Abstract
Current treatment for serious infections caused by methicillin-resistant Staphylococcus aureus relies heavily upon the glycopeptide antibiotic vancomycin. Unfortunately, this practice has led to an intermediate resistance phenotype that is particularly difficult to treat in invasive staphylococcal diseases, such as septicemia and its metastatic complications, including endocarditis. Although the vancomycin-intermediate resistance phenotype has been linked to abnormal cell wall structures and autolytic rates, the corresponding genetic changes have not been fully elucidated. Previously, whole-genome array studies listed numerous genes that are overexpressed in vancomycin-intermediate sensitive strains, including graRS (SACOL0716 to -0717), encoding a two-component regulatory system (TCRS), as well as the adjacent vraFG (SACOL0718 to -0720), encoding an ATP-binding cassette (ABC) transporter; but the exact contribution of these genes to increased vancomycin resistance has not been defined. In this study, we showed that isogenic strains with mutations in genes encoding the GraRS TCRS and the VraFG ABC transporter are hypersensitive to vancomycin as well as polymyxin B. Moreover, GraRS regulates the expression of the adjacent VraFG pump, reminiscent of gram-positive bacteriocin-immunity regulons. Mutations of graRS and vraFG also led to increased autolytic rates and a more negative net surface charge, which may explain, in part, to their increased sensitivity to cationic antimicrobial peptides. Taken together, these data reveal an important genetic mediator to the vancomycin-intermediate S. aureus phenotype and may hold clues to the selective pressures on staphylococci upon exposure to selective cationic peptide antibiotics used in clinical practice.
High-level vancomycin (VAN) resistance, encoded by the vanA operon transmitted from Enterococcus species, is quite rare in Staphylococcus aureus (49). In contrast, intermediate levels of VAN resistance are rising among methicillin-resistant S. aureus (MRSA) strains; these are designated VAN-intermediate S. aureus (VISA) strains (7, 49). VISA strains arise by increasing basal resistance to VAN without acquisition of any clearly defined resistance determinants (7). Although many genetic alterations enabling intermediate VAN resistance have been identified (9, 24, 32, 37, 42), the functional implications of many of these genetic changes are not yet known, nor have their association with VISA been genetically proven. Staphylococcus aureus has 17 chromosomally encoded two-component regulatory systems (TCRS), excluding systems in SSC-mec elements (25), and overexpression of one TCRS (SACOL0716 to -0717, also called graRS) has been linked with the VISA phenotype, yet its function remains undefined (9).
The VISA phenotype is of increasing concern because MRSA strains are now found outside the hospital setting in the community (community-acquired MRSA), potentially increasing the likelihood of community-acquired VISA. VAN binds the d-alanyl-d-alanine moiety on the peptidoglycan murein monomer and prevents cross-linking of VAN-bound murein chains with other peptidoglycan chains, thereby leading to inhibition of transpeptidation (40). Many studies investigating VISA strains have described common phenotypic traits associated with increased VAN resistance. These include a thickening of the cell wall peptidoglycan (10), increasing synthesis of peptidoglycan (18), decreasing levels of autolytic enzymes involved in cell wall turnover (3), reduced levels of cross-linking (4, 19, 46), diminishing levels of the low-molecular-weight penicillin-binding protein PBP4 (13, 45), and decreasing muropeptide amidation (4, 19). One proposed mechanism linking these observable phenotypes to VAN resistance is a clogging mechanism whereby the thickened cell wall of VISA strains may restrict diffusion of VAN, thereby preventing drug binding to the target site of transpeptidation (8). Despite the aforementioned findings, specific genetic mechanisms underlying these phenotypic alterations have not been well established.
TCRS are widely utilized by bacteria to monitor an external stimulus, using a sensor protein and a phosphorelay cascade through a response regulator protein to alter specific target gene transcription (53). The genetic organization of a TCRS and an adjacent ATP-binding cassette (ABC) transporter system, repeatedly found in the low-G+C-content gram-positive bacterial species (Firmicutes), has in some cases been associated with self-immunity against secreted bacteriocins and antimicrobial peptide resistance (5, 21, 30). Given that secreted bacteriocins can damage the host bacterium, it has been proposed that ABC exporters likely provide a self-immunity function by transporting bacteriocin compounds out of the cell. It is important to note that the genetic linkage of a TCRS and an ABC transporter was found only in the Bacillus/Clostridium group (Firmicutes), and not in other gram-positive bacteria, gram-negative bacteria, or archaea, by the Denizot group using bioinformatics identifying only the OmpR-type response regulator, and not other types such as CheA, to adjacent ABC transporters (21). Interestingly, Staphylococcus aureus is classified in the Firmicutes phylum and is not known to secrete a chromosomally encoded bacteriocin (33), yet there are four chromosomal loci where TCRS lie adjacent to ABC transporters (25).
We hypothesized that the four genetic loci linking TCRS and ABC transporters in S. aureus may represent evolutionary events that confer resistance against exogenous antimicrobial peptides, including bacteriocins (e.g., VAN, a glycopeptide produced by Streptomyces orientalis). In this study, we began to address this hypothesis by demonstrating that S. aureus has basal resistance mechanisms, encoded partially by a bacteriocin-like immunity system comprising of the graRS TCRS (9) and the vraFG ABC transporter genes (24), that function to increase resistance against VAN and polymyxin B (PMB). Accordingly, our data demonstrate an active process by which S. aureus protects itself against VAN and PMB and further suggest that the other three TCRS/ABC-transporter loci may also function in resistance mechanisms.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Molecular cloning experiments utilized Escherichia coli TOP10 (Invitrogen). The wild-type and mutant S. aureus strains used in this study are listed in Table 1. Luria-Bertani medium (Becton Dickinson) was used for culture of E. coli, while S. aureus was cultured in tryptic soy broth (TSB) (Becton Dickinson). For certain assays (see below), S. aureus was grown in brain heart infusion broth (Becton Dickinson) or Mueller-Hinton broth (Becton Dickinson) supplemented with 25 μg/ml Ca2+ and 12.5 μg/ml Mg2+ (designated CSMHB). When appropriate, antibiotics were added to the media at the following concentrations: ampicillin at 100 μg/ml for E. coli and chloramphenicol at 10 μg/ml, erythromycin at 2.5 μg/ml, and tetracycline at 5 μg/ml for S. aureus. Chloramphenicol was routinely used to maintain selection for pEPSA5-based plasmids (14).
TABLE 1.
Strains and plasmids
| Strain or plasmid | Comment | Reference or source |
|---|---|---|
| S. aureus strains | ||
| RN4220 | MSSA, derivative of 8325-4, acceptor of foreign DNA | 23 |
| RN6390 | MSSA, derivative of 8325-4, 11-bp deletion in rsbU | 39 |
| RN6390 graR | ΔgraR in-frame deletion mutant of parental strain RN6390 | This study |
| RN6390 graS | ΔgraS in-frame deletion mutant of parental strain RN6390 | This study |
| RN6390 vraG | ΔvraG in-frame deletion mutant of parental strain RN6390 | This study |
| COL | MRSA, wild-type strain | 16 |
| COL graR | ΔgraR in-frame deletion mutant of parental strain COL | This study |
| COL graS | ΔgraS in-frame deletion mutant of parental strain COL | This study |
| COL vraG | ΔvraG in-frame deletion mutant of parental strain COL | This study |
| Mu50 | MRSA, VISA, wild-type strain | 25 |
| Mu50 graR | ΔgraR in-frame deletion mutant of parental strain Mu50 | This study |
| Mu50 graR+ WTR | Wild-type revertant obtained from Mu50 graR mutant cycling | This study |
| Mu50 vraG | ΔvraG in-frame deletion mutant of parental strain Mu50 | This study |
| Mu50 vraG+ WTR | Wild-type revertant obtained from Mu50 vraG mutant cycling | This study |
| E. coli TOP10 | General lab cloning strain | Invitrogen (CA) |
| Plasmids | ||
| pSK236 | Shuttle vector for E. coli and S. aureus | 15 |
| pMAD | Allelic replacement vector to generate S. aureus mutant strains | 1 |
| pMADgraR | pMAD containing 1-kb up- and downstream COL fragments of graR | This study |
| pMADgraS | pMAD containing 1-kb up- and downstream COL fragments of graS | This study |
| pMADvraG | pMAD containing 1-kb up- and downstream COL fragments of vraG | This study |
| pMAD-CM | pMAD containing cat gene from pSK236 using NaeI | This study |
| pMAD-CMgraR | pMAD containing 1-kb up- and downstream Mu50 fragments of graR | This study |
| pMAD-CMvraG | pMAD containing 1-kb up- and downstream Mu50 fragments of vraG | This study |
| pEPSA5 | Expression vector (pEPSA5) for ectopic genes in S. aureus | 14 |
| pEPSA5graR | pEPSA5 containing graR gene amplified from COL | This study |
| pEPSA5graRS1 | pEPSA5 containing graRS genes amplified from Mu50 | This study |
| pEPSA5graRS2 | pEPSA5 containing graRS genes amplified from COL | This study |
| pEPSA5vraFG1 | pEPSA5 containing vraFG genes amplified from Mu50 | This study |
| pEPSA5vraFG2 | pEPSA5 containing vraFG genes amplified from COL | This study |
DNA and computational techniques.
Plasmid DNA was isolated by standard techniques (QIAGEN) and used to transform chemically competent E. coli (44) or electrocompetent S. aureus (41). Restriction endonucleases, ligases, and polymerases were used according to the manufacturers' recommendations. The fidelity of all DNA sequences generated by PCR was verified using fluorescently labeled dideoxynucleotides (BigDye Terminators; PE Applied Biosystems) in DNA sequencing reactions.
Construction of S. aureus mutants.
All mutants were generated with in-frame deletion of target genes by allelic replacement, using the temperature-sensitive plasmid pMAD (1). Briefly, 1-kb PCR products upstream and downstream of targeted sequences were generated and ligated using XmaI. The resulting 2-kb product was inserted into pMAD using BamHI and transformed into E. coli. Plasmids were isolated from E. coli and used to transform S. aureus RN4220. Plasmid from RN4220 was then transformed into S. aureus strains RN6390, COL, and Mu50 for which the gene was to be deleted. The process of allelic replacement was described previously (1). After the excision of pMAD from the merodiploid intermediate strain, either a deletion mutant or a wild-type revertant strain was generated, depending on the location of the DNA crossover event. Wild-type revertant strains have a wild-type genotype but were passaged in conditions identical to those used for the mutant strains. For Mu50, which is erythromycin resistant, a modified pMAD vector (pMAD-CM) was constructed by cloning the chloramphenicol acetyltransferase (cat194) gene from pSK236 into the NaeI site of pMAD (15). Protocols to generate mutants of the VISA strain Mu50 were not performed with low levels of VAN, so wild-type revertant strains were used as controls to ensure that the effect of culturing conditions on mutant phenotypes was minimal. For each in-frame deletion mutant strain, the chromosomal deletion was verified by PCR and DNA sequencing. The resulting deletion strains encoded proteins with the amino acid deletions GraRΔ10-186, GraSΔ21-335, and VraGΔ20-613; two residues (Pro and Gly) were added at the junction sites of all deletions from the XmaI enzyme site. All primers utilized for mutant strain generation are designated by gene name followed by either “MADUP” or “MADDOWN” in Table 2.
TABLE 2.
Primers
| Primer | Sequencea |
|---|---|
| catup | AAAGCCGGCGGATTTTTCGCTACGCTCAAA |
| catdown | AAAGCCGGCAAAGTACAGTCGGCATTATCTCA |
| graRMADUPF | AAAGGATCCGTAACTAAAAGGTGGAGTAATATG |
| graRMADUPR | AAACCCGGGGTCATCTTCTACTAGTAGTATTTG |
| graRMADDOWNF | AAACCCGGGGTTAGTGATAATACGTTAACAG |
| graRMADDOWNR | AAAGGATCCCGTCGTTTCATTTCTGTTAG |
| graSMADUPF | AAAGGATCCGGTGCGACCGCTCGCAG |
| graSMADUPR | AAACCCGGGCCAAAATATCCAGTTCATGCG |
| graSMADDOWNF | AAACCCGGGGAACGCATGTCGGAAGTGAC |
| graSMADDOWNR | AAAGGATCCCAATACTACACTCGTAATTAACG |
| vraGMADUPF | AAAGGATCCGGGGACAACTGTCAGATTG |
| vraGMADUPR | AAACCCGGGGGCATAATGTGATAAATTTTGACG |
| vraGMADDOWNF | AAACCCGGGGCAGTGACGGCTTATAATCAT |
| vraGMADDOWNR | AAAGGATCCCATCATCAATTGCATCACATAATG |
| graSProbeF | GAGCAACAGTTGCAATTGAAC |
| graSProbeR | CTCACTATATTTCAATGCGTTTG |
| vraFProbeF | CAAGGAATTATCTGATATACGC |
| vraFProbeR | CGATTTGCATAGCTTGCTGC |
| gyrBsybrF | GGTGCTGGGCAAATACAAGT |
| gyrBsybrR | TGGGATACCACGTCCGTTAT |
| graSsybrF | TGAAATCTCGCATGAACTGG |
| graSsybrR | ATGGCGTTTCCGCTAAATCT |
| vraFsybrF | GTCTGGTGGACAAAGGCAAC |
| vraFsybrR | TCGATTTGCATAGCTTGCTG |
| graREctopicF | AAAAAAGAGCTCTGATATTGGGTGATATGGATGC |
| graREctopicR | AAAAAATCTAGACAAATTATTCATGAGCCATATATCC |
| graSEctopicR | AAAAAATCTAGAGTTTAAAATGACAAATTTGTCAC |
| vraFEctopicF | AAAAAAGAGCTCTAGATAAATTATAGGAGTGTTAAAGTG |
| vraGEctopicR | AAAAAATCTAGATTATATGGAATGTCTAATTGTTCG |
Sequences are listed in the 5′-to-3′ orientation. Underlined regions represent restriction enzyme sites.
Isolation of RNA and Northern blot hybridization.
Overnight cultures of S. aureus were diluted 1:100 in brain heart infusion broth and grown to exponential (A650 = 0.7 in 18-mm glass tube in a Spectronic 20), late exponential (A650 = 1.1), and postexponential (A650 = 1.7) phases. Total RNA from S. aureus strains was prepared by using the Trizol-glass bead method as described previously (28). The concentration of total RNA was determined by measuring the absorbance at 260 nm. Ten micrograms each of total RNA was analyzed by Northern blotting as described previously (28). Fragments of graS (400 bp) and vraF (400 bp) were amplified by PCR from chromosomal templates and used as probes. For detection of specific transcripts, gel-purified DNA probes were radiolabeled with [α-32P]dCTP by using the random-primed DNA labeling kit (Roche Diagnostics GmbH) and hybridized under aqueous-phase conditions at 65°C. The blots were subsequently washed and bands visualized by autoradiography. Primers used to generate probes in Northern blots are designated by gene name followed by “Probe” in Table 2.
Quantification of transcript levels by real-time PCR.
RNA was isolated as described above except that purified RNA was resuspended in diethyl pyrocarbonate-treated double-distilled water. To remove residual DNA, 10 μg of RNA was treated with RNase-free DNase according to the manufacturer's instructions (Turbo DNA-free; Ambion). One microgram of DNase-treated RNA was reverse transcribed using random hexamer primers in the Transcriptor First Strand cDNA synthesis kit (Roche). Quantification of cDNA levels was performed following the instructions of the LightCycler FastStart DNA MasterPLUS SYBR green I kit (Roche) on a LightCycler 1.5 system (Roche). Analysis was performed using the relative quantification method in the LightCycler software version 4.0 platform (Roche). Primers used in PCR were generated using the Custom Primers-OligoPerfect software (Invitrogen) and designated by gene name followed by “sybr” in Table 2.
Ectopic expression of genes in S. aureus.
To complement the mutant strains, we utilized the pEPSA5 expression plasmid (14). Genes were amplified by PCR (Vent polymerase; New England Biolabs) from multiple strains of S. aureus as noted in Results. The resulting PCR products were gel purified and ligated into pEPSA5 using restriction endonucleases SacI and XbaI. After transformation into E. coli TOP10 and S. aureus RN4220, plasmids were electroporated into final strains, as described above. Genes cloned into pEPSA5 can be induced for expression with xylose (0.5%) or repressed by glucose (0.5%) (14); however, a basal level of expression was always observed without adding exogenous sugars to culturing media. Primers used to construct pEPSA5-based plasmids are designated by gene name followed by “Ectopic” in Table 2.
Antibiotic sensitivity assays.
Antibiotic growth curves were determined according to the following protocol. Strains grown overnight in CSMHB were adjusted to an A650 of 1.1 (corresponding to ∼109 CFU/ml) using sterile CSMHB. Dilutions were then made (200 μl per 7 ml sterile CSMHB) in media with and without 1 μg/ml VAN. Growth curves were generated by plotting A650 over time. For strains carrying pEPSA5-based plasmids, 10 μg/ml of chloramphenicol was included in the culture to maintain the plasmid. To determine the MICs of antibiotics to S. aureus strains, we utilized the standardized microdilution CLSI protocol in CSMHB medium (6), using 105 CFU/ml in 96-well microtiter plates with scoring at 48 h. MIC data are reported as median values from at least three independent experiments for each antibiotic. For the antibiotic daptomycin, we utilized Etest strips (provided by Cubist Pharmaceuticals), rather than microdilution MIC, and followed the supplied protocol. Also, strains containing pEPSA5-based plasmids grew slower in the presence of chloramphenicol (data not shown), so MICs were determined at 72 h to detect growth at various concentrations of VAN and PMB. Median MICs for pEPSA5-based complementing strains were determined using media with and without sugar supplementation (0.5% xylose or glucose).
Determination of Triton X-100-induced autolysis.
The autolysis assay was performed as described previously (20). Briefly, strains grown overnight in TSB were diluted and grown to mid-logarithmic phase (A650 = 0.7). Cells were washed twice in cold sterile distilled water and resuspended in 10 ml of 0.05 M Tris-HCl, pH 7.2, containing 0.05% Triton X-100. Cells were incubated at 30°C, and the A600 was measured every 30 minutes. Data are expressed as percent loss of A600 at the indicated times compared to the zero time point. Each data point represents the mean and standard deviation from three independent experiments.
Zymogram assay.
Zymogram analysis was conducted to detect alterations in autolysin activity as previously described (47) but with minor alterations. Heat-killed RN4220 cells were incorporated into an 8% sodium dodecyl sulfate (SDS)-polyacrylamide gel at 10 mg (wet weight)/ml. Autolytic enzymes were extracted from 10 ml of culture grown to an A650 of 0.7 using 100 μl of 4% SDS, and equivalent protein levels were loaded in the SDS-polyacrylamide gel. After proteins were renatured overnight in water, the gel was incubated with 0.1% methylene blue to visualize clear bands, representing an area of RN4220 cell lysis. The assay was repeated three times, with results from a representative experiment shown.
Determination of whole-cell surface charge.
We sought to determine the whole-cell surface charge by using a cytochrome c binding assay (17, 35). Cytochrome c (Sigma) is a highly positively charged protein (pI = 10; 12 kDa) and can be detected by A530, binding of which is dependent on the net negative surface charge of S. aureus cells (17, 35). Briefly, cells grown overnight in TSB were pelleted and washed twice with 20 mM MOPS (morpholinepropanesulfonic acid), pH 7. Cell densities were measured by A650, and an amount equivalent to 15 absorbance units (e.g., 5 ml of a culture with an A650 of 3.0) was pelleted and incubated with 0.25 mg/ml cytochrome c for 10 min at room temperature. The mixture was centrifuged twice, and the amount of cytochrome c in the supernatant fraction was determined by A530 using a standard curve as the reference. For complementing strains carrying pEPSA5, no exogenous sugar was added to the culturing medium, while chloramphenicol was added to maintain selection of the plasmid. Results shown are the means and standard deviations from three independent experiments.
RESULTS
Four uncharacterized TCRS in S. aureus lie adjacent to ABC transporter genes.
The genome of Staphylococcus aureus strain COL encodes 16 chromosomally encoded TCRS (25), four of which are located next to ABC transporter systems (Table 3) that include an ATPase and a predicted efflux permease. Physically associated TCRS and ABC transporter systems in gram-positive bacteria have been associated with antimicrobial peptide resistance, especially for self-resistance needed for bacteriocin production (21, 30). Three loci (SACOL0716 to -0720, SACOL2356 to -2359, and SACOL2643 to -2646) had at least 100 bp between the two sets of genes, signifying that they may represent two distinct transcripts, while the fourth locus (SACOL1352 to -1355) had a minimal distance between the TCRS and the ABC transporter genes, suggesting that all four genes may be cotranscribed (Table 3). Genomic analyses revealed only one of the four loci (SACOL2356 to -2359) with divergently transcribed TCRS and ABC transporter genes. Interestingly, the SACOL2359 sensor histidine kinase protein is the only “classical” sensor kinase that contains a large extracellular sensor domain, while the rest have truncated sensing domains, as indicated by the total protein size in Table 3. It has been proposed that sensor kinase proteins with truncated sensing domains may monitor intramembrane conditions (29).
TABLE 3.
Genetic linkage of TCRS and ABC transporters in the S. aureus COL genome
| Locus | Arrangementa | Distance (bp) | SK length (amino acids) |
|---|---|---|---|
| SACOL0716 to -0720b | RR-SK-ATP-Perm | 143 | 346 |
| SACOL1352 to -1355 | ATP-Perm-SK-RR | 3 | 363 |
| SACOL2356 to -2359 | ATP-Perm-RR-HK | 134c | 457 |
| SACOL2643 to -2646 | Perm-ATP-SK-RR | 107 | 295 |
RR, response regulator (TCRS); SK, sensor kinase (TCRS); ATP, ATPase (ABC); Perm, permease (ABC).
Renamed graRS (RR-SK) and vraFG (ATP-Perm).
Indicates that the ATP-Perm genes and RR-HK genes are divergently transcribed.
The TCRS GraRS provides basal levels of VAN resistance.
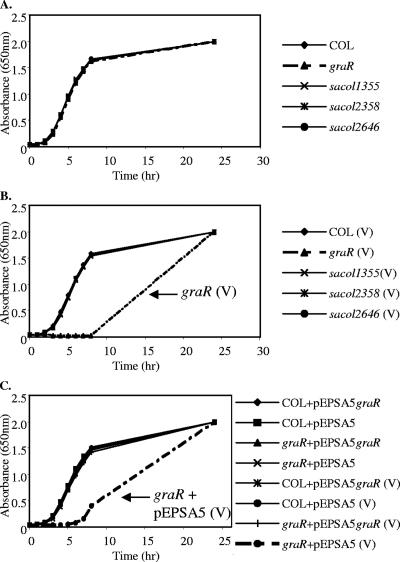
To determine the functions of these four two-component systems with adjacent ABC transporter genes, we utilized a reverse genetic approach whereby the response regulator gene was mutated by an in-frame deletion in each of these four loci of parental strains RN6390 (methicillin-susceptible S. aureus [MSSA]) and COL (MRSA). A recent paper reported higher transcript levels of the TCRS genes SACOL0716 to -0717 in VISA and renamed them graRS (9). While none of these mutations affected growth in CSMHB (Fig. 1A), hypersensitivity to a clinically relevant antibiotic, VAN, at 1 μg/ml was detected in the COL graR mutant but not in other TCRS mutants (Fig. 1B). This phenotype was restored to wild-type levels with ectopic expression of the wild-type graR gene in the graR mutant but not with empty vector pEPSA5 (14) (Fig. 1C), thus establishing the genetic linkage between graRS and baseline resistance to VAN in S. aureus. Similar results showing graR hypersusceptibility to VAN were obtained using mutations in the RN6390 strain background (data not shown). We denote such baseline resistance as the intrinsic level of resistance, even in strains classified as VAN sensitive (e.g., COL). Using this definition, one can speculate that VISA strains (e.g., Mu50) overexpress genes responsible for such basal resistance, as was found with graRS by another group (9).
FIG. 1.
Hypersensitivity of a COL graR mutant to VAN. (A) Growth curves of wild-type COL and four response regulator mutants in CSMHB medium without VAN. (B) Cells grown with 1 μg/ml VAN. Note that only the COL graR mutant exhibited a growth defect. (C) All strains carry a pEPSA5-based plasmid (see Materials and Methods). Cells grown with VAN are indicated by (V). Note that only the noncomplemented COL graR mutant retains a growth defect in the presence of VAN. Similar results were obtained with mutants in the RN6390 background (not shown).
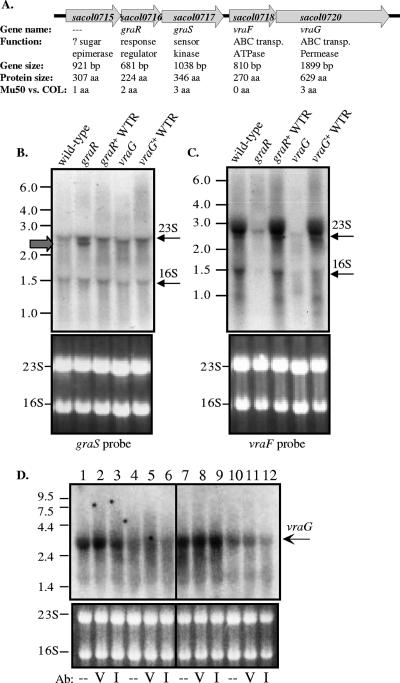
GraRS induces expression of the adjacent vraFG ABC transporter genes.
The genetic arrangement of the graRS locus adjacent to vraFG, which are known to be overexpressed in strains exhibiting increased VAN resistance (24, 31), led us to consider the possibility that vraFG may be a key mediator of GraRS-dependent resistance to VAN (Fig. 2A). The VISA strain Mu50 was utilized for these expression studies due to higher transcript levels of graRS and vraFG as well as their relevance in a VISA strain. While the intergenic distances between the upstream gene SACOL0715 and graR (SACOL0716) and between graR and graS (SACOL0717) are 6 bp and 0 bp, respectively, the intergenic distance between graS and vraF (SACOL0718) is 143 bp (Table 3), indicating that the vraFG operon may be separate from the SACOL0715-graRS transcriptional operon. Northern blot data supported this conjecture, since a probe directed against the sensor kinase gene graS revealed a transcript size of ∼2.7 kb in the wild-type VISA strain Mu50, corresponding to the size of the SACOL0715-graRS transcript (Fig. 2b). However, we observed potential cross-hybridization of the probe with the 23S and 16S rRNA bands as indicated in Fig. 2B, with the SACOL0715-graRS transcript size nearly identical to that of the 23S rRNA band (∼2.7 kb). However, in the Mu50 graR mutant strain, but not in the graR+ wild-type revertant strain, there was an additional transcript corresponding to a reduction in size as expected from a deletion of the graR gene (525 bp deleted). Wild-type revertant strains have a wild-type genotype but were passaged in identical conditions necessary to generate in-frame deletion mutant strains. As such, these strains are useful as controls to ensure that the culture conditions for mutant construction do not affect the experimental result (see Materials and Methods). As predicted, the size of the SACOL0715-graRS transcript (∼2.7 kb) in the Mu50 vraG mutant strain was not diminished (Fig. 2B). The 2.7-kb size of the SACOL0715-graRS transcript in the Mu50 wild-type, graR mutant, and vraG mutant strains supports the hypothesis that the graRS TCRS transcript does not contain the vraFG ABC transporter genes. Moreover, this blot also revealed no significant positive autoregulation by the graR gene, as the graS transcript intensity was not reduced in the Mu50 graR mutant.
FIG. 2.
Regulation of the graRS locus. (A) Diagram of the 5,800-bp chromosomal locus including the graRS and vraFG genes. Shown are the SA open reading frame designation of the COL genome, secondary gene names, proposed functions as given in Entrez Gene (27), gene sizes, translated protein sizes, and amino acid changes between proteins of the COL and Mu50 strains. (B, C, and D) Shown in the upper panels are autoradiographs from the Northern blot assay. To the left of the images are RNA size standards (designated in kb), while to the right of the images are arrows indicating likely cross-reactivity of the labeled probe to rRNA bands (B and C). The lower panels contain the ethidium bromide-stained RNA gels of 23S and 16S rRNA prior to Northern blotting to indicate comparable loading between strains. WTR, wild-type revertant. (B) Northern analysis using the indicated Mu50-derived RNA that was probed with a 400-bp 32P-labeled DNA fragment internal to the graS gene. Note the presence of a transcript, albeit expectedly smaller (∼2.2 kb), in the Mu50 graR mutant strain (gray arrow). (C) Northern analysis using the indicated Mu50-derived RNA that was probed with a 400-bp 32P-labeled DNA fragment internal to the vraG gene. Note the greatly reduced level of vraFG transcript in the Mu50 graR mutant strain. (D) Northern analysis using the indicated COL-derived RNA that was probed with a 400-bp 32P-labeled DNA fragment internal to the vraG gene. RNA was isolated from wild-type COL (lanes 1 to 3), the graR mutant (lanes 4 to 6), the graR mutant with graR expressed from pEPSA5 (lanes 7 to 9), and the graR mutant with empty vector pEPSA5 (lanes 10 to 12). Cells were treated with an antibiotic (Ab) 10 minutes prior to RNA isolation: V, VAN (20 μg/ml); I, imipenem (20 μg/ml); -, no treatment.
To determine if vraFG are regulated by the adjacent TCRS genes graRS, we performed Northern blot analysis using a probe directed against vraF, encoding the putative ATPase. As shown in Fig. 2C, a strong 2.7-kb band was visualized in the wild-type and wild-type revertant strains, again indicating that only vraFG are transcribed in the transcript. In the Mu50 graR mutant, the vraF transcript level was markedly reduced, indicating that vraFG are dependent on GraR for expression. We also observed potential cross-hybridization of the 23S and 16S rRNA bands as indicated in Fig. 2C, but to a lesser extent than when probing against graS. Surprisingly, the Mu50 vraG mutant also exhibited a highly reduced level of vraF transcript even though the vraG deletion was in frame and downstream of vraF. While it is unlikely that the VraG permease has a regulatory role in its own expression, we speculate that a 1.8-kb deletion in a 2.7-kb full-length transcript from the Mu50 vraG mutant may render the transcript unstable. As such, the genotype of the Mu50 vraG mutant is akin to a vraF vraG mutant. Similar results for vraG regulation by GraR were also found in COL strains, where the wild-type strain had a 2.7-kb band (Fig. 2D, lanes 1 to 3) but the band was absent in the COL graR mutant strain (Fig. 2D, lanes 4 to 6). Cross-hybridization of the vraG probe with 23S and 16S rRNAs appeared to be much less in strain COL. When the graR gene was ectopically expressed from pEPSA5 in the COL graR mutant (Fig. 2D, lanes 7 to 9), but not with the empty vector (Fig. 2D, lanes 10 to 12), the graR-complemented strain revealed a strong vraG transcript. When COL wild-type cells were incubated with a bolus of VAN (Fig. 2D, lane 2) but not with imipenem (Fig. 2D, lane 3), there was a potentially mild increase in vraG transcript intensity compared to no treatment (Fig. 2D, lane 1). Unfortunately, the expression level of the SACOL0715-graRS transcript visualized by Northern analysis was too low for reproducible analysis in strain COL (data not shown).
To verify this mode of regulation, independent quantitative real-time reverse transcriptase PCR was performed (Table 4). These data mirrored the Northern analysis, where (i) GraR does not autoregulate the SACOL0715-graRS transcript (graR mutant and wild-type revertant strain transcript levels of graS are nearly equivalent), (ii) VraFG mutations do not affect graS transcription (vraG mutant and wild-type revertant transcript levels of graS are nearly equivalent), (iii) GraR does regulate vraFG (there is more than a 3.5-fold reduction in transcript level in the graR mutant compared to the wild-type revertant strain), and (iv) vraG mutations affect vraF transcript levels (there is a dramatic reduction of the vraF transcript level in the vraG mutant relative to the wild-type revertant strain).
TABLE 4.
Relative transcript levels in Mu50 strains
| Strain | Relative transcript level (mean ± SD)
|
|
|---|---|---|
| graS/gyrB | vraF/gyrB | |
| graR mutant | 0.71 ± 0.34 | 0.25 ± 0.02 |
| graR+ WTRa | 0.67 ± 0.23 | 0.89 ± 0.16 |
| vraG mutant | 0.49 ± 0.11 | 0.13 ± 0.04 |
| vraG+ WTR | 0.73 ± 0.33 | 0.82 ± 0.01 |
WTR, wild-type revertant.
graR and vraG mutants exhibit a limited range of antibiotic resistance defects.
VISA strain Mu50 mutants were tested for increased sensitivities to a wide range of antibiotics to define their antibiotic susceptibility phenotype. Antibiotic sensitivities were determined by the standardized MIC microdilution assay (6). Among the different agents tested, only two antibiotics were more effective in inhibiting mutant growth (Table 5). The MICs of VAN are reduced from 8 μg/ml for wild-type Mu50 to 1 μg/ml for the Mu50 graR mutant and to 2 μg/ml for the Mu50 vraG mutant. The resistance to PMB, a membrane-damaging agent, was reduced from the wild-type level of 128 μg/ml to 16 μg/ml in both mutant strains. Although both of these antibiotics are cationic peptides, they inhibit bacterial growth by various mechanisms, with VAN inhibiting transpeptidation reactions in the cell wall (40) and PMB increasing membrane permeability (12). The remaining 20 antibiotics did not exhibit variations in sensitivity of more than twofold, including those affecting cell wall synthesis, cell membrane stability, and protein synthesis and other, miscellaneous inhibitors. The MICs for wild-type revertant strains were subsequently found to be identical to those for the parental Mu50 (not shown). To verify the phenotypes of VAN and PMB mutants in other strains, we assessed the MICs for COL- and RN6390-derived mutants. As expected, the COL graR, graS, and vraG mutants were more sensitive to VAN (MIC of 1 μg/ml) and PMB (MIC of 8 μg/ml) than the COL wild-type strain (VAN MIC of 2 μg/ml and PMB MIC of 64 μg/ml) (Table 6); similarly, the RN6390 graR, graS, and vraG mutants were more sensitive to VAN (MIC of 0.5 μg/ml) and PMB (MIC of 4 μg/ml) than the RN6390 wild-type strain (VAN MIC of 1 μg/ml and PMB MIC of 32 μg/ml). While the reductions in VAN levels for both COL and RN6390 mutants were only twofold, larger changes may not have been expected, as the wild-type strains are VAN sensitive. Nevertheless, the changes of VAN sensitivity in COL were consistent and led to significant growth alterations in the presence of VAN (Fig. 1).
TABLE 5.
MICs for Mu50 strains
| Category and antibiotic | MIC (μg/ml) for:
|
||
|---|---|---|---|
| Wild-type | graR mutant | vraG mutant | |
| Cell wall agents | |||
| Bacitracin | 64 | 32 | 64 |
| d-Cycloserine | 128 | 64 | 64 |
| Fosfomycin | 1,024 | 1,024 | 1,024 |
| Oxacillin | 512 | 256 | 1,024 |
| VANa | 8 | 1 | 2 |
| Cell membrane agents | |||
| Daptomycinb | 2 | 1 | 2 |
| Gramicidin | ≥512 | ≥512 | ≥512 |
| Lactoferricin | ≥512 | ≥512 | ≥512 |
| Nisin | ≥1,024 | ≥1,024 | ≥1,024 |
| PMBa | 128 | 16 | 16 |
| Protein synthesis agents | |||
| Chloramphenicol | 8 | 8 | 8 |
| Erythromycin | ≥2,048 | 1,024 | ≥2,048 |
| Gentamicin | 256 | 256 | 128 |
| Linezolid | 2 | 4 | 4 |
| Mupirocin | 0.25 | 0.25 | 0.25 |
| Neomycin | 128 | 64 | 128 |
| Tetracycline | 64 | 128 | 64 |
| Tobramycin | ≥2,048 | ≥2,048 | ≥2,048 |
| Miscellaneous agents | |||
| Ciprofloxacin | 32 | 64 | 64 |
| Novobiocin | 0.125 | 0.25 | 0.125 |
| Rifampin | 1,024 | 1,024 | 1,024 |
| Trimethoprim | 4 | 2 | 4 |
Antibiotic showing a ≥4-fold change in MIC.
MIC determined by Etest.
TABLE 6.
VAN and PMB MICs for COL strains
| Strain | Plasmid | DNAa | MIC (μg/ml)
|
|
|---|---|---|---|---|
| VAN | PMB | |||
| Wild type | NAb | NA | 2 | 64 |
| graR mutant | NA | NA | 1 | 8 |
| graS mutant | NA | NA | 1 | 8 |
| vraG mutant | NA | NA | 1 | 8 |
| Wild type | pEPSA5 | 2 | 64 | |
| pEPSA5graRS2 | COL | 4 | 64 | |
| pEPSA5vraFG2 | COL | 2 | 64 | |
| pEPSA5graRS1 | Mu50 | 8 | 128 | |
| pEPSA5vraFG1 | Mu50 | 2 | 64 | |
Genomic DNA from the indicated strain was used as a template in PCR.
NA, not applicable. The strain did not harbor a pEPSA5-derived plasmid.
Multiple GraRS-regulated genes contribute to the Mu50 graR mutant sensitivity.
Initial results demonstrated that complementation of the COL graR mutant with graR expressed in trans (Fig. 1C) restored the basal VAN resistance. Using a genetic approach, we wanted to determine if vraFG expression was the sole determinant in mediating GraRS resistance to VAN and PMB. We utilized pEPSA5-based expression plasmids (14) containing wild-type vraFG and graRS in a complementation experiment with the Mu50 mutants as measured by microdilution MIC. We did not see reproducible differences in MICs when strains were grown with or without induction (0.5% xylose or glucose) in each of the three independent experiments, so the data presented in Table 7 represent the median MIC for all combined conditions and experiments. We subsequently transformed the plasmids containing vraFG or graRS genes from Mu50 DNA, and also one containing vraFG genes from COL DNA, into wild-type strain Mu50 and the graR and vraG mutants. The levels of resistance did not increase in the parent Mu50 containing the recombinant pEPSA5 plasmids, suggesting that additional copies of either graRS or vraFG could not make Mu50 more resistant to VAN or PMB (8 and 256 μg/ml, respectively). When the recombinant pEPSA5graRS1 plasmid (Mu50 DNA) was added to the Mu50 graR mutant, there was an appreciable increase in resistance to VAN and PMB (4 and 128 μg/ml, respectively), albeit not a complete restoration to wild-type levels. In contrast, when vraFG was ectopically expressed from pEPSA5 (pEPSA5vraFG1) in the Mu50 graR mutant, we did not observe a similar increase in resistance levels compared to those with the empty plasmid pEPSA5 (2 and 16 μg/ml, respectively). This indicates that while vraFG is one effector of GraRS-based resistance, it is not the sole determinant, and suggests that other GraR-regulated genes may mediate the observed VAN and PMB resistance. This premise was supported by the finding that ectopic graRS expression in the Mu50 vraG mutant conferred resistance to VAN and PMB relative to the empty vector strain (8 and 256 μg/ml compared to 2 and 16 μg/ml, respectively).
TABLE 7.
VAN and PMB MICs for Mu50 complementing strains
| Strain | Plasmid | DNAa | MIC (μg/ml)
|
|
|---|---|---|---|---|
| VAN | PMB | |||
| Wild type | pEPSA5 | 8 | 256 | |
| pEPSA5graRS1 | Mu50 | 8 | 256 | |
| pEPSA5vraFG1 | Mu50 | 8 | 128 | |
| pEPSA5graRS2 | COL | 8 | 128 | |
| graR mutant | pEPSA5 | 1 | 16 | |
| pEPSA5graRS1 | Mu50 | 4 | 128 | |
| pEPSA5vraFG1 | Mu50 | 2 | 16 | |
| pEPSA5graRS2 | COL | 2 | 64 | |
| vraG mutant | pEPSA5 | 2 | 16 | |
| pEPSA5graRS1 | Mu50 | 8 | 256 | |
| pEPSA5vraFG1 | Mu50 | 4 | 128 | |
| pEPSA5graRS2 | COL | 4 | 64 | |
Genomic DNA from the indicated strain was used as a template in PCR.
In a previous report, Cui et al. suggested that a critical amino acid substitution in the GraR protein (Ser197 in Mu50 to Asn197 in N315) may affect its function (9). To test this further, we used COL DNA, which also contains Asn197 in GraR, in place of Mu50 DNA for complementation in the graR mutant. However, it should be stressed that besides the substitution at position 197 of GraR, there is one additional amino acid changes in GraR and three in GraS between the COL and Mu50 sequences (Fig. 2A). As seen in Table 7, pEPSA5graRS2 from COL DNA increased the resistances to VAN and PMB in both mutant strains but at slightly lower levels compared to the Mu50 graRS genes. It is thus likely that both graRS expression levels and GraRS primary sequences may contribute to resistance levels in Mu50. This hypothesis is further supported by the observation that wild-type COL (VAN sensitive at 2 μg/ml) can be converted to a VISA strain by overexpression of the graRS genes (Table 6). Importantly, the levels of resistance to VAN and PMB were higher in the COL strain that overexpressed Mu50-derived graRS genes (VAN at 8 μg/ml) than in the strain that overexpressed COL-derived graRS genes (VAN at 4 μg/ml). These data further support the notion that some or all of the two amino acid substitutions in GraR and three in GraS are important for full conversion to the VISA phenotype.
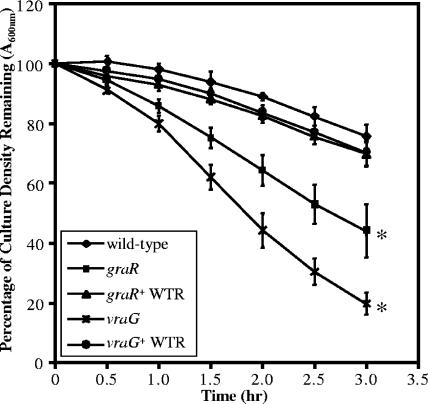
Autolytic activity is increased in graRS mutants of S. aureus.
To help define the mechanism of GraRS-mediated antibiotic resistance, we sought to evaluate the autolytic activity of the Mu50 strains. VISA strains (including Mu50) have been shown to have greatly reduced levels of autolysis (3, 34, 47, 50). We considered the possibility that the reduction in VAN resistance in the Mu50 graR and vraG mutants may be associated with increased autolytic activity. Accordingly, autolysis was measured by lysis of cells in a Triton X-100 buffer over time (Fig. 3) (20). While the wild-type Mu50 and both wild-type revertants had minimal autolysis over the course of these experiments, both mutants had decreased optical densities (P < 0.01) after 3 hours of incubation, indicating augmented autolysis compared to the wild type. The differences between mutant and revertant strains indicated that the culturing conditions associated with allelic replacement did not cause the large increase in autolysis, even though there were slight changes between the wild-type parental and revertant strains attributable to repeated culturing without VAN selection (see Materials and Methods). We next sought to determine if the increased autolytic rate in the mutant strains could be visualized by zymogram analysis (data not shown). We observed that only in the Mu50 vraG mutant strain was there a more intense lytic band, migrating at approximately 60 kDa, compared to the Mu50 wild type, capable of lysing heat-killed RN4220 S. aureus cells. While the Mu50 graR mutant possesses an increased autolytic rate, the change in autolysin activity was not detectable by routine zymogram analysis.
FIG. 3.
Autolysis of Mu50 wild-type and mutant strains. The graph reveals the reduction of bacterial density over time in the presence of buffer containing 0.05% Triton X-100 at 30°C (see Materials and Methods). The mean percent loss of cell density at the indicated times are overlaid with standard deviations from three independent experiments. *, statistical significance of the indicated strain with respect to the Mu50 parental wild-type strain at the 3-hour time point, as determined by the paired Student t test (P < 0.01). WTR, wild-type revertant.
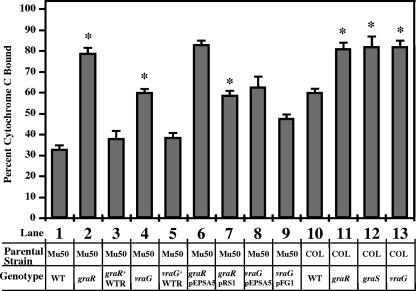
A less negative bacterial surface charge is produced by graRS expression.
As VAN and PMB are both cationic peptides produced by bacteria, we sought to determine if the overall charge of the bacteria is affected by these mutations and hence alters the sensitivity of the mutants to cationic antimicrobial peptides. Previous studies on teichoic acid synthesis revealed that the dlt mutant, which fails to incorporate d-alanine residues into the negatively charged lipoteichoic acid (LTA) molecule, has a more net negative charge than the parental wild-type strain, which consequently renders cells more susceptible to cationic peptides (17, 35). The net surface charge was determined by an assay for binding to cytochrome c, a highly charged molecule (pI = 10) that is readily detectable by A530 (17, 35) (Fig. 4). With this assay, we found that mutations in the graR response regulator gene (Fig. 4, bar 2) and the vraG permease gene (bar 4) led to statistically significant (P < 0.01) more binding by cytochrome c than in the parental wild-type strain Mu50 (bar 1) as well as the wild-type revertant strains (bars 3 and 5). Complementation of these two mutant strains conferred less binding (less negatively charged) to cytochrome c, as expected (bars 6 to 9). Furthermore, graR, graS, or vraG mutants of the MRSA strain COL also bound higher levels of cytochrome c (P < 0.01), consistent with a more negative surface charge (bars 10 to 13). Overall, these results imply that a functional graRS TCRS may render the bacterial cells more positively charged; this alteration in surface charge may, in part, be due to the expression of the GraR-regulated ABC transporter VraFG.
FIG. 4.
Binding of cytochrome c to whole S. aureus cells. The graph shows percent binding of cytochrome c after 10 min of incubation with S. aureus at room temperature. Detailed below the graph are the parental strain (Mu50 or COL) and genotypes of various genetic constructs. Data represent the means and standard deviations from three independent experiments. *, statistical significance of the indicated strain compared to the respective control strain (lane 1 versus lanes 2 to 5, lane 6 versus lane 7, lane 8 versus lane 9, and lane 10 versus lanes 11 to 13), as determined by the paired Student t test (P < 0.01). WTR, wild-type revertant.
DISCUSSION
In this study, we have described two genes that likely mediate the VAN-intermediate resistance phenotype in S. aureus. Although graRS and vraFG have been previously linked to overexpression in VISA strains by microarray analyses, we have genetically linked their activity to VAN resistance in this report. More specifically, we found that the isogenic mutants of graR and vraG in the VISA strain Mu50 had increased susceptibility to the clinically relevant antibiotics VAN and PMB (both having a net positive charge). These mutant strains exhibited increased autolysis rates and enhanced net negative surface charges compared to the wild-type and wild-type revertant strains. The observation that GraRS regulated the expression of vraFG links their phenotypes to an expression cascade, but partial phenotypes observed in Mu50 graR mutants complemented with vraFG genes suggest that other genetic elements controlled by GraRS may affect VISA phenotypes. Of note, we did not visualize considerable alterations in RNA transcript levels of sarA, RNAIII of agr, pbp2, or pbp4 using Northern analysis (data not shown). Moreover, the mutant phenotypes in non-VISA strains (COL and RN6390) also confirmed our observation that these genes are important in mediating basal resistance to VAN and PMB.
The classic examples of genetic linkage between a TCRS and an adjacent ABC transporter in gram-positive bacteria are the bacteriocin immunity regulons (21, 30). Strains producing bacteriocins (bacterially derived antimicrobial peptides) must protect themselves from autologous peptide killing (e.g., by nisin or bacitracin), whereby a TCRS senses the bacteriocin and activates transcription of an adjacent operon encoding an ABC transporter, which then exports the peptide externally to prevent binding at its intended target site. Unlike the classical immunity pumps, which are usually located near the bacteriocin biosynthesis genes on the chromosome, the genetic locus of SACOL0716 to -0720 (encoding GraRS and VraFG) is not located at a region of bacteriocin production. It is interesting that the sensor domain of GraS is much smaller than the typical sensor histidine kinases. This type of truncated membrane sensor domain has been postulated to represent an intramembrane sensor (29). Whether this pertains to GraS as a potential membrane sensor is not clear. Computational analyses of the vraFG locus indicated that it likely acts as an exporter pump, rather than as an importer, since most import ABC transporters contain a high-affinity solute-binding domain, which is apparently lacking in either protein (11, 51). We found that among the many antibiotics tested, the vraG mutants are sensitive to only two tested cationic peptides (VAN and PMB).
PMB is generally thought to be ineffective against gram-positive bacteria due in part to the cell wall, which limits diffusion of PMB to the cell membrane (26, 48), and also to the absence of lipopolysaccharide, the target of PMB, thus reducing binding affinities of PMB to the gram-positive cell (43). In place of lipopolysaccharide, many gram-positive bacteria incorporate LTA and wall teichoic acid into the cell membrane and cell wall, respectively. The increased sensitivities to PMB in the graR and vraG mutants suggest that PMB may gain greater access to the cell membrane, bind with higher affinity, or increase the membrane permeability to PMB in these mutants. The results shown here indicate that the overall surface charge is more negative in these mutants, possibly implying that PMB-mediated inhibition may be at the level of binding affinity, but further experimentation is clearly needed to validate this hypothesis. We also studied the extent of peptidoglycan cross-linking in the Mu50 graR and vraG mutants and discovered no significant differences compared with the parental strain (data not shown). We thus speculate that rates of diffusion of PMB through the cell wall may be unaffected in the hypersensitive mutants. Indeed, preliminary studies in our laboratory showed that a mutation in ypfP (22), which reduces LTA attachment to the cell surface, but not in a cell wall teichoic acid tagO mutation (52), led to hypersusceptibility to PMB compared to that of wild-type parental strains, suggesting that the presence of anchored LTA increases protection of S. aureus from PMB action (data not shown). Furthermore, recent microarray data from Friedrich Götz's group reveal that GraR positively regulates the dlt operon (S. Herbert et al., submitted for publication). The contribution of d-alanylation via the dlt operon has been well established as generating a more net positive bacterial surface charge that alters levels of resistance to cationic antimicrobial peptides (35) and VAN (36) in S. aureus. The observed phenotypes of the Mu50 graR mutant strain may therefore have been affected by altered expression levels of both the vraFG transporter and the dlt d-alanylation operons.
Based on data presented in this study, one mechanism of VAN-intermediate resistance involves the regulation of multiple GraR-mediated genes. We speculate that on one level, the VraFG ABC transporter potentially enhances export of cell wall/teichoic acid precursors or modifying subunits rather than export of VAN or PMB, since the molecular structures of these antibiotics are quite dissimilar. Meanwhile, other GraR-mediated genes may encode transporter proteins or modifying enzymes that could potentially provide redundancy for VraFG or function downstream of VraFG. Further research will delineate the specific transport functions of VraFG, the signaling molecule initiating TCRS phosphotransfer in GraS, and the nature of genes other than vraFG that provide resistance against VAN and PMB. Furthermore, the nature of the observed overexpression of graRS in strain Mu50 remains mysterious, as we have not yet uncovered potential regulators for the graRS operon. A comparison of the putative promoter upstream of the SACOL0715 gene that drives the graRS operon revealed four nucleotide substitutions between strains Mu50 and COL (not shown). All of these substitutions are upstream of the −35 binding region of the RNA polymerase but within 300 base pairs of the start ATG codon, indicating that these substitutions may be included in the promoter, but we are unsure if they have an effect on gene expression.
The discovery that a bacterial sensor histidine kinase recognizes mammalian antimicrobial peptides (2) is of interest in that the system studied here may have bearing on bacterial basal resistance against mammalian and other microbial peptides in human hosts and in diverse polymicrobial environments. This conjecture was supported by a recent report demonstrating that Bacillus subtilis ABC transporter genes are upregulated via an adjacent TCRS in the presence of human antimicrobial peptide LL-37 (38). Indeed, some of our long-term objectives are to characterize the four linked loci in S. aureus (Table 3) for resistance phenotypes against a broad range of bacterially and mammalian-derived antimicrobial peptides. This would advance our understanding of the role of these TCRS transporter loci in VAN-intermediate strains and other intermediate phenotypes arising from selection in the presence of antibacterial peptides. An appreciation of these processes will enable microbiologists to predict patterns of resistance against current and future antimicrobial peptides that are being developed against staphylococci. Additionally, these studies may provide an impetus to search for novel agents to increase the sensitivity of S. aureus to VAN and other related peptide antibiotics. As these TCRS-ABC transporter loci are also present in other pathogenic bacteria, these studies may apply to other pathogens as well.
Acknowledgments
We thank Guido Memmi for construction of the pMAD-CM vector, Mariana Pinho for experimental contributions to cell wall cross-linking analysis, and Hoiwan Cheung for technical assistance. We also thank Cubist Pharmaceutics for supplying daptomycin Etest strips.
A.L.C. is supported by NIH grants (AI37142 and AI56114).
Footnotes
Published ahead of print on 14 May 2007.
REFERENCES
- 1.Arnaud, M., A. Chastanet, and M. Debarbouille. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ Microbiol. 70:6887-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bader, M. W., S. Sanowar, M. E. Daley, A. R. Schneider, U. Cho, W. Xu, R. E. Klevit, H. Le Moual, and S. I. Miller. 2005. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122:461-472. [DOI] [PubMed] [Google Scholar]
- 3.Boyle-Vavra, S., M. Challapalli, and R. S. Daum. 2003. Resistance to autolysis in vancomycin-selected Staphylococcus aureus isolates precedes vancomycin-intermediate resistance. Antimicrob. Agents Chemother. 47:2036-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyle-Vavra, S., H. Labischinski, C. C. Ebert, K. Ehlert, and R. S. Daum. 2001. A spectrum of changes occurs in peptidoglycan composition of glycopeptide-intermediate clinical Staphylococcus aureus isolates. Antimicrob. Agents Chemother. 45:280-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chatterjee, C., M. Paul, L. Xie, and W. A. van der Donk. 2005. Biosynthesis and mode of action of lantibiotics. Chem. Rev. 105:633-684. [DOI] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 7th ed. Document M7-A7. Clinical and Laboratory Standards Institute, Wayne, PA.
- 7.Cosgrove, S. E., K. C. Carroll, and T. M. Perl. 2004. Staphylococcus aureus with reduced susceptibility to vancomycin. Clin. Infect. Dis. 39:539-545. [DOI] [PubMed] [Google Scholar]
- 8.Cui, L., A. Iwamoto, J. Q. Lian, H. M. Neoh, T. Maruyama, Y. Horikawa, and K. Hiramatsu. 2006. Novel mechanism of antibiotic resistance originating in vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 50:428-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cui, L., J. Q. Lian, H. M. Neoh, E. Reyes, and K. Hiramatsu. 2005. DNA microarray-based identification of genes associated with glycopeptide resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 49:3404-3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui, L., X. Ma, K. Sato, K. Okuma, F. C. Tenover, E. M. Mamizuka, C. G. Gemmell, M. N. Kim, M. C. Ploy, N. El-Solh, V. Ferraz, and K. Hiramatsu. 2003. Cell wall thickening is a common feature of vancomycin resistance in Staphylococcus aureus. J. Clin. Microbiol. 41:5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davidson, A. L., and J. Chen. 2004. ATP-binding cassette transporters in bacteria. Annu. Rev. Biochem. 73:241-268. [DOI] [PubMed] [Google Scholar]
- 12.Evans, M. E., D. J. Feola, and R. P. Rapp. 1999. Polymyxin B sulfate and colistin: old antibiotics for emerging multiresistant gram-negative bacteria. Ann. Pharmacother. 33:960-967. [DOI] [PubMed] [Google Scholar]
- 13.Finan, J. E., G. L. Archer, M. J. Pucci, and M. W. Climo. 2001. Role of penicillin-binding protein 4 in expression of vancomycin resistance among clinical isolates of oxacillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:3070-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forsyth, R. A., R. J. Haselbeck, K. L. Ohlsen, R. T. Yamamoto, H. Xu, J. D. Trawick, D. Wall, L. Wang, V. Brown-Driver, J. M. Froelich, K. G. C., P. King, M. McCarthy, C. Malone, B. Misiner, D. Robbins, Z. Tan, Z. Y. Zhu Zy, G. Carr, D. A. Mosca, C. Zamudio, J. G. Foulkes, and J. W. Zyskind. 2002. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol. Microbiol. 43:1387-1400. [DOI] [PubMed] [Google Scholar]
- 15.Garcia, C., C. Briggs, L. Zhang, L. Guan, J. L. Gabriel, and T. J. Rogers. 1998. Molecular characterization of the putative T-cell receptor cavity of the superantigen staphylococcal enterotoxin B. Immunology 94:160-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gill, S. R., D. E. Fouts, G. L. Archer, E. F. Mongodin, R. T. Deboy, J. Ravel, I. T. Paulsen, J. F. Kolonay, L. Brinkac, M. Beanan, R. J. Dodson, S. C. Daugherty, R. Madupu, S. V. Angiuoli, A. S. Durkin, D. H. Haft, J. Vamathevan, H. Khouri, T. Utterback, C. Lee, G. Dimitrov, L. Jiang, H. Qin, J. Weidman, K. Tran, K. Kang, I. R. Hance, K. E. Nelson, and C. M. Fraser. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187:2426-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamilton, A., D. L. Popham, D. J. Carl, X. Lauth, V. Nizet, and A. L. Jones. 2006. Penicillin-binding protein 1a promotes resistance of group B streptococcus to antimicrobial peptides. Infect. Immun. 74:6179-6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanaki, H., K. Kuwahara-Arai, S. Boyle-Vavra, R. S. Daum, H. Labischinski, and K. Hiramatsu. 1998. Activated cell-wall synthesis is associated with vancomycin resistance in methicillin-resistant Staphylococcus aureus clinical strains Mu3 and Mu50. J. Antimicrob. Chemother. 42:199-209. [DOI] [PubMed] [Google Scholar]
- 19.Hanaki, H., H. Labischinski, Y. Inaba, N. Kondo, H. Murakami, and K. Hiramatsu. 1998. Increase in glutamine-non-amidated muropeptides in the peptidoglycan of vancomycin-resistant Staphylococcus aureus strain Mu50. J. Antimicrob. Chemother. 42:315-320. [DOI] [PubMed] [Google Scholar]
- 20.Ingavale, S. S., W. Van Wamel, and A. L. Cheung. 2003. Characterization of RAT, an autolysis regulator in Staphylococcus aureus. Mol. Microbiol. 48:1451-1466. [DOI] [PubMed] [Google Scholar]
- 21.Joseph, P., G. Fichant, Y. Quentin, and F. Denizot. 2002. Regulatory relationship of two-component and ABC transport systems and clustering of their genes in the Bacillus/Clostridium group, suggest a functional link between them. J. Mol. Microbiol. Biotechnol. 4:503-513. [PubMed] [Google Scholar]
- 22.Kiriukhin, M. Y., D. V. Debabov, D. L. Shinabarger, and F. C. Neuhaus. 2001. Biosynthesis of the glycolipid anchor in lipoteichoic acid of Staphylococcus aureus RN4220: role of YpfP, the diglucosyldiacylglycerol synthase. J. Bacteriol. 183:3506-3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 24.Kuroda, M., K. Kuwahara-Arai, and K. Hiramatsu. 2000. Identification of the up- and down-regulated genes in vancomycin-resistant Staphylococcus aureus strains Mu3 and Mu50 by cDNA differential hybridization method. Biochem. Biophys. Res. Commun. 269:485-490. [DOI] [PubMed] [Google Scholar]
- 25.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 26.LaPorte, D. C., K. S. Rosenthal, and D. R. Storm. 1977. Inhibition of Escherichia coli growth and respiration by polymyxin B covalently attached to agarose beads. Biochemistry 16:1642-1648. [DOI] [PubMed] [Google Scholar]
- 27.Maglott, D., J. Ostell, K. D. Pruitt, and T. Tatusova. 2005. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 33:D54-D58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manna, A. C., S. S. Ingavale, M. Maloney, W. van Wamel, and A. L. Cheung. 2004. Identification of sarV (SA2062), a new transcriptional regulator, is repressed by SarA and MgrA (SA0641) and involved in the regulation of autolysis in Staphylococcus aureus. J. Bacteriol. 186:5267-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mascher, T. 2006. Intramembrane-sensing histidine kinases: a new family of cell envelope stress sensors in Firmicutes bacteria. FEMS Microbiol. Lett. 264:133-144. [DOI] [PubMed] [Google Scholar]
- 30.Mascher, T., N. G. Margulis, T. Wang, R. W. Ye, and J. D. Helmann. 2003. Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol. Microbiol. 50:1591-1604. [DOI] [PubMed] [Google Scholar]
- 31.McAleese, F., S. W. Wu, K. Sieradzki, P. Dunman, E. Murphy, S. Projan, and A. Tomasz. 2006. Overexpression of genes of the cell wall stimulon in clinical isolates of Staphylococcus aureus exhibiting vancomycin-intermediate-S. aureus-type resistance to vancomycin. J. Bacteriol. 188:1120-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mongodin, E., J. Finan, M. W. Climo, A. Rosato, S. Gill, and G. L. Archer. 2003. Microarray transcription analysis of clinical Staphylococcus aureus isolates resistant to vancomycin. J. Bacteriol. 185:4638-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nascimento Jdos, S., M. Giambiagi-deMarval, S. S. de Oliveira, H. Ceotto, K. R. dos Santos, and C. Bastos Mdo. 2005. Genomic fingerprinting of bacteriocin-producer strains of Staphylococcus aureus. Res. Microbiol. 156:837-842. [DOI] [PubMed] [Google Scholar]
- 34.Nelson, J. L., K. C. Rice, S. R. Slater, P. M. Fox, G. L. Archer, K. W. Bayles, P. D. Fey, B. N. Kreiswirth, and G. A. Somerville. 2007. Vancomycin-intermediate Staphylococcus aureus strains have impaired acetate catabolism: implications for polysaccharide intercellular adhesin synthesis and autolysis. Antimicrob. Agents Chemother. 51:616-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peschel, A., M. Otto, R. W. Jack, H. Kalbacher, G. Jung, and F. Gotz. 1999. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 274:8405-8410. [DOI] [PubMed] [Google Scholar]
- 36.Peschel, A., C. Vuong, M. Otto, and F. Gotz. 2000. The d-alanine residues of Staphylococcus aureus teichoic acids alter the susceptibility to vancomycin and the activity of autolytic enzymes. Antimicrob. Agents Chemother. 44:2845-2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pieper, R., C. L. Gatlin-Bunai, E. F. Mongodin, P. P. Parmar, S. T. Huang, D. J. Clark, R. D. Fleischmann, S. R. Gill, and S. N. Peterson. 2006. Comparative proteomic analysis of Staphylococcus aureus strains with differences in resistance to the cell wall-targeting antibiotic vancomycin. Proteomics 6:4246-4258. [DOI] [PubMed] [Google Scholar]
- 38.Pietiainen, M., M. Gardemeister, M. Mecklin, S. Leskela, M. Sarvas, and V. P. Kontinen. 2005. Cationic antimicrobial peptides elicit a complex stress response in Bacillus subtilis that involves ECF-type sigma factors and two-component signal transduction systems. Microbiology 151:1577-1592. [DOI] [PubMed] [Google Scholar]
- 39.Recsei, P., B. Kreiswirth, M. O'Reilly, P. Schlievert, A. Gruss, and R. P. Novick. 1986. Regulation of exoprotein gene expression in Staphylococcus aureus by agar. Mol. Gen. Genet. 202:58-61. [DOI] [PubMed] [Google Scholar]
- 40.Reynolds, P. E. 1989. Structure, biochemistry and mechanism of action of glycopeptide antibiotics. Eur. J. Clin. Microbiol. Infect. Dis. 8:943-950. [DOI] [PubMed] [Google Scholar]
- 41.Schenk, S., and R. A. Laddaga. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 73:133-138. [DOI] [PubMed] [Google Scholar]
- 42.Scherl, A., P. Francois, Y. Charbonnier, J. M. Deshusses, T. Koessler, A. Huyghe, M. Bento, J. Stahl-Zeng, A. Fischer, A. Masselot, A. Vaezzadeh, F. Galle, A. Renzoni, P. Vaudaux, D. Lew, C. G. Zimmermann-Ivol, P. A. Binz, J. C. Sanchez, D. F. Hochstrasser, and J. Schrenzel. 2006. Exploring glycopeptide-resistance in Staphylococcus aureus: a combined proteomics and transcriptomics approach for the identification of resistance-related markers. BMC Genomics 7:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scott, M. G., M. R. Gold, and R. E. Hancock. 1999. Interaction of cationic peptides with lipoteichoic acid and gram-positive bacteria. Infect. Immun. 67:6445-6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seidman, C. E., K. Struhl, J. Sheen, and T. Jessen. 1997. Introduction of plasmid DNA into cells, p. 1.8.1-1.8.10. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, Hoboken, NJ. [DOI] [PubMed]
- 45.Sieradzki, K., M. G. Pinho, and A. Tomasz. 1999. Inactivated pbp4 in highly glycopeptide-resistant laboratory mutants of Staphylococcus aureus. J. Biol. Chem. 274:18942-18946. [DOI] [PubMed] [Google Scholar]
- 46.Sieradzki, K., and A. Tomasz. 1999. Gradual alterations in cell wall structure and metabolism in vancomycin-resistant mutants of Staphylococcus aureus. J. Bacteriol. 181:7566-7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sieradzki, K., and A. Tomasz. 2006. Inhibition of the autolytic system by vancomycin causes mimicry of vancomycin-intermediate Staphylococcus aureus-type resistance, cell concentration dependence of the MIC, and antibiotic tolerance in vancomycin-susceptible S. aureus. Antimicrob. Agents Chemother. 50:527-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Storm, D. R., K. S. Rosenthal, and P. E. Swanson. 1977. Polymyxin and related peptide antibiotics. Annu. Rev. Biochem. 46:723-763. [DOI] [PubMed] [Google Scholar]
- 49.Tenover, F. C., and L. C. McDonald. 2005. Vancomycin-resistant staphylococci and enterococci: epidemiology and control. Curr. Opin. Infect. Dis. 18:300-305. [DOI] [PubMed] [Google Scholar]
- 50.Utaida, S., R. F. Pfeltz, R. K. Jayaswal, and B. J. Wilkinson. 2006. Autolytic properties of glycopeptide-intermediate Staphylococcus aureus Mu50. Antimicrob. Agents Chemother. 50:1541-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van der Heide, T., and B. Poolman. 2002. ABC transporters: one, two or four extracytoplasmic substrate-binding sites? EMBO Rep. 3:938-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weidenmaier, C., J. F. Kokai-Kun, S. A. Kristian, T. Chanturiya, H. Kalbacher, M. Gross, G. Nicholson, B. Neumeister, J. J. Mond, and A. Peschel. 2004. Role of teichoic acids in Staphylococcus aureus nasal colonization, a major risk factor in nosocomial infections. Nat. Med. 10:243-245. [DOI] [PubMed] [Google Scholar]
- 53.West, A. H., and A. M. Stock. 2001. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 26:369-376. [DOI] [PubMed] [Google Scholar]