Abstract
The epidemiology of clavulanic acid-inhibited extended-spectrum β-lactamases (ESBLs) was investigated among infection-associated enterobacterial isolates at the University Hospital in Lausanne, Switzerland, from January 2004 to June 2005. Out of 57 nonrepetitive ESBL producers (prevalence rate of 0.7%), 45 produced CTX-M-like ESBLs. CTX-M enzymes were mostly from clonally nonrelated Escherichia coli isolates, from urinary infections and community-acquired infections. Pediatric patients (20 out of 57) accounted for a large number of CTX-M producers. CTX-M-15 was the most frequent CTX-M-type enzyme. The plasmid-located blaCTX-M genes were associated with either ISEcp1 or ISCR1 insertion sequences. This study is the first published report of CTX-M-type β-lactamases in Switzerland.
Plasmid-mediated extended-spectrum β-lactamases (ESBLs) were first identified in a Klebsiella pneumoniae isolate in Germany in 1983 (14). Since then, ESBL-positive Enterobacteriaceae have been isolated worldwide mostly from hospitalized patients (18). These enzymes hydrolyze significantly expanded-spectrum cephalosporins such as cefotaxime and ceftazidime and the monobactam aztreonam, sparing carbapenems. Their activity is inhibited in vitro by clavulanic acid. Until the 2000s, most of the ESBLs were structurally related to the narrow-spectrum TEM- and SHV-type β-lactamases, with one to several amino acid substitutions surrounding their active site (18). Beginning in the late 1990s, novel types of ESBLs, the CTX-M enzymes, emerged worldwide, mostly from Escherichia coli. The CTX-Ms are mostly from community-acquired isolates (2, 22). The over 50 CTX-M enzymes so far reported may be grouped into five main subgroups according to amino acid sequence identity (CTX-M-1, -M-2, -M-8, -M-9, and -M-25) (2). Most of the CTX-Ms hydrolyze cefotaxime better than ceftazidime. However, several CTX-Ms including CTX-M-15 (2, 12, 25), which is now the most widespread CTX-M enzyme worldwide (6), hydrolyze ceftazidime efficiently (25).
Since those CTX-Ms are reported increasingly in France (8, 15, 17, 26), Italy (4, 20), and recently Austria (9), it was interesting to search for those enzymes in Switzerland, a country known to have a strict policy of antibiotic prescription and to have an overall low level of multidrug resistance in bacteria (10).
The aim of the present study was to estimate the prevalence and the type of the ESBLs produced by enterobacterial isolates among nonrepetitive clinical isolates over an 18-month period from January 2004 to June 2005 at the University Hospital of Lausanne, Switzerland.
MATERIALS AND METHODS
Bacterial isolates.
ESBL-producing enterobacterial isolates resulted from the screening of 8,259 enterobacterial isolates obtained from infection samples that were sent to the Department of Microbiology of the Lausanne University Hospital, Switzerland, from January 2004 to June 2005. Isolates were first identified by using the Vitek2 system (bioMérieux SA, Marcy-l'Etoile, France). Electrocompetent E. coli TOP10 (Invitrogen, Cergy Pontoise, France) was used as a recipient strain in transformation experiments. E. coli NCTC 50192, harboring 154-, 66-, 38-, and 7-kb plasmids, was used as a plasmid-containing reference strain (27).
Susceptibility testing and screening for ESBL-producing isolates.
The antibiotic susceptibility of enterobacterial clinical isolates was determined with the Vitek2 Advance Expert System (bioMerieux) and by the disk diffusion method on Mueller-Hinton (MH) agar plates with β-lactam and non-β-lactam antibiotic-containing disks (Sanofi Diagnostics Pasteur, Marnes-La-Coquette, France), according to the Clinical and Laboratory Standards Institute guidelines (7). The double-disk synergy test was performed with ceftriaxone, ceftazidime, aztreonam, cefpodoxime, and amoxicillin-clavulanic acid disks using different disk spacings (20, 25, 30, and 40 mm) on MH agar plates, and the results were interpreted as described previously (11). The Etest strips containing cefepime plus clavulanic acid (AB Biodisk, Solna, Sweden) were also used. Then, MICs were determined for selected β-lactams by an agar dilution technique on MH agar with an inoculum of 104 CFU per spot, as described previously (23). MICs of several β-lactams were determined alone or in combination with a fixed concentration of either clavulanic acid (4 μg/ml) or tazobactam (4 μg/ml). MICs were interpreted according to the guidelines of the Clinical and Laboratory Standards Institute (7).
PCR amplification for detection of ESBL genes, analysis of their genetic environment, and sequencing.
Under standard PCR conditions (27), a series of primers was used for detection of several Ambler class A β-lactamase genes (Table 1). Detection was performed for genes encoding TEM (PRETEM-1 and PRETEM-2), SHV (OS5 and OS6), and CTX-M (CTX-MA1 and CTX-MA2) (8, 25). For each reaction, 0.5 μg of whole-cell DNA of the ESBL-possessing enterobacterial isolates or 0.5 μg of plasmid DNA from E. coli TOP10 electroporants was used.
TABLE 1.
Sequences of primers used for detection of blaCTX-M genes and their genetic environment
| Primer name | Primer sequence | Location |
|---|---|---|
| CTX-MA1 | 5′-SCSATGTGCAGYACCAGTAA-3′ | blaCTX-M gene |
| CTX-MA2 | 5′-CCG CRA TAT GRT TGG TGG TG-3′ | blaCTX-M gene, reverse primer |
| CTXM2A | 5′-GCC GCT CAA TGT TAA CGG-3′ | blaCTX-M gene (CTX-M-2 group) |
| CTXM2B | 5′-GAA ACC GTG GGT TAC GAT-3′ | blaCTX-M gene (CTX-M-2 group), reverse primer |
| CTXM9A | 5′-CTG ATG TAA CAC GGA TTG AC-3′ | blaCTX-M gene (CTX-M-9 group) |
| CTXM9C rev | 5′-AGC GCC CCA TTA TTG AGA G-3′ | blaCTX-M gene (CTX-M-9 group), reverse primer |
| TOHO2b rev | 5′-TTA CAG CCC TTC GGC GAT-3′ | blaCTX-M gene (CTX-M-9 group), reverse primer |
| CTXMpréB | 5′-CAC TTT GCC GTC GTC TAA GGC G-3′ | blaCTX-M gene (CTX-M-1 group), reverse primer |
| ISEcpPROM+ | 5′-TGC TCT GTG GAT AAC TTG C-3′ | ISEcp1 upstream of the promoter |
| ISEcpPROM− | 5′-GCA GTC TAA ATT CTT CGT G-3′ | ISEcp1 downstream of the promoter |
| IS903Bint | 5′-GCT TTT TGA CTT TCC ACT CGC-3′ | IS903B transposase, reverse primer |
| Orf513-D3 | 5′-CTC ACG CCC TGG CAA GGT TT-3′ | Orf513 |
| Orf513-D5 | 5′-CTT TTG CCC TAG CTG CGG T-3′ | Orf513, reverse primer |
| Orf513-5′ext | 5′-CAG CTG GTA GAG CAG CGT C-3′ | 5′ end of Orf513, reverse primer |
| OS5 | 5′-TTA TCT CCC TGT TAG CCA CC-3′ | blaSHV gene |
| OS6 | 5′-GAT TTG CTG ATT TCG CCG G-3′ | blaSHV gene, reverse primer |
| PRETEM-1 | 5′-GTA TCC GCT CAT GAG ACA ATA-3′ | blaTEM gene |
| PRETEM-2 | 5′-TCT AAA GTA TAT ATG AGT AAA CTT GGT CTG-3′ | blaTEM gene, reverse primer |
The genetic environment of the blaCTX-M genes was characterized by PCR (16) since different genetic elements are associated with the blaCTX-M genes such as ISEcp1-like insertion sequences and the ISCR1 element comprising orf513 which is embedded in a sul1-type integron (6). Whole-cell DNA of the isolates was extracted as described previously (12). The regions located upstream of the blaCTX-M genes were amplified with primers annealing to ISEcp1 and to orf513 of ISCR1 together with primers for the blaCTX-M genes (26). The sequences located downstream of the blaCTX-M genes were studied by PCR experiments with the forward primer CTX-MA1 and the reverse primer CTXMpreB or IS903Bint. When the orf513 gene was found upstream of blaCTX-M, a PCR experiment was performed with primer CTX-MA1 and reverse primer qacED-1B in order to search for a sul1-type integron structure as previously described with a duplication (even partial) of the ISCR1 element (16, 26).
For direct DNA sequencing, PCR products were purified using PCR purification columns (QIAGEN, Courtaboeuf, France). The sizes of the sequenced blaSHV, blaTEM, and blaCTX-M genes were 795, 861, and 876 bp, respectively. Sequencing reactions were performed using specific primers and an automated sequencer (ABI 377; Applied Biosystems, Foster City, CA). The nucleotide and deduced protein sequences were analyzed with software available over the Internet at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov).
Plasmid transfer and analysis.
Plasmid DNAs of enterobacterial isolates were extracted using the Kieser technique (13). They were then electroporated into E. coli TOP10, and recombinant strains were selected onto cefotaxime-containing (1 μg/ml) Trypticase soy agar plates. Plasmid DNAs of these transformants were detected by electrophoresis on a 0.7% agarose gel.
Hybridization.
DNA-DNA hybridizations were performed as described previously with a Southern transfer of an agarose gel containing plasmid DNA from E. coli TOP10 electroporants (25). The probes consisted of a ca.-500-bp PCR fragment generated from isolate 23 producing CTX-M-15, isolate 8 producing CTX-M-14, and isolate 25 producing CTX-M-2. They were internal to the blaCTX-M genes (15). Labeling of the probe and signal detection were carried out using a nonradioactive labeling and detection kit according to the manufacturer's instructions (Amersham Pharmacia Biotech).
PFGE.
Whole-cell DNAs embedded in 1% agarose plugs (Bio-Rad) were digested with XbaI restriction enzyme (Amersham Pharmacia Biotech) and separated in a 1% pulsed-field certified agarose gel (Bio-Rad) by using a CHEF DRII system (Bio-Rad), as described previously (24). Pulsed-field gel electrophoresis (PFGE) was performed at 14°C, with a 6-V/cm current, a switch angle of 120°, and for E. coli, a run time of 12 h followed by a run time of 12 h, with two linear switch ramps of 4 and 12 s and 15 to 36 s, and for K. pneumoniae, switch times of 2 to 32 s for 20 h. After migration, gels were stained in an 0.5-mg/ml ethidium bromide solution, and PFGE results were analyzed according to the criteria of Tenover et al. (28).
RESULTS
Epidemiology and PCR detection of β-lactamase genes.
A total of 57 nonrepetitive ESBL-positive isolates were collected from 54 patients. The prevalence of ESBL producers was 0.7%. Any enterobacterial isolate flagged as a possible ESBL producer by the Vitek2 Advance Expert System was tested by the double-disk test and by the Etest strip containing cefepime/cefepime plus clavulanic acid. The isolates were E. coli (n = 31), Klebsiella pneumoniae (n = 17), Enterobacter cloacae (n = 6), Proteus mirabilis (n = 2), and Klebsiella oxytoca (n = 1). The sex ratio of patients was 32/22 (female/male). The isolates were from hospitalized patients (43/54), outpatients (11/54), and significantly from pediatric patients (19/54). Out of the 43 hospitalized patients, nine isolates were obtained within 48 h of their hospitalization. Therefore, community-acquired isolates were from 20 patients (nine hospitalized patients plus 11 outpatients).
Three patients had two different ESBL-positive isolates (E. coli and K. pneumoniae in two cases and E. coli and P. mirabilis in one case) that were collected at different times. Moreover, the same ESBL-positive isolate was identified from a urinary sample from a woman and from the eyes of her twins.
The ESBL-positive isolates were mostly from urine (63%) and from pus (24%) but also from respiratory tract or blood. Most of the patients were treated successfully with a carbapenem-containing antibiotic regimen (data not shown).
PCR experiments with primers specific for the blaCTX-M, blaTEM, and blaSHV genes yielded blaCTX-M-positive results for 45 out of the 57 isolates (Table 2). A blaCTX-M gene was identified in 14 out of the 20 community-acquired isolates. The blaTEM and blaSHV genes encoding ESBLs were identified in all the 12 blaCTX-M-negative isolates. A single isolate (E. coli 14) expressed two different ESBLs.
TABLE 2.
Characterization of acquired β-lactamases in ESBL-positive isolates
| Isolate | β-Lactamase type(s)a:
|
||
|---|---|---|---|
| CTX-M | TEM | SHV | |
| E. cloacae | |||
| 1 | CTX-M-9 | ||
| 2 | TEM-1 | SHV-5 | |
| 3 | SHV-5 | ||
| 4 | SHV 5 | ||
| 5 | TEM-1 | SHV-5 | |
| 6 | TEM-1 | SHV-5 | |
| E. coli | |||
| 7 | CTX-M-15 | TEM-1 | |
| 8 | CTX-M-14 | TEM-1 | |
| 9 | CTX-M-15 | ||
| 10 | CTX-M-14 | TEM-1 | |
| 11 | CTX-M-15 | ||
| 12 | CTX-M-14 | ||
| 13 | CTX-M-1 | ||
| 14 | CTX-M-15, CTX-M-14 | TEM-1 | |
| 15 | CTX-M-15 | ||
| 16 | CTX-M-15 | TEM-1 | |
| 17 | CTX-M-15 | TEM-1 | |
| 18 | CTX-M-1 | TEM-1 | |
| 19 | TEM-53 | ||
| 20 | CTX-M-15 | TEM-1 | |
| 21 | CTX-M-14 | TEM-1 | |
| 22 | CTX-M-15 | ||
| 23 | CTX-M-15 | TEM-1 | |
| 24 | TEM-1 | SHV-5 | |
| 25 | CTX-M-2 | ||
| 26 | CTX-M-15 | ||
| 27 | CTX-M-15 | TEM-1 | |
| 28 | CTX-M-15 | TEM-1 | |
| 29 | CTX-M-15 | TEM-1 | |
| 30 | CTX-M-14 | ||
| 31 | TEM-52 | ||
| 32 | CTX-M-15 | ||
| 33 | CTX-M-15 | ||
| 34 | CTX-M-15 | ||
| 35 | CTX-M-15 | TEM-1 | |
| 36 | CTX-M-15 | ||
| 37 | CTX-M-15 | ||
| K. oxytoca | |||
| 38 | CTX-M-15 | TEM-1 | |
| K. pneumoniae | |||
| 39 | CTX-M-15 | TEM-1 | |
| 40 | CTX-M-9 | ||
| 41 | TEM-1 | SHV-5 | |
| 42 | TEM-1/TEM-4 | SHV-11 | |
| 43 | SHV-5 | ||
| 44 | CTX-M-15 | TEM-1 | |
| 45 | SHV-2a | ||
| 46 | CTX-M-15 | ||
| 47 | CTX-M-15 | TEM-1 | |
| 48 | CTX-M-15 | ||
| 49 | CTX-M-15 | ||
| 50 | CTX-M-15 | TEM-1 | |
| 51 | CTX-M-15 | TEM-1 | |
| 52 | CTX-M-15 | TEM-1 | |
| 53 | CTX-M-15 | ||
| 54 | CTX-M-15 | TEM-1 | |
| 55 | CTX-M-15 | TEM-1 | |
| P. mirabilis | |||
| 56 | CTX-M-15 | TEM-1 | |
| 57 | CTX-M-14 | TEM-1 | |
The ESBL names are in bold.
Molecular identification of blaCTX-M, blaTEM, and blaSHV genes and genetic environment of blaCTX-M genes.
Seventy-nine percent of ESBLs were CTX-M enzymes (46/57) (Table 2). Most of them (78%, 36/46) belonged to the CTX-M-1 group, CTX-M-15 being predominant (34/36). Twenty percent of the CTX-Ms were CTX-M-9-like (9/46), being either CTX-M-14 or CTX-M-9. A single isolate produced CTX-M-2. A single E. coli isolate produced both CTX-M-14 and CTX-M-15. Sixty percent of the CTX-M producer isolates produced also the narrow-spectrum β-lactamase TEM-1.
E. coli was the main ESBL producer. It was identified for 63% of the CTX-M-positive isolates, and CTX-M enzymes accounted for 91% of the ESBLs in that species. The prevalence of CTX-M among ESBL types was also high in K. pneumoniae (76%). Only one out of the six E. cloacae isolates produced a CTX-M enzyme. The other ESBLs were mostly of the SHV type in E. cloacae and K. pneumoniae (Table 2).
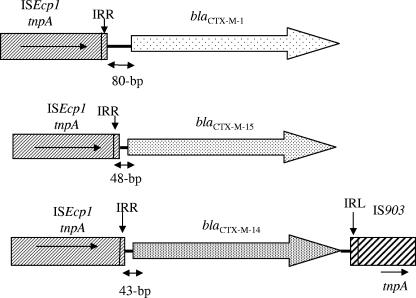
The genetic structure surrounding the blaCTX-M genes was then determined. ISEcp1 was identified upstream of 42 out of the 46 blaCTX-M genes (91%). The right boundary of ISEcp1 was located between 43 and 80 bp upstream of the start codon of blaCTX-M genes. ISEcp1 was identified at 80 bp upstream of the blaCTX-M-1 gene and at 48 bp upstream of blaCTX-M-15 genes. A 43-bp region with an identical sequence was found between ISEcp1 and the start codon of blaCTX-M-9-like genes (blaCTX-M-9 and blaCTX-M-14). Downstream of four out of six blaCTX-M-14 genes (14, 12, 8, and 21), an IS903-like element was found as already described for this gene (24). ISEcp1 was not identified upstream of the blaCTX-M-2 and blaCTX-M-9 genes, as previously reported (21). ISCR1 was found in isolates harboring blaCTX-M-2 and blaCTX-M-9 genes (Fig. 1).
FIG. 1.
Surrounding DNA sequences for the blaCTX-M genes for the 42 isolates. Row 1, E. coli 13 and 18; row 2, E. coli 7, 9, 11, 14, 15, 16, 17, 20, 22, 23, 26, 28, 29, 32, 33, 34, 35, 36, and 37; K. oxytoca 38; K. pneumoniae 39, 44, 46, 47, 48, 49, 50, 51, 52, 53, 54, and 55; P. mirabilis 56 and 57; row 3, E. coli 8, 10, 12, 14, 21, and 30 and P. mirabilis 57. IRR and IRL, inverted repeat (right) and inverted repeat (left), respectively.
Antibiotic susceptibility results.
The isolates were resistant to amino-, ureido-, and carboxypenicillins, and clavulanic acid and tazobactam addition partially restored the activity of those antibiotics (Table 3). Isolates producing CTX-Ms were resistant to cefotaxime whereas this was not always the case for TEM and SHV producers. The CTX-M-15 producers were resistant also to ceftazidime except for two isolates (E. coli 37 and P. mirabilis 56). Susceptibilities to cefepime and cefpirome varied whereas susceptibilities to carbapenems were constant. Disk diffusion susceptibility testing indicated that overall resistance rates of ESBL producers were 2, 63, 50, 75, and 30% for amikacin, sulfonamides, ciprofloxacin, tetracyclines, and chloramphenicol, respectively, with no significant difference between CTX-M and non-CTX-M producers (data not shown).
TABLE 3.
MICs of β-lactams for ESBL-positive isolates
| β-Lactam(s) | MIC range (μg/ml) for isolates producing ESBL:
|
||||||
|---|---|---|---|---|---|---|---|
| CTX-M-1 | CTX-M-15 | CTX-M-2 | CTX-M-9 | CTX-M-14 | TEM-ES | SHV-ES | |
| Amoxicillin | >256 | >256 | >256 | >256 | >256 | >256 | >256 |
| Amoxicillin and clavulanic acida | 16 | 8-32 | 8 | 8->256 | 4-8 | 2-8 | 4-256 |
| Ticarcillin | >256 | >256 | >256 | >256 | >256 | >256 | >256 |
| Ticarcillin and clavulanic acid | 128 | 32-128 | 64 | 32 | 32-64 | 16-256 | 16-256 |
| Piperacillin | >256 | 128-256 | 128 | 64-128 | 32-256 | 32->256 | 32->256 |
| Piperacillin and tazobactamb | 8 | 2-32 | 2 | 2-16 | 1-16 | 1-16 | 1-64 |
| Cephalothin | >256 | >256 | >256 | >256 | >256 | 256 | >256 |
| Cefoxitin | 8 | 4-32 | 4 | 8-32 | 4-16 | 4-16 | 4-256 |
| Ceftazidime | 4-8 | 1->256 | 1 | 1 | 0.5-2 | 4 | 0.25-128 |
| Cefotaxime | >256 | 128-256 | 128 | 32-128 | 32-64 | 0.12-128 | 2-128 |
| Cefepime | 32-128 | 8-128 | 16 | 2-8 | 2-8 | 0.25-32 | 0.25-8 |
| Aztreonam | 64-256 | 2-256 | 16 | 2-4 | 2-8 | 2-16 | 2-128 |
| Imipenem | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 |
| Ertapenem | 0.12 | 0.003-0.5 | 0.06 | 0.06-1 | 0.003-0.12 | 0.06 | 0.06-0.5 |
Clavulanic acid was at a fixed concentration of 4 μg/ml.
Tazobactam was at a fixed concentration of 4 μg/ml.
PFGE.
Analysis of XbaI-restricted DNA of CTX-M-producing E. coli and K. pneumoniae isolates showed that the blaCTX-M-positive isolates were mostly nonclonally related. Only three E. coli isolates (isolates 11, 15, and 36) were clonally related or identical. Those isolates produced CTX-M-15, and two of them (isolates 11 and 15) were identified from patients hospitalized in the same ward. Analysis of the PFGE profiles of CTX-M-15-producing K. pneumoniae isolates showed that three isolates (50, 51, and 52) were undistinguishable. They were from urine from a mother of premature twins and from conjunctivitis of her twins.
Plasmid analysis and hybridization.
Plasmid DNAs of the CTX-M-producing isolates were extracted and analyzed by gel electrophoresis. Their sizes ranged from 7 to 170 kb with one to five plasmids per isolate. The plasmid DNAs hybridized with any of the three internal probes for blaCTX-M-1-like, blaCTX-M-2, and blaCTX-M-9-like (data not shown). A single hybridization signal was evidenced in each isolate, except for E. coli isolate 14, which exhibited two hybridization signals. These results showed that all CTX-M producers except one contained a single blaCTX-M gene. Those β-lactamase genes were located on 50- and 170-kb plasmids. Four out of the seven blaCTX-M-14 genes were located on similarly sized plasmids (ca. 80 kb) although the CTX-M producers were not clonally related. The blaCTX-M-15 genes were located mostly on plasmids varying in size. The blaCTX-M-14 and blaCTX-M-15 genes of E. coli isolate 14 were located on two different plasmids of 70 kb and 155 kb, respectively. These results indicated that the spread of blaCTX-M genes resulted from both clonal and plasmid spread.
DISCUSSION
The results of this work provided insights into the molecular epidemiology of the spread of ESBLs in enterobacterial isolates responsible for infections at a university hospital in Switzerland. The prevalence rate of ESBL producers was 0.7%, which is very similar to that reported in the nearby country Austria in 2004 (9). However, this rate is lower than that reported in The Netherlands (7.8% in Amsterdam, 2004 [1]), in France (2.3% at Bicêtre Hospital, 2004 [P. Nordmann, personal data]), Italy (1.6%, 2003 [4]), and the United States (4.9%, 2001 to 2002 [19]). Variable prevalence rates of ESBL producers may be related in part to differences in antibiotic policy (more quinolone used than β-lactams for treating community patients in Switzerland [10]) but also to differences in urban sizes (Paris area or Amsterdam versus Lausanne, for example). The blaCTX-M genes were widespread among ESBL producers, 79%, which was a slightly higher value than that reported at Bicêtre Hospital (65%) (personal data), in Amsterdam (64%) (1), and in Austria (58%) (9). A recent study performed in several Swiss laboratories between 2001 and 2003 identified 63.4% of ESBL producers as producing CTX-Ms (5).
ESBLs of E. coli were mostly CTX-Ms at Lausanne (91%), at Bicêtre (84%, personal data), in Austria (85%) (9), and in Amsterdam (63%) (1). Moreover, two/three of the CTX-M producers were E. coli. The prevalence of CTX-Ms among ESBL-positive K. pneumoniae strains was also similarly high in Lausanne (76%) and in Paris (83%), whereas this value was much lower in Austria (30%) and in Italy (12.3%) (20).
Most ESBL-producing E. coli isolates (62%) have been isolated from urinary tract infections, as found in other studies (4, 23, 29). More than one-third of CTX-M-producing isolates were from community-acquired infections, as reported from other European countries (3, 4, 22).
Most of the CTX-Ms identified in Switzerland belonged to the CTX-M group 1 (CTX-M-1, … .) as reported in France (Bicêtre Hospital [personal data]) and especially in central and southern France (17), in Austria (9), and in Amsterdam (1). CTX-M-15 was the predominant CTX-M in Lausanne (94%), as elsewhere in Western Europe (4, 17, 20, 29). Twenty percent of the CTX-M enzymes were CTX-M-9-like in Lausanne, 26% were CTX-M-9-like in Austria, and only 11% were CTX-M-9-like in Bicêtre Hospital. Moreover, CTX-M-9 and the genetically related CTX-M-14 were the most prevalent CTX-M types isolated from clinical samples in Spain until 2004 (3).
PFGE analysis showed that the spread of blaCTX-M-positive E. coli and K. pneumoniae isolates was not related to the spread of single clones as reported in several studies from other countries (United Kingdom, France, Italy, and Spain) (8, 20, 21, 29).
Concerning the non-β-lactam antimicrobial susceptibilities of CTX-M producers, high rates of resistance were observed especially for ciprofloxacin (50%), as reported in other countries such as Austria (49% [9]), Canada (66% [22]), and Italy (73% [20]). However, the rate of resistance to ciprofloxacin of isolates from pediatric patients was lower (11%) than that of isolates from adults (59%), which is consistent with a low usage of fluoroquinolones in pediatric cases.
We identified here the spread of community-acquired CTX-M producers in Switzerland, with CTX-M-15 being the most prevalent type as observed now in other European countries. One of the most interesting findings of this study may be the size of the reservoir of CTX-M producers in pediatric patients. Finally, the spread of CTX-M producers in community-acquired E. coli infections in Switzerland in a manner similar to that observed in neighboring countries may indicate difficulty in controlling these emerging resistance determinants whatever the antibiotic policy is.
Acknowledgments
This work was funded by a grant from the Ministère de l'Education Nationale et de la Recherche (UPRES-EA3539), Université Paris XI, France, and by the European Community (6th PCRD, LSHM-CT-2005-018705). L.P. is a researcher from the INSERM, France.
We thank Cristina Bellini for help in reviewing the medical charts.
Footnotes
Published ahead of print on 30 April 2007.
REFERENCES
- 1.Al Naiemi, N., A. Bart, M. D. de Jong, C. M. Vandenbroucke-Grauls, P. J. Rietra, Y. J. Debets-Ossenkopp, P. C. Wever, L. Spanjaard, A. J. Bos, and B. Duim. 2006. Widely distributed and predominant CTX-M extended-spectrum β-lactamases in Amsterdam, The Netherlands. J. Clin. Microbiol. 44:3012-3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonnet, R. 2004. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bou, G., M. Cartelle, M. Tomas, D. Canle, F. Molina, R. Moure, J. M. Eiros, and A. Guerrero. 2002. Identification and broad dissemination of the CTX-M-14 β-lactamase in different Escherichia coli strains in the northwest area of Spain. J. Clin. Microbiol. 40:4030-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brigante, G., F. Luzzaro, M. Perilli, G. Lombardi, A. Coli, G. M. Rossolini, G. Amicosante, and A. Toniolo. 2005. Evolution of CTX-M-type β-lactamases in isolates of Escherichia coli infecting hospital and community patients. Int. J. Antimicrob. Agents 25:157-162. [DOI] [PubMed] [Google Scholar]
- 5.Bruderer, T., M. Jutzi, H. Adler, and R. Frei. 2006. The CTX-M is the most predominant group of extended-spectrum β-lactamases (ESBL) in Switzerland, p. 130. Swiss Soc. Microbiol. 65th Annu. Assembly. Lausanne, Switzerland, 7 and 8 March 2006.
- 6.Canton, R., and T. M. Coque. 2006. The CTX-M β-lactamase pandemic. Curr. Opin. Microbiol. 9:466-475. [DOI] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute. 2005. Performance standards for antimicrobial susceptibility testing; 15th informational supplement. M100-S15. Clinical and Laboratory Standards Institute, Wayne, PA.
- 8.Eckert, C., V. Gautier, M. Saladin-Allard, N. Hidri, C. Verdet, Z. Ould-Hocine, G. Barnaud, F. Delisle, A. Rossier, T. Lambert, A. Philippon, and G. Arlet. 2004. Dissemination of CTX-M-type β-lactamases among clinical isolates of Enterobacteriaceae in Paris, France. Antimicrob. Agents Chemother. 48:1249-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eisner, A., E. J. Fagan, G. Feierl, H. H. Kessler, E. Marth, D. M. Livermore, and N. Woodford. 2006. Emergence of Enterobacteriaceae isolates producing CTX-M extended-spectrum β-lactamase in Austria. Antimicrob. Agents Chemother. 50:785-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Filippini, M., G. Masiero, and K. Moschetti. 2006. Socioeconomic determinants of regional differences in outpatient antibiotic consumption: evidence from Switzerland. Health Policy 78:77-92. [DOI] [PubMed] [Google Scholar]
- 11.Jarlier, V., M. H. Nicolas, G. Fournier, and A. Philippon. 1988. Extended broad-spectrum β-lactamases conferring transferable resistance to newer β-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev. Infect. Dis. 10:867-878. [DOI] [PubMed] [Google Scholar]
- 12.Karim, A., L. Poirel, S. Nagarajan, and P. Nordmann. 2001. Plasmid-mediated extended-spectrum β-lactamase (CTX-M-3 like) from India and gene association with insertion sequence ISEcp1. FEMS Microbiol. Lett. 201:237-241. [DOI] [PubMed] [Google Scholar]
- 13.Kieser, T. 1984. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid 12:19-36. [DOI] [PubMed] [Google Scholar]
- 14.Knothe, H., P. Shah, V. Krcmery, M. Antal, and S. Mitsuhashi. 1983. Transferable resistance to cefotaxime, cefoxitin, cefamandole and cefuroxime in clinical isolates of Klebsiella pneumoniae and Serratia marcescens. Infection 11:315-317. [DOI] [PubMed] [Google Scholar]
- 15.Lartigue, M. F., N. Fortineau, and P. Nordmann. 2005. Spread of novel expanded-spectrum β-lactamases in Enterobacteriaceae in a university hospital in the Paris area, France. Clin. Microbiol. Infect. 11:588-591. [DOI] [PubMed] [Google Scholar]
- 16.Lartigue, M. F., L. Poirel, and P. Nordmann. 2004. Diversity of genetic environment of blaCTX-M genes. FEMS Microbiol. Lett. 34:201-207. [DOI] [PubMed] [Google Scholar]
- 17.Lavigne, J. P., H. Marchandin, J. Delmas, N. Bouziges, E. Lecaillon, L. Cavalie, H. Jean-Pierre, R. Bonnet, and A. Sotto. 2006. qnrA in CTX-M-producing Escherichia coli from France. Antimicrob. Agents Chemother. 50:4224-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Medeiros, A. A. 1997. Evolution and dissemination of β-lactamases accelerated by generations of β-lactam antibiotics. Clin. Infect. Dis. 24(Suppl.):S19-S45. [DOI] [PubMed] [Google Scholar]
- 19.Moland, E. S., N. D. Hanson, J. A. Black, A. Hossain, W. Song, and K. S. Thomson. 2006. Prevalence of newer β-lactamases in gram-negative clinical isolates collected in the United States from 2001 to 2002. J. Clin. Microbiol. 44:3318-3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mugnaioli, C., F. Luzzaro, F. De Luca, G. Brigante, M. Perilli, G. Amicosante, S. Stefani, A. Toniolo, and G. M. Rossolini. 2006. CTX-M-type extended-spectrum β-lactamases in Italy: molecular epidemiology of an emerging countrywide problem. Antimicrob. Agents Chemother. 50:2700-2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oteo, J., C. Navarro, E. Cercenado, A. Delgado-Iribarren, I. Wilhelmi, B. Orden, C. Garcia, S. Miguelanez, M. Perez-Vazquez, S. Garcia-Cobos, B. Aracil, V. Bautista, and J. Campos. 2006. Spread of Escherichia coli strains with high-level cefotaxime and ceftazidime resistance between the community, long-term facilities, and hospital institutions. J. Clin. Microbiol. 44:2359-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pitout, J. D., P. Nordmann, K. B. Laupland, and L. Poirel. 2005. Emergence of Enterobacteriaceae producing extended-spectrum β-lactamases (ESBLs) in the community. J. Antimicrob. Chemother. 56:52-59. [DOI] [PubMed] [Google Scholar]
- 23.Poirel, L., M. Gniadkowski, and P. Nordmann. 2002. Biochemical analysis of the ceftazidime-hydrolysing extended-spectrum β-lactamase CTX-M-15 and of its structurally related β-lactamase CTX-M-3. J. Antimicrob. Chemother. 50:1031-1034. [DOI] [PubMed] [Google Scholar]
- 24.Poirel, L., O. Menuteau, N. Agoli, C. Cattoen, and P. Nordmann. 2003. Outbreak of extended spectrum β-lactamase VEB-1-producing isolates of Acinetobacter baumannii in a French hospital. J. Clin. Microbiol. 41:3542-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poirel, L., T. Naas, I. Le Thomas, A. Karim, E. Bingen, and P. Nordmann. 2001. CTX-M-type extended-spectrum β-lactamase that hydrolyzes ceftazidime through a single amino acid substitution in the omega loop. Antimicrob. Agents Chemother. 45:3355-3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saladin, M., V. T. Cao, T. Lambert, J. L. Donay, J. L. Herrmann, Z. Ould-Hocine, C. Verdet, F. Delisle, A. Philippon, and G. Arlet. 2002. Diversity of CTX-M β-lactamases and their promoter regions from Enterobacteriaceae isolated in three Parisian hospitals. FEMS Microbiol. Lett. 209:161-168. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook, J. E., F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 28.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woodford, N., M. E. Ward, M. E. Kaufmann, J. Turton, E. J. Fagan, D. James, A. P. Johnson, R. Pike, M. Warner, T. Cheasty, A. Pearson, S. Harry, J. B. Leach, A. Loughrey, J. A. Lowes, R. E. Warren, and D. M. Livermore. 2004. Community and hospital spread of Escherichia coli producing CTX-M extended-spectrum β-lactamases in the UK. J. Antimicrob. Chemother. 54:735-743. [DOI] [PubMed] [Google Scholar]