into quantitative terms and to study individual events that lead to the rescue of RNA synthesis, we modified the template sequence and introduced a unique site for the incorporation of cytidine analogues under elongation mode at position +16 (Fig. 4A). First, we analyzed the stability of the complex to assess whether pyrophosphorolysis can occur under single-turnover conditions. For this purpose, we added heparin as a trap under the following different conditions: (i) heparin was preincubated together with the apoenzyme prior to the start of the reaction; (ii) heparin was preincubated together with HCV NS5B bound to the template and the GG primer, again prior to the start of the chain termination reaction; or (iii) heparin was added after primer elongation and chain termination at position +16. The data show that the combined excision/rescue reaction is insensitive to the addition of heparin once the arrested elongation complex is formed (Fig. 4). In contrast, the initiation step was inhibited 50% with 0.15 μM heparin. This result demonstrates that HCV NS5B does not dissociate from a stalled elongation complex, which is likewise the case for the BVDV enzyme (6). Thus, the rates of excision at this stage of the reaction are not influenced by the dissociation of the protein nucleic acid complex.
Pyrimidines are more sensitive to excision than are purines.
We next analyzed whether the efficiency of excision depends on differences in the sugar moiety of the incorporated nucleotide analogue. Thus, we compared the rates of excision and rescue of 3′-dCMP-, 2′-O-Me-CMP-, and 2′-C-Me-CMP-terminated DNA synthesis and found that all three chain terminators are sensitive to pyrophosphorolysis (Fig. 5A). The rates of excision were literally identical, which suggests that the chemical modifications in the sugar moiety presented herein have little effect on the reaction.
We next looked at the influence of the base moiety on rates of excision. In order to measure excision and primer rescue under the same conditions with adenosine analogues (A analogues), we designed a template with a single uridine at position +16. We found that the levels of excision of the A analogues were reduced five- to eightfold compared to the rates obtained with C analogues (Fig. 5B; Table 1). This result corroborates the data shown in Fig. 3B and shows that the efficiency of rescue is primarily driven by the nature of the nucleobase. Very similar trends were observed when the chain-terminated primers were rescued in the presence of Mn2+, Mg2+, or a mixture of both divalent metal ions (Fig. 6), although 3′-dCMP primer rescue with Mg2+ alone was significantly slower than that when Mn2+ was present in the reaction.
TABLE 1.
Kinetics of primer rescue catalyzed by wild-type and S282T HCV NS5B
| Enzyme | C analogue | krescue(C) (min−1)a | A analogue | krescue(A) (min−1)a | krescue(C)/krescue(A) |
|---|---|---|---|---|---|
| Wild-type NS5B | 3′-Deoxy | 0.151 ± 0.027 | 3′-Deoxy | 0.019 ± 0.002 | 7.8 |
| 2′-O-Me | 0.168 ± 0.035 | 2′-O-Me | 0.033 ± 0.007 | 5.1 | |
| 2′-C-Me | 0.160 ± 0.001 | 2′-C-Me | 0.027 ± 0.006 | 5.8 | |
| S282T mutant | 3′-Deoxy | 0.141 ± 0.007 | 3′-Deoxy | 0.019 ± 0.004 | 7.5 |
| 2′-O-Me | 0.160 ± 0.040 | 2′-O-Me | 0.035 ± 0.008 | 4.6 | |
| 2′-C-Me | 0.169 ± 0.071 | 2′-C-Me | 0.031 ± 0.015 | 5.5 |
krescue was determined as described in Materials and Methods. Data are means ± standard deviations determined for three independent experiments.
We next compared the efficiency of pyrophosphorolysis with that of the combined excision/rescue reaction, using 2′-C-Me-CMP-terminated primers (Fig. 7A, left panel). Increasing concentrations of PPi led to the formation of a shorter product, which unambiguously shows that the incorporated nucleotide analogue can be excised through pyrophosphorolysis. However, the combined excision/rescue reaction was much more effective (Fig. 7A, right panel). Together, these findings suggest that the excised chain terminator can be reincorporated in the absence of the competing natural counterpart that is only present in the excision/rescue reaction. At the same time, these data help to explain why pyrophosphorolysis is sometimes difficult to observe when the particular sequence context diminishes the reaction per se (11).
Effects of S282T mutation on excision and nucleotide selectivity.
Drug susceptibility studies and selection experiments with the replicon system have shown that a single mutation, i.e., S282T, confers reduced susceptibility to 2′-C-Me-adenosine. The S282T mutation confers cross-resistance to other 2′-C-Me nucleotides (18, 24). Modeling studies and enzyme kinetic analysis suggested that this mutation can affect binding of the inhibitor to the active site of NS5B. Here we asked whether this mutation may likewise affect the excision of incorporated nucleotide analogues. However, a comparison of rates of rescue of different chain-terminated primers did not reveal any significant difference between WT NS5B and the mutated enzyme (Table 1). In agreement with previous reports, we found that the S282T mutation discriminates the 2′-C-Me-nucleotide analogue. In a single-nucleotide competition experiment, the mutant enzyme showed increases in IC50s of between 9.5-fold and >24-fold for 2′-C-Me-CTP and 2′-C-Me-ATP, respectively (Table 2). Increases in Km values suggest that the mutated enzyme displays decreased inhibitor binding (11).
TABLE 2.
Inhibitory activity of nucleotide analogues in a single-nucleotide competition assay
| Enzyme | Nucleotide | IC50 (μM)a | Selectivityb |
|---|---|---|---|
| Wild-type NS5B | 3′-dCTP | 0.46 ± 0.064 | |
| 2′-C-Me-CTP | 6.3 ± 1.6 | ||
| 3′-dATP | 4.4 ± 0.14 | ||
| 2′-C-Me-ATP | 4.2 ± 0.7 | ||
| S282T mutant | 3′-dCTP | 0.77 ± 0.036 | 1.7 |
| 2′-C-Me-CTP | 60 ± 20 | 9.5 | |
| 3′-dATP | 8.8 ± 2.2 | 2.0 | |
| 2′-C-Me-ATP | >100 | >24 |
Determined as described in Materials and Methods. Data are means ± standard deviations determined for three independent experiments.
Selectivity was calculated as the ratio of the IC50 of the S282T mutant to the IC50 of the wild type.
Formation of DEC.
Previous biochemical studies with DNA and RNA polymerases have shown that the presence of the next complementary nucleotide upstream of the 3′ end of the chain-terminated primer can compromise excision (6, 23, 39). For HIV-1 RT, nucleotide binding is associated with the formation of a DEC that provides a certain degree of protection from excision (22). The complex is inactive with respect to both forward and back reactions provided that the chain terminator lacks the 3′-hydroxyl group. Nonobligate chain terminators contain the 3′-hydroxyl group, which theoretically permits attack on the α-phosphate of an incoming nucleotide. However, modeling studies with 2′-C-Me-AMP at the 3′ end of the primer suggested that the methyl group may cause steric conflicts with the sugar moiety of an incoming nucleotide (24). This model helps to explain why the incorporation of the next nucleotide is severely compromised. A steric clash between the 2′-C-modified primer terminus and the incoming nucleotide can diminish the stability of a ternary complex. The possible consequences are twofold. First, the incoming nucleotide is not incorporated, which leads to inhibition of RNA synthesis. However, an unstable DEC may not provide protection from excision, which counteracts the inhibitory effects.
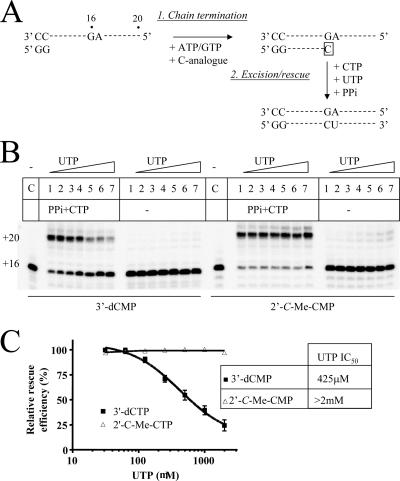
To test these hypotheses, we looked at the efficiency of the combined excision/rescue reaction in the presence of increasing concentrations of the next complementary nucleotide (Fig. 8A). For the obligate chain terminator 3′-dCMP, we found that the reaction was sensitive to the presence of the next nucleotide substrate (Fig. 8B). The rescue of RNA synthesis was literally blocked in the high micromolar range (Fig. 8C). Thus, the incorporated obligate chain terminator allows binding of the next nucleotide and, in turn, formation of a DEC that blocks excision. In contrast, the excision/rescue reaction with the nonobligate chain terminator 2′-C-Me-CMP was not sensitive to the presence of the templated nucleotide. The control reaction in the absence of PPi showed only minimal increases in product formation, which is indicative of efficient chain termination. Rescue of RNA synthesis was evident following the addition of PPi at the full range of concentrations of the next complementary nucleotide. The presence of 2 mM UTP did not reduce the efficiency of the reaction, which suggests that the formation of a ternary DEC is severely compromised in this case. Thus, 2′-C-methylated, nonobligate chain terminators diminish binding of the next complementary nucleotide, which provides a mechanism of action. However, the diminished binding of the next nucleotide prevents the formation of a stable DEC, which renders the incorporated inhibitor vulnerable to excision at the same time.
FIG. 8.
DEC formation with cytidine analogues. (A) The combined excision/rescue reaction was monitored in the presence of increasing concentrations of the templated nucleotide UTP. (B) Following chain termination with 3′-dCMP or 2′-C-Me-CMP (lanes C), excision/rescue was initiated with 125 μM PPi, 10 μM CTP, and increasing concentrations of UTP. Lanes 1 to 7, 31, 62.5, 125, 250, 500, 1,000, and 2,000 μM UTP, respectively. (C) Results were expressed as percentages of rescue relative to that under conditions with the lowest UTP concentration (lanes 1 in panel B). IC50 values were determined from at least three independent experiments, with error bars displaying the standard deviation for each concentration point.
DISCUSSION
We analyzed the pyrophosphorolytic excision of chain-terminating nucleotides by HCV NS5B, using a broad panel of adenosine and cytidine analogues, including the clinically relevant nonobligate chain terminator 2′-C-Me-CTP. We found that incorporated pyrimidines can be excised in the presence of physiologically relevant concentrations of PPi. The efficiency of excision depends critically on the nucleobase of the chain terminator. Our experiments show that adenosine analogues are by far not as sensitive to excision as cytidine analogues. This trend was observed with the obligate chain terminators 3′-dNTPs as well as with the nonobligate chain terminators 2′-O-Me-NTPs and 2′-C-Me-NTPs. We also tested the effect of the S282T mutant that confers decreased susceptibility to 2′-C-methylated nucleotides. Rates of primer rescue were not increased with the mutant enzyme, which indicates that other mechanisms are likely to be involved in resistance to this class of compounds. In agreement with previous studies, we found that the selectivity with respect to nucleotide incorporation was markedly increased (11, 24). Thus, resistance to 2′-C-Me-NTPs is based on substrate discrimination, not on differences with respect to rates of excision.
Based on previous studies on the removal of obligate chain terminators by BVDV NS5B and HIV-1 RT (6, 13, 31, 32), we propose a model that helps to explain the requirements for excision of nonobligate chain terminators by HCV NS5B (Fig. 9). The incorporation of obligate or nonobligate chain terminators must take place at the nucleotide binding site (N site) of the enzyme (Fig. 9A). Following nucleotide binding and catalysis, PPi is released from the complex, which is a prerequisite for subsequent polymerization events. Alternatively, PPi can rebind and promote excision of the incorporated nucleotide (Fig. 9B). HIV-1 RT was shown to be able to recruit ATP as a PPi donor (for recent reviews, see references 8, 12, and 14). However, unlike the retroviral DNA polymerase, viral RNA-dependent RNA polymerases utilize natural NTP substrates for the forward reaction, not as pyrophosphate donor molecules (6).
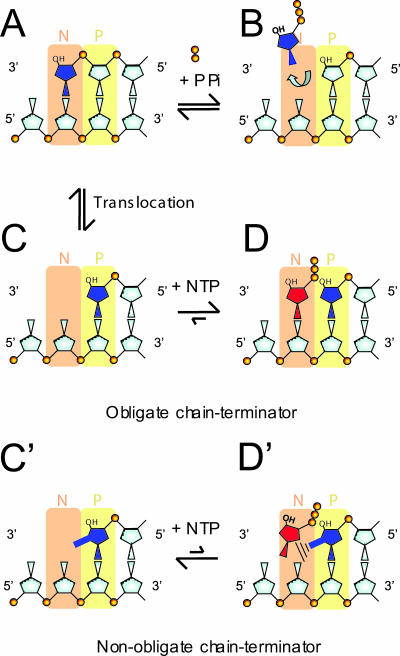
FIG. 9.
Parameters affecting the excision of nonobligate chain terminators. The nucleotide binding site of HCV NS5B is shown in orange (N site [N]), and the priming site is shown in yellow (P site [P]). Natural nucleotides are highlighted in gray, and the nonobligate chain terminator is shown in blue. The methyl group at the 2′ position of the sugar moiety is shown enlarged. (A) Immediately following nucleotide incorporation, the chain terminator still occupies the N site. (B) Excision can occur only in this configuration. (C) Binding of the next nucleotide requires polymerase translocation, which clears the nucleotide binding site. (C′) The methyl group at the 2′ position of the sugar moiety is shown enlarged. The chain terminator is now located in the P site. (D) In this position, binding of the next correct nucleotide blocks the enzyme in a ternary DEC that is catalytically inactive. (D′) Nucleotide binding and DEC formation are severely compromised with 2′-methylated chain terminators.
It was recently demonstrated that the intracellular concentrations of PPi, ATP, and GTP that are present in both quiescent and activated lymphocytes are sufficiently high to catalyze the excision of chain-terminating nucleotides by HIV-1 RT (37). Activated T cells contain 55 to 79 μM PPi, which is in the range of concentrations used in our experiments. Thus, it would be interesting to conduct similar experiments with cytoplasmic extracts from human hepatocytes to further assess the biological significance of the excision reaction. Such experiments may be conducted with purified HCV NS5B or with isolated, membrane-associated ribonucleoprotein complexes, which may better mimic physiologically relevant conditions (1). For HIV, a combination of clinical studies, in vitro selection experiments, and biochemical data provided strong evidence to suggest that the excision of nucleoside analogue RT inhibitors can occur in vivo. The ATP-dependent excision reaction represents an important mechanism for resistance (8, 12, 14). For HCV, it remains to be seen whether novel nucleoside analogue inhibitors can select for mutations that increase the rate of excision.
The efficiency of the primer rescue reaction is approximately 25-fold lower than that of the forward reaction when we compare our measurements with kinetic parameters for nucleotide incorporation events (11). This difference points to a slow, rate-limiting pyrophosphorolysis step. Thus, as for DNA synthesis in HIV replication, pyrophosphorolysis may play only a minor role in editing HCV RNA synthesis; however, more-detailed fidelity measurements with mispaired primers are required to address this issue (19).
Although the detailed requirements for excision can differ among the various polymerases, binding of the next nucleotide requires that the enzyme must translocate relative to its primer/template substrate to clear the N site. This movement brings the 3′ end of the primer to the priming site (P site) (Fig. 9C), which places the chain terminator away from the active site. As a consequence, excision cannot occur in the context of this posttranslocated complex. For HIV-1 RT, there is strong evidence to suggest that the enzyme can rapidly oscillate between pre- and posttranslocated conformations (13, 20, 21). The next nucleotide binds to and selectively traps the posttranslocational state, which provides protection from excision. Our experiments with HCV NS5B show that the next correct templated nucleotide can block excision of the obligate chain terminator 3′-dCMP. Thus, like HIV-1 RT and BVDV NS5B, the HCV enzyme appears to be capable of forming a stable ternary DEC with the incoming nucleotide (Fig. 9D).
In contrast to the removal of the obligate chain terminator 3′-dCMP, excision of 2′-C-Me-CMP is not compromised in the presence of high millimolar concentrations of the next complementary nucleotide. This indicates that the templated nucleotide does not trap the posttranslocational configuration (Fig. 9D′). Thus, the primer terminus can freely shuttle between the N and P sites, and access to the N site in the pretranslocational complex allows excision to occur. The putative steric clash between the methyl group of the incorporated 2′-C-Me-CMP and the sugar ring of the incoming nucleotide may therefore be sufficient to prevent nucleotide binding (5). This model helps to explain the favorable properties of 2′-methylated compounds as de facto chain terminators; however, at the same time, little or no nucleotide binding renders the incorporated inhibitor vulnerable to excision. This model does not explain why purines are less efficiently excised than pyrimidines. A similar observation has been made with HIV RT, which suggests a related common mechanism (36). At this point, it is tempting to propose that pyrimidines have better access to the pretranslocated complex, although this remains to be shown.
Taken together, our biochemical data provide strong support for the notion that nonobligate, 2′-methylated chain terminators impede binding of the next nucleotide. This provides a mechanism of action; however, the diminished ability to bind the next nucleotide and to form a stable ternary DEC limits protection from excision. The excision of incorporated nonobligate chain terminators may diminish their inhibitory effects. In vivo, other factors, such as metabolic activation of nucleoside analogue prodrugs and rates of incorporation of the monophosphate, are important parameters that determine the potency of these compounds (40). Based on our results, we propose that the phosphorolytic attack at the ultimate phosphodiester bond of the primer is perhaps an additional parameter that could account for the increased antiviral activity of 2′-C-Me-adenosine over that of 2′-C-Me-cytidine in cell-based replicon systems (26, 40). Overall, these findings warrant further investigation towards the development of novel purine analogues, despite the fact that these compounds compete with relatively high intracellular concentrations of ATP and GTP.
Acknowledgments
We thank Steven S. Carroll, Daria J. Hazuda, and David B. Olsen (Merck, West Point, PA) for kindly providing the 2′-C-methyl-modified NTPs used in this study. We are grateful to Craig E. Cameron (Pennsylvania State University, PA), who provided the plasmid for the expression of HCV NS5B, and to Suzanne McCormick for excellent technical assistance.
J.D. is the recipient of a postdoctoral research award from the Canadian Institutes of Health Research (CIHR). M.H.P. is the recipient of a student stipend from the National Canadian Training Program in Hepatitis C (NCRTP). C.M.D. is the recipient of a doctoral research award from the Fonds de la Recherche en Santé du Québec (FRSQ). This research was funded by a grant from the Cancer Research Society, Inc. (to M.G.). M.G. is the recipient of a national career award from the Canadian Institutes of Health Research (CIHR).
Footnotes
Published ahead of print on 14 May 2007.
REFERENCES
- 1.Ali, N., K. D. Tardif, and A. Siddiqui. 2002. Cell-free replication of the hepatitis C virus subgenomic replicon. J. Virol. 76:12001-12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alter, H. J., and L. B. Seeff. 2000. Recovery, persistence, and sequelae in hepatitis C virus infection: a perspective on long-term outcome. Semin. Liver Dis. 20:17-35. [DOI] [PubMed] [Google Scholar]
- 3.Arion, D., N. Kaushik, S. McCormick, G. Borkow, and M. A. Parniak. 1998. Phenotypic mechanism of HIV-1 resistance to 3′-azido-3′-deoxythymidine (AZT): increased polymerization processivity and enhanced sensitivity to pyrophosphate of the mutant viral reverse transcriptase. Biochemistry 37:15908-15917. [DOI] [PubMed] [Google Scholar]
- 4.Biswal, B. K., M. M. Cherney, M. Wang, L. Chan, C. G. Yannopoulos, D. Bilimoria, O. Nicolas, J. Bedard, and M. N. James. 2005. Crystal structures of the RNA-dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors. J. Biol. Chem. 280:18202-18210. [DOI] [PubMed] [Google Scholar]
- 5.Carroll, S. S., J. E. Tomassini, M. Bosserman, K. Getty, M. W. Stahlhut, A. B. Eldrup, B. Bhat, D. Hall, A. L. Simcoe, R. LaFemina, C. A. Rutkowski, B. Wolanski, Z. Yang, G. Migliaccio, R. De Francesco, L. C. Kuo, M. MacCoss, and D. B. Olsen. 2003. Inhibition of hepatitis C virus RNA replication by 2′-modified nucleoside analogs. J. Biol. Chem. 278:11979-11984. [DOI] [PubMed] [Google Scholar]
- 6.D'Abramo, C. M., L. Cellai, and M. Gotte. 2004. Excision of incorporated nucleotide analogue chain-terminators can diminish their inhibitory effects on viral RNA-dependent RNA polymerases. J. Mol. Biol. 337:1-14. [DOI] [PubMed] [Google Scholar]
- 7.De Francesco, R., and G. Migliaccio. 2005. Challenges and successes in developing new therapies for hepatitis C. Nature 436:953-960. [DOI] [PubMed] [Google Scholar]
- 8.Deval, J., J. Courcambeck, B. Selmi, J. Boretto, and B. Canard. 2004. Structural determinants and molecular mechanisms for the resistance of HIV-1 RT to nucleoside analogues. Curr. Drug Metab. 5:305-316. [DOI] [PubMed] [Google Scholar]
- 9.Di Marco, S., C. Volpari, L. Tomei, S. Altamura, S. Harper, F. Narjes, U. Koch, M. Rowley, R. De Francesco, G. Migliaccio, and A. Carfi. 2005. Interdomain communication in hepatitis C virus polymerase abolished by small molecule inhibitors bound to a novel allosteric site. J. Biol. Chem. 280:29765-29770. [DOI] [PubMed] [Google Scholar]
- 10.Dutartre, H., J. Boretto, J. C. Guillemot, and B. Canard. 2005. A relaxed discrimination of 2′-O-methyl-GTP relative to GTP between de novo and elongative RNA synthesis by the hepatitis C RNA-dependent RNA polymerase NS5B. J. Biol. Chem. 280:6359-6368. [DOI] [PubMed] [Google Scholar]
- 11.Dutartre, H., C. Bussetta, J. Boretto, and B. Canard. 2006. General catalytic deficiency of hepatitis C virus RNA polymerase with an S282T mutation and mutually exclusive resistance towards 2′-modified nucleotide analogues. Antimicrob. Agents Chemother. 50:4161-4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldschmidt, V., and R. Marquet. 2004. Primer unblocking by HIV-1 reverse transcriptase and resistance to nucleoside RT inhibitors (NRTIs). Int. J. Biochem. Cell Biol. 36:1687-1705. [DOI] [PubMed] [Google Scholar]
- 13.Gotte, M. 2006. Effects of nucleotides and nucleotide analogue inhibitors of HIV-1 reverse transcriptase in a ratchet model of polymerase translocation. Curr. Pharm. Des. 12:1867-1877. [DOI] [PubMed] [Google Scholar]
- 14.Gotte, M. 2004. Inhibition of HIV-1 reverse transcription: basic principles of drug action and resistance. Expert Rev. Anti Infect. Ther. 2:707-716. [DOI] [PubMed] [Google Scholar]
- 15.Gu, B., V. K. Johnston, L. L. Gutshall, T. T. Nguyen, R. R. Gontarek, M. G. Darcy, R. Tedesco, D. Dhanak, K. J. Duffy, C. C. Kao, and R. T. Sarisky. 2003. Arresting initiation of hepatitis C virus RNA synthesis using heterocyclic derivatives. J. Biol. Chem. 278:16602-16607. [DOI] [PubMed] [Google Scholar]
- 16.Hong, Z., C. E. Cameron, M. P. Walker, C. Castro, N. Yao, J. Y. Lau, and W. Zhong. 2001. A novel mechanism to ensure terminal initiation by hepatitis C virus NS5B polymerase. Virology 285:6-11. [DOI] [PubMed] [Google Scholar]
- 17.Kao, C. C., X. Yang, A. Kline, Q. M. Wang, D. Barket, and B. A. Heinz. 2000. Template requirements for RNA synthesis by a recombinant hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 74:11121-11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klumpp, K., V. Leveque, S. Le Pogam, H. Ma, W. R. Jiang, H. Kang, C. Granycome, M. Singer, C. Laxton, J. Q. Hang, K. Sarma, D. B. Smith, D. Heindl, C. J. Hobbs, J. H. Merrett, J. Symons, N. Cammack, J. A. Martin, R. Devos, and I. Najera. 2006. The novel nucleoside analog R1479 (4′-azidocytidine) is a potent inhibitor of NS5B-dependent RNA synthesis and hepatitis C virus replication in cell culture. J. Biol. Chem. 281:3793-3799. [DOI] [PubMed] [Google Scholar]
- 19.Marchand, B., and M. Gotte. 2004. Impact of the translocational equilibrium of HIV-1 reverse transcriptase on the efficiency of mismatch extensions and the excision of mispaired nucleotides. Int. J. Biochem. Cell Biol. 36:1823-1835. [DOI] [PubMed] [Google Scholar]
- 20.Marchand, B., and M. Gotte. 2003. Site-specific footprinting reveals differences in the translocation status of HIV-1 reverse transcriptase. Implications for polymerase translocation and drug resistance. J. Biol. Chem. 278:35362-35372. [DOI] [PubMed] [Google Scholar]
- 21.Marchand, B., E. P. Tchesnokov, and M. Gotte. 2007. The pyrophosphate analogue foscarnet traps the pre-translocational state of HIV-1 reverse transcriptase in a Brownian ratchet model of polymerase translocation. J. Biol. Chem. 282:3337-3346. [DOI] [PubMed] [Google Scholar]
- 22.Meyer, P. R., S. E. Matsuura, R. F. Schinazi, A. G. So, and W. A. Scott. 2000. Differential removal of thymidine nucleotide analogues from blocked DNA chains by human immunodeficiency virus reverse transcriptase in the presence of physiological concentrations of 2′-deoxynucleoside triphosphates. Antimicrob. Agents Chemother. 44:3465-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyer, P. R., S. E. Matsuura, A. G. So, and W. A. Scott. 1998. Unblocking of chain-terminated primer by HIV-1 reverse transcriptase through a nucleotide-dependent mechanism. Proc. Natl. Acad. Sci. USA 95:13471-13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Migliaccio, G., J. E. Tomassini, S. S. Carroll, L. Tomei, S. Altamura, B. Bhat, L. Bartholomew, M. R. Bosserman, A. Ceccacci, L. F. Colwell, R. Cortese, R. De Francesco, A. B. Eldrup, K. L. Getty, X. S. Hou, R. L. LaFemina, S. W. Ludmerer, M. MacCoss, D. R. McMasters, M. W. Stahlhut, D. B. Olsen, D. J. Hazuda, and O. A. Flores. 2003. Characterization of resistance to non-obligate chain-terminating ribonucleoside analogs that inhibit hepatitis C virus replication in vitro. J. Biol. Chem. 278:49164-49170. [DOI] [PubMed] [Google Scholar]
- 25.Oh, J. W., T. Ito, and M. M. Lai. 1999. A recombinant hepatitis C virus RNA-dependent RNA polymerase capable of copying the full-length viral RNA. J. Virol. 73:7694-7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olsen, D. B., A. B. Eldrup, L. Bartholomew, B. Bhat, M. R. Bosserman, A. Ceccacci, L. F. Colwell, J. F. Fay, O. A. Flores, K. L. Getty, J. A. Grobler, R. L. LaFemina, E. J. Markel, G. Migliaccio, M. Prhavc, M. W. Stahlhut, J. E. Tomassini, M. MacCoss, D. J. Hazuda, and S. S. Carroll. 2004. A 7-deaza-adenosine analog is a potent and selective inhibitor of hepatitis C virus replication with excellent pharmacokinetic properties. Antimicrob. Agents Chemother. 48:3944-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ranjith-Kumar, C. T., L. Gutshall, R. T. Sarisky, and C. C. Kao. 2003. Multiple interactions within the hepatitis C virus RNA polymerase repress primer-dependent RNA synthesis. J. Mol. Biol. 330:675-685. [DOI] [PubMed] [Google Scholar]
- 28.Ray, A. S., E. Murakami, A. Basavapathruni, J. A. Vaccaro, D. Ulrich, C. K. Chu, R. F. Schinazi, and K. S. Anderson. 2003. Probing the molecular mechanisms of AZT drug resistance mediated by HIV-1 reverse transcriptase using a transient kinetic analysis. Biochemistry 42:8831-8841. [DOI] [PubMed] [Google Scholar]
- 29.Reardon, J. E. 1989. Herpes simplex virus type 1 and human DNA polymerase interactions with 2′-deoxyguanosine 5′-triphosphate analogues. Kinetics of incorporation into DNA and induction of inhibition. J. Biol. Chem. 264:19039-19044. [PubMed] [Google Scholar]
- 30.Russo, M. W., and M. W. Fried. 2003. Side effects of therapy for chronic hepatitis C. Gastroenterology 124:1711-1719. [DOI] [PubMed] [Google Scholar]
- 31.Sarafianos, S. G., A. D. Clark, Jr., K. Das, S. Tuske, J. J. Birktoft, P. Ilankumaran, A. R. Ramesha, J. M. Sayer, D. M. Jerina, P. L. Boyer, S. H. Hughes, and E. Arnold. 2002. Structures of HIV-1 reverse transcriptase with pre- and post-translocation AZTMP-terminated DNA. EMBO J. 21:6614-6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarafianos, S. G., A. D. Clark, Jr., S. Tuske, C. J. Squire, K. Das, D. Sheng, P. Ilankumaran, A. R. Ramesha, H. Kroth, J. M. Sayer, D. M. Jerina, P. L. Boyer, S. H. Hughes, and E. Arnold. 2003. Trapping HIV-1 reverse transcriptase before and after translocation on DNA. J. Biol. Chem. 278:16280-16288. [DOI] [PubMed] [Google Scholar]
- 33.Shepard, C. W., L. Finelli, and M. J. Alter. 2005. Global epidemiology of hepatitis C virus infection. Lancet Infect. Dis. 5:558-567. [DOI] [PubMed] [Google Scholar]
- 34.Shim, J., G. Larson, V. Lai, S. Naim, and J. Z. Wu. 2003. Canonical 3′-deoxyribonucleotides as a chain terminator for HCV NS5B RNA-dependent RNA polymerase. Antivir. Res. 58:243-251. [DOI] [PubMed] [Google Scholar]
- 35.Shim, J. H., G. Larson, J. Z. Wu, and Z. Hong. 2002. Selection of 3′-template bases and initiating nucleotides by hepatitis C virus NS5B RNA-dependent RNA polymerase. J. Virol. 76:7030-7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sluis-Cremer, N., D. Arion, U. Parikh, D. Koontz, R. F. Schinazi, J. W. Mellors, and M. A. Parniak. 2005. The 3′-azido group is not the primary determinant of 3′-azido-3′-deoxythymidine (AZT) responsible for the excision phenotype of AZT-resistant HIV-1. J. Biol. Chem. 280:29047-29052. [DOI] [PubMed] [Google Scholar]
- 37.Smith, A. J., P. R. Meyer, D. Asthana, M. R. Ashman, and W. A. Scott. 2005. Intracellular substrates for the primer-unblocking reaction by human immunodeficiency virus type 1 reverse transcriptase: detection and quantitation in extracts from quiescent- and activated-lymphocyte subpopulations. Antimicrob. Agents Chemother. 49:1761-1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith, I. L., J. M. Cherrington, R. E. Jiles, M. D. Fuller, W. R. Freeman, and S. A. Spector. 1997. High-level resistance of cytomegalovirus to ganciclovir is associated with alterations in both the UL97 and DNA polymerase genes. J. Infect. Dis. 176:69-77. [DOI] [PubMed] [Google Scholar]
- 39.Tabor, S., and C. C. Richardson. 1990. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Effect of pyrophosphorolysis and metal ions. J. Biol. Chem. 265:8322-8328. [PubMed] [Google Scholar]
- 40.Tomassini, J. E., K. Getty, M. W. Stahlhut, S. Shim, B. Bhat, A. B. Eldrup, T. P. Prakash, S. S. Carroll, O. Flores, M. MacCoss, D. R. McMasters, G. Migliaccio, and D. B. Olsen. 2005. Inhibitory effect of 2′-substituted nucleosides on hepatitis C virus replication correlates with metabolic properties in replicon cells. Antimicrob. Agents Chemother. 49:2050-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomei, L., S. Altamura, L. Bartholomew, M. Bisbocci, C. Bailey, M. Bosserman, A. Cellucci, E. Forte, I. Incitti, L. Orsatti, U. Koch, R. De Francesco, D. B. Olsen, S. S. Carroll, and G. Migliaccio. 2004. Characterization of the inhibition of hepatitis C virus RNA replication by nonnucleosides. J. Virol. 78:938-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wasley, A., and M. J. Alter. 2000. Epidemiology of hepatitis C: geographic differences and temporal trends. Semin. Liver Dis. 20:1-16. [DOI] [PubMed] [Google Scholar]
- 43.Zeuzem, S., S. V. Feinman, J. Rasenack, E. J. Heathcote, M. Y. Lai, E. Gane, J. O'Grady, J. Reichen, M. Diago, A. Lin, J. Hoffman, and M. J. Brunda. 2000. Peginterferon alfa-2a in patients with chronic hepatitis C. N. Engl. J. Med. 343:1666-1672. [DOI] [PubMed] [Google Scholar]
- 44.Zhong, W., E. Ferrari, C. A. Lesburg, D. Maag, S. K. Ghosh, C. E. Cameron, J. Y. Lau, and Z. Hong. 2000. Template/primer requirements and single nucleotide incorporation by hepatitis C virus nonstructural protein 5B polymerase. J. Virol. 74:9134-9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhong, W., A. S. Uss, E. Ferrari, J. Y. Lau, and Z. Hong. 2000. De novo initiation of RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase. J. Virol. 74:2017-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]