Abstract
In 1992, Xie et al. identified a cDNA sequence in the expression cloning search for the κ opioid receptor. When the cDNA was expressed in Cos-7 cells, binding of opioid compounds was observed to be of low affinity and without κ, μ, or δ selectivity [Xie, G.-X., Miyajima, A. and Goldstein, A. (1992) Proc. Natl. Acad. Sci. USA 89, 4124–4128]. This cDNA was highly homologous to the human neurokinin-3 (NK-3) receptor sequence, and displayed lower homology to NK-1 and NK-2 sequences. This sequence was stably expressed in Chinese hamster ovary cells, which do not express neurokinin receptors naturally, and ligand binding and second messenger characteristics were compared with a human NK-3 receptor. The NK-3 receptor homolog bound [3H]senktide with a Kd of 39 nM, similar to that of the NK-3 receptor. The rank order of tachykinin peptides competing for [3H]senktide binding at the NK-3 receptor homolog was [MePhe7]neurokinin B > senktide > substance P = neurokinin A > neurokinin B. This cell line also bound [125I-MePhe7]neurokinin B; however, neurokinin B was an effective competitor. Tachykinin peptides stimulated both inositol phospholipid hydrolysis and arachidonic acid release at NK-3 and NK-3 receptor homolog cell lines, with similar rank orders of potency of [MePhe7]neurokinin B = neurokinin B = senktide > NKA = substance P. These results indicate that expression of the NK-3 receptor homolog cDNA in the Chinese hamster ovary cell system induces the expression of a receptor site with many similarities but certain key differences from that of the human NK-3 receptor. The results are discussed with reference to the existence of a novel human tachykinin receptor.
Over the past several years, much information has become available on the primary structures and functions of G-protein-coupled peptide hormone receptors due to advances in molecular cloning strategies and in the development of potent nonpeptide antagonists. One exemplary system in which this has occurred is that of the tachykinin peptide family. It is now widely appreciated that the major tachykinin peptides, substance P (SP), neurokinin A (NKA), and neurokinin B (NKB), are the primary agonists at three distinct pharmacologically and molecularly characterized receptor types, called neurokinin-1 (NK-1), NK-2, and NK-3 (1–3). These receptors appear to mediate the functions of the tachykinin peptides on diverse biological processes including smooth muscle contraction, blood pressure regulation, neuronal communication, and endocrine and exocrine gland secretions. These peptides also appear to be involved in the regulation of certain immune and inflammatory states.
Due to the homology among receptors in the G-protein-coupled receptor superfamily, several putative receptor sequences have been cloned in which the natural ligand has not been identified (4). In addition, various genetic strategies have resulted in the identification of receptor-like sequences in which no known ligand has been determined (e.g., see ref. 5). The identification of these so-called “orphan” receptors has also come about in the search for other receptors using more directed methodologies. In 1992, Xie and coworkers attempted to characterize, using an affinity enrichment procedure, the κ opioid receptor from a human placental cDNA library (6). They identified a cDNA clone, called “hKIR,” which encoded a 440 residue protein consistent with the structure of a 7 transmembrane-containing G-protein-coupled receptor (hereafter called NK-3 receptor homolog). This sequence was highly homologous to the NK-3 subfamily of tachykinin receptors, and had a lower identity to NK-1 and NK-2 receptors (7). The sequence, when expressed in Cos-7 cells, induced binding of the opiate alkaloid bremazocine with a Kd of 87 nM. [3H]Bremazocine binding was displaced similarly by μ, δ, and κ selective compounds, but not substantially by the nonselective antagonists naloxone or naltrexone. This relatively low affinity for bremazocine, coupled with a lack of appropriate κ ligand selectivity, cast doubt on it being an opioid receptor. Indeed, a G-protein-coupled receptor has since been characterized that had the pharmacology consistent with that of a κ opioid receptor (8). Moreover, Xie and coworkers did not detect binding of the tachykinin peptide agonist [3H]eledoisin in Cos-7 cells transfected with hKIR, but could detect specific binding in cells transfected with the rat NK-3 receptor. Consequently, the receptor properties of this NK-3 receptor homolog were uncertain and as such it was considered to be an “orphan” receptor. To clarify the biological relevance of this NK-3 receptor homolog, we examined the radioligand binding and response properties of this cDNA after stable expression in Chinese hamster ovary (CHO) cells. The results indicate that this cDNA sequence encodes a receptor with agonist binding properties and activities similar to, but not identical with, the previously characterized human NK-3 receptor.
EXPERIMENTAL PROCEDURES
Materials.
Most of the reagents used have been described (9, 10). Oligonucleotides were synthesized by the Washington University Protein and Nucleic Acid Chemistry Laboratory. The PCR II vector was from Invitrogen. Maloney murine leukemia RNase H− reverse transcriptase, called SuperScript II, was from GIBCO/BRL. NKB (Asp-Met-His-Asp-Phe-Phe-Val-Gly-Leu-Met-NH2), senktide (succinyl-Asp-Phe-MePhe-Gly-Leu-Met-NH2), and [MePhe7]NKB (Asp-Met-His-Asp-Phe-Phe-MePhe-Gly-Leu-Met-NH2) were from Bachem, and four batches of [3H]senktide (66, 66, 69, and 74 Ci/mmol; 1 Ci = 37GBq) were from DuPont/New England Nuclear. [125I-MePhe7]NKB was prepared by oxidative iodination of [MePhe7]NKB with chloramine-T and Na125I, and the monoiodo form was purified by high performance liquid chromatography. Na125I (2175 Ci/mmol) was from DuPont/New England Nuclear. Klentaq and Klentaq long and accurate were obtained from W. Barnes (Washington University School of Medicine) (11).
Molecular Cloning and Sequence Analysis of NK-3 and NK-3 Homolog cDNAs.
The human NK-3 receptor cDNA was cloned using a reverse transcriptase–polymerase chain reaction (PCR) protocol with hypothalamic RNA as the message source. Human hypothalamic tissue was obtained at autopsy (2 hr from death) from an adult who died from cardiac arrest, and was provided by the Washington University Department of Pathology. The cDNA was generated as two fragments, using 5′-CCACCATGGCCACTCTCCCAGCAGCA-3′ and 5′-CCATGACGGCCATTGCGGTGGAC-3′ as forward primers and 5′-TTAAGAATATTCATCCACAGAGGT-3′ and 5′-GGAAGGCAAGTAGAAATGCTAG-3′ as reverse primers (12, 13). For the first primer, an optimal Kozak sequence CCACC (14) was inserted upstream of the initiator methionine to enhance 40s ribosomal subunit binding. The PCR products were cloned into the PCR II vector and characterized by restriction and nucleotide sequence analysis. Due to the presence of a BstXI site at nucleotide 540 of the NK-3 cDNA coding region, the two partial cDNAs could be restricted and ligated to generate the full coding region. This ligated NK-3 cDNA was subcloned into the mammalian expression vector, pM2 (15), for transfection studies. The human NK-3 receptor homolog was obtained from Guo-xi Xie and Robert Thompson (University of Michigan). XhoI and BamHI sites were inserted onto the 5′ and 3′ portions of the cDNA, respectively, using a PCR procedure and the cDNA was subcloned into the pM2 expression vector. Both the original cDNA and that cloned into the mammalian expression vector were sequenced. Analysis was performed on an Applied Biosystems model 373A automated sequencer. The human NK-3 coding sequence was identical to that previously reported (12, 13). Sequence analysis of the NK-3 receptor homolog cDNA identified at nucleotides 175 and 176, an inversion of the previously reported GC pair, which changed the coding sequence at amino acid 59 from an alanine to an arginine. This arginine residue is conserved in all NK-3 receptors characterized to date (7). Also, at coding region base 552 a cytidine was identified as compared with the previously reported thymidine, which was silent regarding the amino acid sequence but resulted in the presence of a BamHI site in the cDNA. PCR procedures were performed with Klentaq 1, which is devoid of 5′ exonuclease activity, or with the LA system (15), the enzyme system that combines Klentaq 1 with the proofreading enzyme Pfu DNA polymerase, which is devoid of 5′ exonuclease activity. The use of human tissues for RNA isolation was performed under institutional approval, protocol number 95–0363.
Generation of Stably Transfected CHO Cell Lines.
The procedures used were those previously described (9, 10). Stably transfected cells were cloned after isolation of individual colonies, and were screened for receptor expression by radioligand binding and by second messenger responsiveness. Cells were routinely grown in minimal essential medium-α, containing 10% fetal bovine serum and 0.8 mg active G418 per ml.
Radioligand Binding.
[3H]Senktide and [125I-MePhe7]NKB binding assays were performed for both the NK-3 receptor and the NK-3 receptor homolog using whole cells in polypropylene tubes at 4°C. No detectable specific binding was observed in nontransfected CHO cells. Cells were rinsed with cold Tris-buffered saline (TBS, 0.05 M Tris·HCl buffer containing 120 mM NaCl, pH 7.4), scraped from the tissue culture dish and resuspended in TBS. The binding reaction in a total volume of 100 μl was initiated by adding 80 μl of the cell suspension to radioligand and test competitors in 20 μl TBS containing 1 mg/ml BSA, 0.2 mg/ml bacitracin, 20 μg/ml leupeptin, and 20 μg/ml chymostatin. After incubation at 4°C for 2 hr, the reactions were terminated by rapid filtration over GF/C filters that had been presoaked for 2 hr in TBS containing 2% BSA/0.1% Tween-20. The filters were rinsed 4 times with 4 ml ice-cold TBS containing 0.01% SDS. Under these conditions [3H]senktide and [125I-MePhe7]NKB-specific binding amounted to 90–95% of the total binding and to less than 5% of the total radioactivity. Nonspecific absorbtion to filters amounted to less than 0.02% of the total radioactivity and to about 5–10% of the total binding. Competition binding experiments were carried out with 4 nM [3H]senktide and with 0.3 nM [125I-MePhe7]NKB. Saturation binding experiments were carried out over a concentration range of 0.1 to 270 nM[3H]senktide. Nonspecific binding was defined with 1000-fold excess of unlabeled senktide or with 1000-fold excess of unlabeled [MePhe7]NKB. In competition binding experiments the competitor concentration producing 50% inhibition (IC50) of the radioligand binding and the Hill coefficient (nH), values were determined from Hill plots of log (B/Bo − B) versus log of the competitor concentration, where Bo and B are specific binding in the absence and in the presence of the competitor, respectively. Equilibrium binding data (Kd and Bmax) were analyzed by a nonlinear least-squares method with the ligand program (16). The data were best fit to a single site model. All experiments were performed in duplicate and the results were confirmed in at least two independent experiments, as specified in the figure legends. The results are provided as the mean ± SEM. For competition binding experiments, IC50 values were generated from experiments using at least six competitor doses. Differences in individual IC50 values between the NK-3 receptor and NK-3 receptor homolog were determined using an unpaired student’s t test on logrithmically transformed data. Differences were considered significant at the P < 0.05 level.
Cell Stimulation and Second Messenger Response Assays.
The accumulation of total [3H]inositol phosphates was measured in the presence of 20 mM LiCl to inhibit inositol monophosphatase as described (10) using a 45-min stimulation time. Agonist-stimulated arachidonic acid release was examined as reported previously using a 15–30 min stimulation period (10). A thin layer chromatographic analysis was performed on the released radioactivity from the NK-3 receptor cell line, and the 3H comigrated with authentic arachidonic acid. This chromatographic system provided baseline separation of arachidonic acid, prostaglandin E2, prostaglandin D2, prostaglandin A2, and prostaglandin F2. NK-3 agonist-stimulated inositol phosphate or arachidonic acid release responses were not detected in untransfected CHO cells. EC50 values were determined from studies examining at least 6 doses of agonist by use of SigmaPlot (San Rafael, CA).
RESULTS
Generation and Characterization of Human NK-3 and NK-3 Receptor Homolog CHO Cell Line.
Stable CHO cell lines expressing the wild-type human NK-3 receptor and the NK-3 receptor homolog were generated using the mammalian expression vector pM2 (14), in which the neomycin resistance gene is controlled by the simian virus V40 early promoter and the cDNA of interest is controlled by the Harvey murine sarcoma virus long terminal repeat. For each receptor cDNA, approximately 30 transformants were cloned and propagated. For the NK-3 receptor bearing cell lines, clonal cell lines were initially examined by radioligand binding using [3H]senktide. For the NK-3 receptor homolog, initially a population of unselected transfectants was assayed for agonist-stimulated arachidonic acid release with various tachykinin agonists. As responses were observed, clonal cell lines were subsequently screened by agonist-stimulated responses and by [3H]senktide binding. Ultimately, NK-3 and the NK-3 receptor homolog cell lines were identified that expressed a similar number of high-affinity sites.
Radioligand Binding Studies.
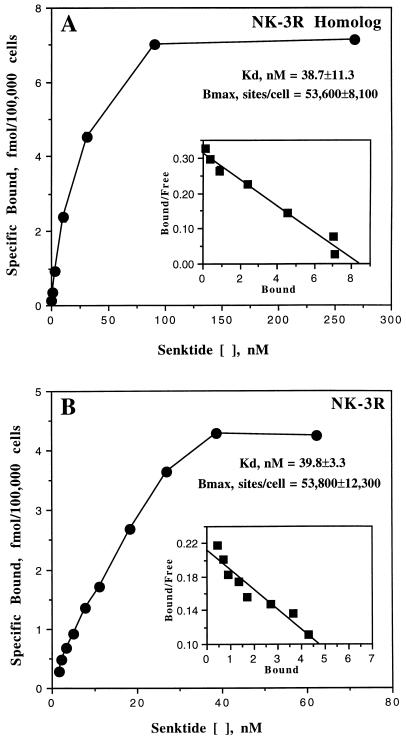
Saturation binding experiments were performed on at least two clones for each receptor type using [3H]senktide as radioligand. As shown in Fig. 1, the cell lines selected for extensive analysis displayed saturable binding. The ligand binding results were analyzed by the nonlinear least-squares method using ligand (16), and binding with both cell lines was consistent with a single affinity site model. The NK-3 and NK-3 receptor homolog had Kd values of 40 and 39 nM, respectively, and both cell lines expressed approximately 54,000 high-affinity sites per cell.
Figure 1.
Saturation of specific [3H]senktide binding to CHO cells expressing the human NK-3 receptor homolog (A) or the human NK-3 receptor (B). Binding assays were performed using increasing concentrations of [3H]senktide for 2 hr at 4°C as described. The saturation results shown are from a representative experiment, and the insets show Scatchard plots of these data. The numbers presented for the Kd and Bmax values are from three independent experiments for the NK-3 receptor homolog and from four independent experiments for the NK-3 receptor.
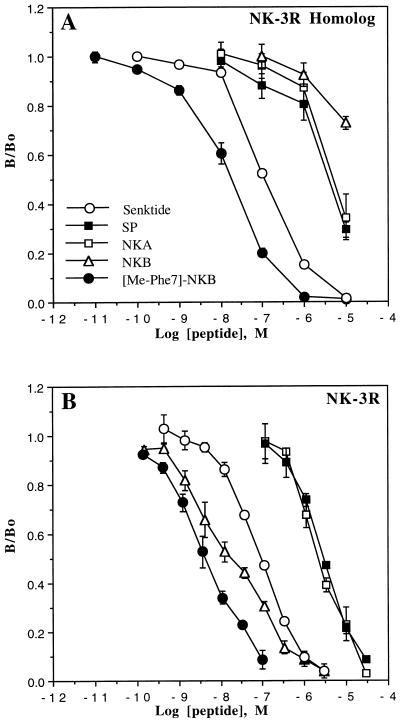
Competition binding studies using [3H]senktide as radioligand were performed with natural tachykinin agonists as well as with NK-3 receptor selective agonists and the results are shown in Fig. 2 and summarized in Table 1. The major difference observed between the two cell lines concerned the ability of NKB to displace [3H]senktide binding, where it was a weak inhibitor at the NK-3 receptor homolog cell line. At the NK-3 receptor, the rank order of natural agonist potency was NKB ≫ NKA = SP, with IC50 values of 13, 2390, and 2960 nM, respectively. The NK-3 receptor-selective agonists senktide and [Me-Phe7]NKB were effective competitors with IC50 values of 84 and 5.3 nM, respectively. The NK-3 receptor homolog cell line displayed a competition binding pharmacology quite similar to the NK-3 receptor, with one striking exception as mentioned above. Using [3H]senktide as the radioligand, the rank order of natural agonist potency was SP = NKA > NKB, with IC50 values of 4,450, 9,150, and >10,000 nM, respectively. Both [Me-Phe7]NKB and senktide were effective competitors, with IC50 values of 12 and 127 nM, respectively. Similar competition binding results to those described above were also obtained using a second independently isolated clonal cell line for each receptor type (data not shown).
Figure 2.
Inhibition of [3H]senktide binding at cloned human NK-3 receptor homolog (A) and NK-3 receptor (B) bearing CHO cell lines by tachykinin peptides. The results shown represent the mean values of three independent experiments performed in duplicate. All IC50 values are provided in Table 1. The Hill coefficients for displacement of [3H]senktide binding at the NK-3 receptor homolog for NKB, senktide, [MePhe7]NKB, SP, and NKA were 0.69, 0.99, 0.83, 0.61, and 0.85, respectively. For the NK-3 receptor, Hill coefficients for the same peptides were 0.61, 0.94, 0.72, 1.07 and 1.08, respectively.
Table 1.
Summary of the IC50 values of peptide agonist displacement of [3H]senktide and [125I-MePhe7]NKB binding at the human NK-3 receptor and human NK-3 receptor homolog
| Peptide | NK-3 receptor
|
NK-3 receptor homolog
|
||
|---|---|---|---|---|
| [3H]senktide | [125I-MePhe7]NKB | [3H]senktide | [125I-MePhe7]NKB | |
| SP | 2960 ± 60* | >10,000 | 4450 ± 390 | >10,000 |
| NKA | 2390 ± 230 | 7250 ± 2300 | 9150 ± 2440 | >10,000 |
| NKB | 13 ± 1* | 17 ± 4** | >10,000 | 126 ± 27 |
| Senktide | 84 ± 5 | 312 ± 81 | 127 ± 26 | 211 ± 14 |
| [MePhe7]NKB | 5.3 ± 0.5* | 12 ± 2* | 12 ± 1 | 45 ± 6 |
The data reported for [3H]senktide competition is derived from the data shown in Fig. 2.
Significantly different at P < 0.05 compared with that obtained at the NK-3 receptor homolog, using unpaired t test on logarithmically transformed data.
Competition binding studies using [125I-MePhe7]NKB as radioligand were also performed and the results are summarized in Table 1 by comparison with those obtained with [3H]senktide. With this iodinated radioligand, the rank order of potencey at the NK-3 receptor homolog was similar to that observed with [3H]senktide, with the major exception being that NKB was an effective competitor. The rank order of potency was [MePhe7] − NKB > senktide = NKB ≫ NKA = SP. At the NK-3 receptor, the rank order of potency was similar to that observed using [3H]senktide as radioligand.
Agonist-Stimulated Second Messenger Responses.
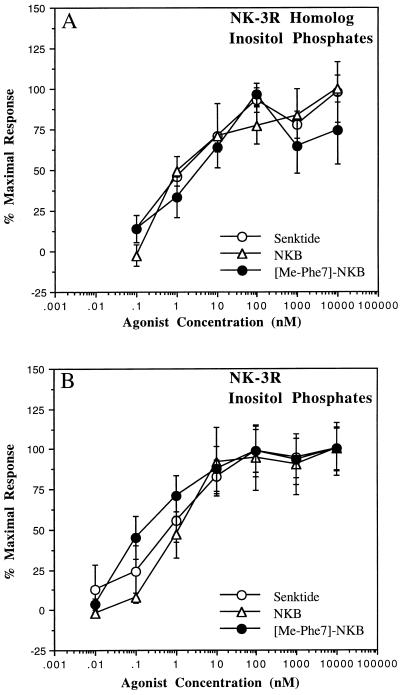
Agonist-stimulated inositol phospholipid hydrolysis and arachidonic acid release were examined at cell lines expressing both receptors using NKB, senktide, and [MePhe7]NKB, as well as SP and NKA. The concentration-response curves obtained for the various tachykinin agonists on inositol phospholipid turnover are shown in Fig. 3 and summarized in Table 2. The time course of the response using a maximal NKB concentration reached a peak level at approximately 60 min and responses were linear with respect to time for 45 min (data not shown, results summarized in Table 2). Consequently, a 45-min time point was routinely used for the concentration response studies. At the NK-3 receptor, NKB, senktide, and [MePhe7]NKB were all full agonists with EC50 values of 1.1, 5.1, and 0.5 nM, respectively. These agonists stimulated inositol phosphate production from 5.6 to 6.7 times over basal levels. SP and NKA stimulated total inositol phosphate formation with EC50 values of 128 and 77 nM, respectively (data not shown, results summarized in Table 2). At the NK-3 receptor homolog cell line, NKB, senktide, and [MePhe7]NKB were all full agonists, with EC50 values of 0.75, 2.3, and 7.1 nM, respectively. At this receptor, these agonists stimulated inositol phosphate levels from 2.14- ([MePhe7]NKB) to 3.82- (NKB) fold above basal levels. In addition, NKA and SP stimulated inositol phospholipid turnover with EC50 values of 54 and 214 nM, respectively (data not shown). Thus, the major difference between the two receptor bearing cell lines in terms of inositol phospholipid hydrolysis was the weaker responses elicited by all agonists examined at the NK-3 receptor homolog cell line.
Figure 3.
Agonist-stimulated inositol phosphate generation at human NK-3 receptor homolog (A) and human NK-3 receptor (B) CHO cell lines. Cells were prelabeled with [3H]myoinositol and agonist-stimulated inositol phosphate production was measured as described. The results shown here represent the mean ± SEM percent of maximum response from three independent experiments performed in duplicate. The maximum responses observed for the human NK-3 receptor homolog cell line ranged from 2.1- to 3.8-fold above basal levels, whereas for the human NK-3 receptor cell line the responses observed ranged from 5.6- to 6.7-fold above basal levels.
Table 2.
Summary of the EC50 values for stimulation of inositol phospholipid turnover and arachidonic acid release in CHO cell lines expressing the human NK-3 receptor and human NK-3 receptor homolog
| Peptide | NK-3 receptor
|
NK-3 receptor homolog
|
||
|---|---|---|---|---|
| Inositol phosphate | Arachidonic acid | Inositol phosphate | Arachidonic acid | |
| SP | 128 ± 38 | 153 ± 43 | 214 ± 143 | 552 ± 244 |
| NKA | 77 ± 11 | 120 ± 37 | 54 ± 16 | 237 ± 67 |
| NKB | 1.1 ± 0.5 | 9.3 ± 0.1 | 0.75 ± 0.02 | 4.2 ± 3.0 |
| Senktide | 5.1 ± 0.7 | 7.1 ± 2.4 | 2.3 ± 2.1 | 2.8 ± 1.3 |
| [MePhe7]NKB | 0.5 ± 0.1 | 0.6 ± 0.2 | 7.1 ± 6.4 | 0.6 ± 0.5 |
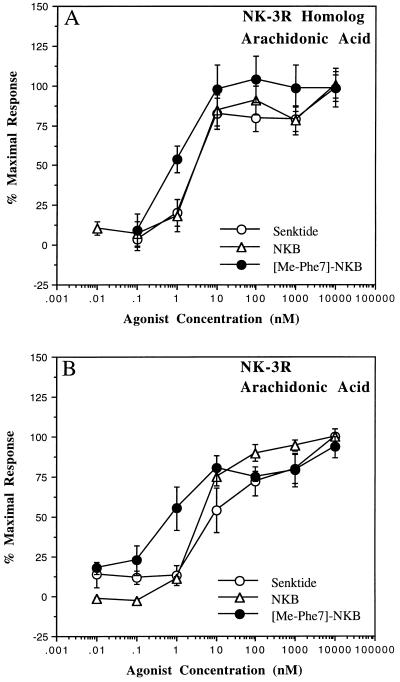
Tachykinin agonists also stimulate the release of arachidonic acid, and this response was examined in the stably transfected CHO cells. The time course of agonist-stimulated arachidonic acid release for each receptor reached a maximum from 15 to 30 min after stimulation (data not shown), consequently a 15-min time period was used to analyze concentration-response relations of various agonists (Fig. 4). At both the NK-3 receptor and NK-3 receptor homolog, the rank order of agonist potency was [MePhe7]NKB = NKB = senktide. EC50 values at the NK-3 receptor for [MePhe7]NKB, NKB, and senktide were 0.6, 9.3, and 7.1 nM, respectively. NKA and SP were weaker agonists with EC50 values of 54 and 214 nM, respectively (data not shown, results summarized in Table 2). At the NK-3 receptor homolog, EC50 values for [MePhe7]NKB, NKB, and senktide were 0.6, 4.2, and 2.8 nM, respectively. NKA and SP were much weaker yet were full agonists with EC50 values of 237 and 552 nM, respectively (data not shown, results summarized in Table 2). Furthermore, the maximum responses of the agonists at the NK-3 receptor were generally 3-fold above basal levels, whereas at the NK-3 receptor homolog the levels of released arachidonic acid were 1.4- to 1.6-fold above basal levels. Thus, the major detectable difference between the two cell lines was the stronger maximal arachidonic acid release by all agonists tested at the classical NK-3 receptor, which was about 2-fold greater than that observed with the NK-3 receptor homolog cell line.
Figure 4.
Agonist-stimulated arachidonic acid release at human NK-3 receptor homolog (A) and human NK-3 receptor (B) CHO cell lines. Cells were prelabeled with [3H]arachidonic acid and agonist-stimulated arachidonic acid release was measured as described. The results shown here represent the mean ± SEM percent of maximum response from three independent experiments performed in duplicate. The maximum responses observed for the human NK-3 receptor homolog cell line ranged from 1.4- to 1.6-fold above basal levels, whereas for the human NK-3 receptor cell line the responses observed ranged from 2.9- to 3.2-fold above basal levels.
DISCUSSION
The results of this study clearly demonstrate that the human NK-3 receptor homolog, originally cloned from a human placental library by Xie and coworkers (6), binds the NK-3 receptor selective radioligands [3H]senktide and [125I-MePhe7]NKB, and is responsive to the NK-3 receptor agonists NKB, [MePhe7]NKB, and senktide, as well as SP and NKA. NKB appears to be the natural agonist for the NK-3 receptor (1–3), whereas [MePhe7]NKB (17) and senktide (18) are NK-3 receptor selective agonists. Side-by-side comparisons with the previously identified human NK-3 receptor in saturation binding, competition binding, and agonist-stimulated inositol phospholipid hydrolysis and arachidonic acid release demonstrate that the receptors are similar in many regards. With regard to agonist interactions and agonist-stimulated second messenger responses, three differences have been observed. First, the most striking difference identified was that NKB was a weak competitor of [3H]senktide binding (IC50 value greater than 10,000 nM), though NKB was a potent, full agonist at the NK-3 receptor homolog for both inositol phosphate and arachidonic acid responses. On the other hand, when [125I-MePhe7]NKB is used as radioligand, NKB was an effective competitor, though it was 10-fold less potent at the NK-3 receptor homolog. Because [MePhe7]NKB is a decapeptide agonist, whereas senktide is a hexapeptide agonist, this result indicates a difference in the interaction of [3H]senktide at each receptor agonist-binding pocket, and the ability of NKB to compete for this site. The second series of differences was related to IC50 values for other agonists at each receptor. With [3H]senktide as radioligand, [MePhe7]NKB and SP had decreased potencies at the NK-3 receptor homolog. With [125I-MePhe7]NKB as radioligand, [MePhe7]NKB had a decreased potency at the NK-3 receptor homolog. The third difference was that the NK-3 receptor homolog CHO cell line, when stimulated with any of the tachykinin agonists, had 2-fold less robust inositol phosphate and arachidonic acid responses than that which expressed the classical NK-3 receptor, even though each cell line expressed the same number of high-affinity binding sites (approximately 55,000 sites/cell).
The NK-3 receptor homolog was originally identified (6) in the search for the κ opioid receptor cDNA as described in the Introduction. We compared the sequence of this putative receptor with previously cloned tachykinin receptor sequences (7) and observed that this NK-3 receptor homolog branched off from that of the NK-3 receptor subfamily, with an overall identity to other NK-3 receptors of approximately 85%. The major sequence differences between the human NK-3 receptor and its homolog occur in the amino terminal extracellular domain, where there exists two major sequence gaps in the NK-3 receptor homolog. Sequences in this NK-3 receptor branch are approximately 70% identical to NK-1 receptor sequences and are approximately 60% identical to NK-2 receptor sequences. Also, sequences within the NK-1 and NK-2 receptor subfamilies are more similar to each other than is the NK-3 receptor homolog sequence to other NK-3 receptor subfamily members.
Xie and coworkers (6) attempted to examine binding of this putative receptor expressed in Cos-7 cells with the tachykinin receptor ligand [3H]eledoisin. They were unable to identify specific binding, whereas the rat NK-3 receptor did bind [3H]eledoisin with the appropriate pharmacology for natural tachykinin ligands in limited studies. Eledoisin is a somewhat nonselective tachykinin radioligand that binds with a relatively high affinity to both NK-2 and NK-3 receptors and generally has high nonspecific binding. Consequently, we used for our radioligand binding studies [3H]senktide, which was developed by Selinger and coworkers (19) as an NK-3 selective agent. Our saturation and competition binding studies at both the previously characterized NK-3 receptor and at the NK-3 receptor homolog demonstrate similar affinities for the NK-3 receptor selective agonists senktide and [MePhe7]NKB. Recent studies with natural ligands compared with synthetic ligands at peptidergic receptors indicate that partially overlapping agonist subsites exist, with key differences having been observed in affinity and the effects of mutations on the respective affinities of individual agonists (20, 21). This may be the case as well for both the NK-3 receptor and the NK-3 receptor homolog. It will be of interest to determine whether NKB is the natural agonist of the novel NK-3 receptor homolog, or whether there exist additional NKB-like peptides that discriminate between these two receptors. Though an initial study questioned the existence of NK-3 receptor binding sites in primate central nervous system (22), this appears to be due to the use of low agonist concentrations in the in situ autoradiographic studies. Since the initial cloning of the rat NK-3 receptor (23), several groups have now cloned the NK-3 receptor from human brain and expressed the receptor in CHO cells to examine inositol phosphate turnover and/or Ca2+ mobilization (12, 13, 24–26) or arachidonic acid release (25, 27). Moreover, to date, there is limited evidence for NK-3 receptor homolog expression in human central nervous system (ref. 6 and this report). A detailed mRNA and protein expression analysis remains to be performed on the NK-3 receptor homolog, and this may prove to be challenging due to the extensive sequence similarities.
The issue of a functional NK-3 receptor homolog raises the question of nomenclature for the novel receptor sequence. The pharmacological classification of receptors involves both molecular recognition events (relative potencies and selectivities of agonists and antagonists) and signal transduction events (28). For the latter, similar signal transduction events and potencies have been detected between the previously characterized NK-3 receptor and its homolog in our studies reported herein, namely inositol phospholipid hydrolysis and arachidonic acid release. This is not surprising since the intracellular sequences expected to be essential for G-alphaq/11 coupling are nearly identical for the two receptors (6, 7). As to the agonist and antagonist molecular recognition events, some agonist binding differences have been noted already and an extensive antagonist analysis remains to be performed. It may be that a classification neutral label such as NK-4 would be appropriate for this NK-3 receptor homolog, since the molecular determinants of nonpeptide antagonist selectivity can be observed in receptor sequences that differ in only a limited number of residues. This has been dramatically shown in the species selectivity of nonpeptide NK-1 receptor antagonists (e.g., ref. 29 and references therein). Nonetheless, the determination of the functions, the molecular genetic mechanisms by which this receptor DNA sequence originated (gene duplication or retroviral reverse transcription event), and the extent to which multiple NK-3-related receptor sequences exist within other mammalian species will be of much interest.
Acknowledgments
We thank Drs. Guo-xi Xie and Robert Thompson for providing the human NK-3 receptor homolog cDNA. This work was supported in part by National Institutes of Health Grant NS21937.
Footnotes
Abbreviations: NKB, neurokinin B; NKA, neurokinin A; SP, substance P; NK-3, neurokinin-3; CHO, Chinese hamster ovary.
References
- 1.Otsuka M, Yoshioka K. Physiol Rev. 1993;73:229–308. doi: 10.1152/physrev.1993.73.2.229. [DOI] [PubMed] [Google Scholar]
- 2.Maggi C A, Patacchini R, Rovero P, Giachetti A. J Autonom Pharmacol. 1993;13:23–93. doi: 10.1111/j.1474-8673.1993.tb00396.x. [DOI] [PubMed] [Google Scholar]
- 3.Regoli D, Boudon A, Fauchere J L. Pharmacol Rev. 1994;46:551–599. [PubMed] [Google Scholar]
- 4.Probst W C, Snyder L A, Schuster D I, Brosius J, Sealfon S C. DNA Cell Biol. 1992;11:1–20. doi: 10.1089/dna.1992.11.1. [DOI] [PubMed] [Google Scholar]
- 5.Young D, Waitcher G, Birchmeier C, Fasano O, Wigler M. Cell. 1986;45:711–719. doi: 10.1016/0092-8674(86)90785-3. [DOI] [PubMed] [Google Scholar]
- 6.Xie G-X, Miyajima A, Goldstein A. Proc Natl Acad Sci USA. 1992;89:4124–4128. doi: 10.1073/pnas.89.9.4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krause J E, Sachais B S, Blount P. In: Handbook of Receptors and Channels. Peroutka S J, editor. Boca Raton, FL: CRC; 1994. pp. 277–298. [Google Scholar]
- 8.Yasuda K, Raynor K, Kong H, Breder C D, Takeda J, Reisine T, Bell G L. Proc Natl Acad Sci USA. 1993;90:6736–6740. doi: 10.1073/pnas.90.14.6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takeda Y, Blount P, Sachais B S, Hershey A D, Raddatz R, Krause J E. J Neurochem. 1992;59:740–745. doi: 10.1111/j.1471-4159.1992.tb09430.x. [DOI] [PubMed] [Google Scholar]
- 10.Brodbeck R M, Sachais B S, Krause J E. Mol Pharmacol. 1995;47:1065–1071. [PubMed] [Google Scholar]
- 11.Barnes W M. Proc Natl Acad Sci USA. 1994;91:2216–2220. doi: 10.1073/pnas.91.6.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang R C, Cheung A H, Mazina K E, Strader C D, Fong T M. Biochem Biophys Res Commun. 1992;184:966–972. doi: 10.1016/0006-291x(92)90685-e. [DOI] [PubMed] [Google Scholar]
- 13.Buell G, Schultz M F, Arkinstall S J, Maury K, Missoten M, Adami N, Talabot F, Kawashima E. FEBS Lett. 1992;299:90–95. doi: 10.1016/0014-5793(92)80107-r. [DOI] [PubMed] [Google Scholar]
- 14.Kozak M. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matzuk M M, Kreiger M, Corless C L, Boime I. Proc Natl Acad Sci USA. 1987;84:6354–6358. doi: 10.1073/pnas.84.18.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Munson P J, Rodbard D. Anal Biochem. 1980;107:220–239. doi: 10.1016/0003-2697(80)90515-1. [DOI] [PubMed] [Google Scholar]
- 17.Drapeau G, D’Orleans-Juste P, Dion S, Rhaleb N E, Rouissi N E, Regoli D. Neuropeptides. 1987;10:43–54. doi: 10.1016/0143-4179(87)90088-6. [DOI] [PubMed] [Google Scholar]
- 18.Wormser U, Laufer R, Hart Y, Chorev M, Gilon C, Selinger Z. EMBO J. 1986;5:2805–2808. doi: 10.1002/j.1460-2075.1986.tb04571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laufer R, Gilon C, Chorev M, Selinger Z. J Biol Chem. 1986;261:10257–10263. [PubMed] [Google Scholar]
- 20.Sagan S, Chassaing G, Pradier L, Lavielle S. J Pharmacol Exp Ther. 1996;270:1039–1048. [PubMed] [Google Scholar]
- 21.Schwartz T W, Rosenkilde M M. Trends Pharmacol Sci. 1996;17:213–216. doi: 10.1016/0165-6147(96)10017-1. [DOI] [PubMed] [Google Scholar]
- 22.Dietl M M, Palacios J M. Brain Res. 1991;539:211–222. doi: 10.1016/0006-8993(91)91623-9. [DOI] [PubMed] [Google Scholar]
- 23.Shigemoto R, Yokota Y, Tsuchida K, Nakanishi S. J Biol Chem. 1990;265:623–628. [PubMed] [Google Scholar]
- 24.Chung F Z, Wu L H, Vartanian M A, Watling K J, Guard S, Woodruff G N, Oxender D L. Biochem Biophys Res Commun. 1994;198:967–972. doi: 10.1006/bbrc.1994.1138. [DOI] [PubMed] [Google Scholar]
- 25.Oury-donat F, Carayon P, Thurneyssen O, Pailhon V, Emonds-Alt X, Soubrie P, de Fur G. J Pharmacol Exp Ther. 1995;274:148–154. [PubMed] [Google Scholar]
- 26.Pinnock R D, Suman-Chauhan N, Chung F Z, Webdale L, Madden Z, Hall D R, Woodruff G. Eur J Pharmacol. 1994;269:73–78. doi: 10.1016/0922-4106(94)90028-0. [DOI] [PubMed] [Google Scholar]
- 27.Stables J M, Beresford I J M, Arkinstall S, Seale P W, Ward P, Hagan R M. Neuropeptides. 1994;27:333–341. doi: 10.1016/0143-4179(94)90059-0. [DOI] [PubMed] [Google Scholar]
- 28.Kenakin T P, Bond R A, Bonner T I. Pharmacol Rev. 1992;44:351–362. [PubMed] [Google Scholar]
- 29.Sachais B S, Krause J E. Mol Pharmacol. 1994;46:122–128. [PubMed] [Google Scholar]