Abstract
Natural isolates of Bacillus subtilis are often difficult to transform due to their low genetic competence levels. Here we describe two methods that stimulate natural transformation. The first method uses plasmid pGSP12, which expresses the competence transcription factor ComK and stimulates competence development about 100-fold. The second method stimulates Campbell-type recombination of DNA ligation mixtures in B. subtilis by the addition of polyethylene glycol. We employed these novel methods to study the regulation of the synthetases for the lipopeptide antibiotics mycosubtilin (myc) and surfactin (srfA) in B. subtilis strain ATCC 6633. By means of lacZ reporter fusions, it was shown that the expression of srfA is >100 times lower in strain ATCC 6633 than in the laboratory strain B. subtilis 168. Expression of the myc operon was highest in rich medium, whereas srfA expression reached maximal levels in minimal medium. Further genetic analyses showed that the srfA operon is mainly regulated by the response regulator ComA, while the myc operon is primarily regulated by the transition-state regulator AbrB. Although there is in vitro evidence for a synergistic activity of mycosubtilin and surfactin, the expression of both lipopeptide antibiotics is clearly not coordinated.
The endospore-forming soil bacterium Bacillus subtilis is able to produce more than two dozen antibiotics with an amazing variety of structures. Most of these components show antimicrobial or antiviral activity (44). B. subtilis is amenable towards genetic manipulations, thanks to its ability to become naturally genetically competent. This feature would facilitate study into the production and engineering of these antibiotics were it not that the highly competent laboratory strains have lost the capacity to produce almost all antibiotics. Unfortunately, natural B. subtilis isolates that do make antibiotics appear to be difficult to transform, due to a much reduced (natural) level of competence. In this study, we describe two methods to facilitate the genetic transformability of B. subtilis strains and show their value by studying the regulation of the mycosubtilin (myc) and surfactin (srfA) synthetase operons in B. subtilis ATCC 6633.
Many bacteria produce small, modified peptides that are synthesized nonribosomally by large multienzyme complexes, i.e., the peptide synthetases (40). Owing to important medical properties of several of these peptides and to the promising engineering prospects of the peptide synthetases (11), there is a growing interest in these multienzyme complexes. Most nonribosomally synthesized peptides produced by B. subtilis are cyclic peptides with a fatty acid modification, such as surfactin, fengycin, and the members of the iturin family, including mycosubtilin. B. subtilis strain ATCC 6633 produces two lipopeptides, surfactin and mycosubtilin (8). Surfactin consists of a cyclic heptapeptide closed into a lactone ring by a β-hydroxy fatty acid. This lipopeptide exhibits strong antiviral and hemolytic activities but only a limited antibacterial activity. Surfactin seems to also be required for gliding motility (20, 30). Mycosubtilin consists of a cyclic heptapeptide closed into an amide ring by a β-amino fatty acid. Mycosubtilin exhibits a strong antifungal activity, especially against filamentous fungi (23).
In contrast to the structure and function of peptide synthetases, not much is known about the regulation of expression of these large enzyme complexes. Only the transcriptional regulation of the surfactin synthetase operon has been studied extensively, because of its role in the development of genetic competence (6, 14). Expression of srfA is medium and growth phase dependent and increases sharply at the transition from exponential to stationary-phase growth. In addition, surfactin production is associated with increased cell densities. Expression of srfA is mainly governed by the two-component regulatory system ComA and ComP (29, 35). Phosphorylation of ComA stimulates binding of this response regulator to the promoter of srfA, which induces the expression of this operon. The membrane kinase ComP senses the accumulation of ComX pheromone in the medium and activates ComA (33). Binding of ComA to the srfA promoter is inhibited by RapC. Like that of most members of the Rap family, the activity of RapC is dependent on the accumulation of a specific secreted pentapeptide, PhrC in this case. PhrC is taken up via oligopeptide permeases and represses the activity of RapC (4, 42). In addition, the srfA promoter is under direct negative control of the transcription factor CodY (39). Several other transcription factors, such as DegU and PerR, influence srfA expression as well, and it is evident that regulation of this antibiotic is part of a complex cascade that governs multiple differentiation pathways in B. subtilis (13, 16, 17).
Biochemical experiments have shown that surfactin displays a synergistic effect on the biological properties of iturin A (25). Mycosubtilin belongs to the iturin family, and B. subtilis strain ATCC 6633 produces both mycosubtilin and surfactin (8). Therefore, we wondered whether the production of these antibiotics is coordinated in this strain. Since ATCC 6633 develops poor levels of genetic competence, methods were developed to facilitate natural transformation. The first method makes use of a plasmid that increases the concentration of the competence transcription factor ComK. The second method is based on a ligation procedure that facilitates Campbell-type recombination in B. subtilis.
MATERIALS AND METHODS
General methods and materials.
Bacterial strains and plasmids used in this study are listed in Table 1. Molecular cloning and PCR procedures were carried out using standard techniques. Plasmids constructed by PCR were verified by sequencing. Oligonucleotides used for PCR are listed in Table S1 in the supplemental material. B. subtilis sporulation and minimal media were prepared as described by Schaeffer et al. (38) and Spizizen (43), respectively, and TY broth was used as rich medium. B. subtilis chromosomal DNA was purified according to the method of Venema et al. (50). Reverse transcription-PCR (RT-PCR) was performed using Superscript reverse transcriptase (Roche Diagnostics) and primers DF1, DF2, and FF2 (see Table S1 in the supplemental material). Total RNA isolations for RT-PCR were performed using a High Pure RNA isolation kit (Roche Diagnostics).
TABLE 1.
Strains and plasmids
| B. subtilis strain or plasmid | Relevant genotype/characteristics | Source or reference |
|---|---|---|
| Strains | ||
| 168-7G5 | Derivative of B. subtilis 168; surfactin positive | 46 |
| 168-8G5 | Derivative of B. subtilis 168; surfactin negative | 2 |
| AG665 | Cmr Δspo0H | 19 |
| ATCC6633 | Mycosubtilin positive; surfactin positive | 9 |
| BD1777 | Cmr ΔcomA | 12 |
| BV12E12 | (ATCC 6633) KmrmycA-lacZ; mycosubtilin negative | This work |
| BV12E13 | (ATCC 6633) KmrsrfAD-lacZ | This work |
| BV12E14 | (ATCC 6633) Kmrmyc-lacZ; mycosubtilin positive | This work |
| BV12E15 | (168-8G5) KmrsrfAD-lacZ; surfactin negative | This work |
| BV12E16 | (ATCC 6633) Cmr KmrmycA-lacZ ΔcomA | This work |
| BV12E18 | (ATCC 6633) Cmr KmrmycA-lacZ ΔabrB | This work |
| BV12E20 | (ATCC 6633) Cmr KmrmycA-lacZ ΔsinR | This work |
| BV12E22 | (ATCC 6633) Spr KmrmycA-lacZ ΔcssS | This work |
| BV12E24 | (ATCC 6633) Cmr KmrmycA-lacZ ΔdegU | This work |
| BV12E25 | (ATCC 6633) Cmr KmrmycA-lacZ ΔsrfAA | This work |
| BV12E27 | (168-8G5) Kmrmycp-lacZ; surfactin negative | This work |
| BV12E28 | (168-7G5) Kmrmycp-lacZ; surfactin positive | This work |
| BV12E29 | (ATCC 6633) Cmr KmrmycA-lacZ Δspo0K | This work |
| BV12E31 | (ATCC 6633) Cmr KmrmycA-lacZ Δspo0H | This work |
| BV12E32 | (ATCC 6633) Cmr KmrsrfAD-lacZ Δspo0K | This work |
| BV12E33 | (ATCC 6633) Cmr KmrsrfAD-lacZ Δspo0H | This work |
| BV12E35 | (168-8G5) Kmrmycp-lacZ ΔcomA; surfactin negative | This work |
| BV12E39 | (168-8G5) Kmrmycp-lacZ ΔabrB; surfactin negative | This work |
| BV12E40 | (168-7G5) KmrsrfAD-lacZ; surfactin positive | This work |
| BV12I11 | (ATCC 6633) Cmr KmrsrfAD-lacZ ΔcomA | This work |
| BV12I37 | (ATCC 6633) Cmr KmrmycA-lacZ ΔcodY | This work |
| BV12I38 | (ATCC 6633) Cmr KmrsrfAD-lacZ ΔcodY | This work |
| BV15D29 | Spr Kmr ΔcssS | 18 |
| IS432 | Cmr ΔsinR | 10 |
| JH12586 | Cmr ΔabrB | 31 |
| KI566 | Cmr ΔSpo0K | 37 |
| Plasmids | ||
| pGSP12 | Emr; contains comK | 47 |
| pLGW300 | Kmr; contains promoterless spo0V-lacZ fusion | 49 |
| pUC19C | Ampr Cmr | Lab collection, unpublished |
Transformations.
Transformation protocols for competent B. subtilis cells were based on those of Spizizen, with some adjustments (15, 43). Protoplast transformation was performed as described by Chang and Cohen (3). Transformation of pGSP12-containing B. subtilis strains was done as follows. An overnight culture was grown in minimal medium with 2.5 μg/ml erythromycin at 37°C, with continuous shaking at 300 rpm. After 100-fold dilution of the overnight culture in minimal medium, incubation was continued, and the optical density at 600 nm (OD600) was monitored. Two hours after the transition from exponential to stationary-phase growth, 1 μg of DNA was added to 0.5 ml of competent cells. Samples were kept at 37°C with shaking. After 20 min, 0.3 ml of TY medium was added, and growth was continued for another 30 min, after which the cells were plated on selective TY-agar plates.
PEG ligation.
All ligation reactions were performed overnight at room temperature, using T4 ligase and buffer from Roche Diagnostics in a total volume of 30 μl. For polyethylene glycol (PEG) ligations, a PEG 8000 solution (heat sterilized) was added to a final concentration of 15% (32).
Reporter gene fusions.
A detailed description of the construction of the different reporter gene fusions can be found in the supplemental material. To measure the expression and regulation of the srfA and myc operons, lacZ reporter gene fusions were made. For the construction of the transcriptional myc-lacZ fusion in B. subtilis ATCC 6633 (strain BV12E12), an internal part of mycA, obtained by PCR, was cloned into pLGW300. This plasmid contains the ribosomal binding site of the B. subtilis spoVG gene fused to a promoterless lacZ gene (49). In order to determine the possible effects of mycosubtilin production on the expression of myc, a transcriptional lacZ fusion was also made downstream of myc, without disrupting the operon (strain BV12E14).
To study the expression and transcriptional regulation of myc in B. subtilis 168, we inserted a transcriptional fusion of the myc promoter region with lacZ into the genome of the B. subtilis 168 derivative strain 8G5 (denoted 168-8G5), resulting in strain BV12E27. The myc promoter-lacZ fusion was inserted between dacC and ppsA, identical to the position occupied by myc in B. subtilis ATCC 6633 (8). Because plasmids containing the mycosubtilin promoter are not stable in Escherichia coli, this organism could not be used as the cloning host, and the ligation products were transformed directly into competent 168-8G5 cells. To increase the efficiency of this process, ligation was performed in the presence of PEG, as described above.
To examine whether the expression and transcriptional regulation of srfA in B. subtilis ATCC 6633 are comparable to those in B. subtilis 168-8G5, transcriptional fusions of srfA with lacZ were made in both strains, resulting in strains BV12E13 and BV12E15, respectively. B. subtilis 168 does not produce surfactin due to a defective phosphopantetheinyl transferase encoded by sfp (28). To test whether surfactin production influences srfA expression, the srfA-lacZ reporter fusion was also introduced into strain 168-7G5, which has an intact sfp gene and produces surfactin (5). Surfactin production of the resulting strain, BV12E40, was confirmed using blood agar plates, as described below.
Mutational analyses.
To determine the possible involvement of the AbrB, CodY, ComA, CssS, DegU, SinR, and Spo0K proteins in the transcriptional regulation of srfA and myc in B. subtilis ATCC 6633, the genes encoding these proteins were mutated. This was done mostly by transformation with chromosomal DNAs of B. subtilis strains already harboring the desired mutations, marked with an antibiotic resistance gene. The resulting strains are listed in Table 1, and a detailed description of the construction of these strains can be found in the supplemental material.
β-Galactosidase assays.
β-Galactosidase activities were used to measure the expression levels of the lipopeptide synthetase operons in strains harboring the lacZ fusions. For this purpose, samples were taken at hourly intervals for OD600 readings and β-galactosidase assays (49). β-Galactosidase activities were expressed as activity units per OD600 unit.
Bioassays for lipopeptide production.
The production of the lipopeptides surfactin and mycosubtilin was measured using bioassays described by Besson et al. and Mulligan and Gibbs, respectively (1, 27). For mycosubtilin production, Saccharomyces cerevisiae G910 was used as an indicator, and for surfactin production, sheep blood was used as an indicator.
RESULTS
Stimulating transformation by a plasmid-located copy of comK.
The genes coding for the DNA uptake and integration machinery are activated by a single transcription factor, the competence transcription factor ComK. Competence is a complicated process of physiological differentiation in which cell division is blocked as well. It is therefore not surprising that ComK expression is tightly regulated (reviewed in reference 16). In fact, the comK promoter is directly controlled by no fewer than five different transcription regulators, namely, ComK, Rok, CodY, AbrB, and DegU. ComK stimulates its own expression with the help of the response regulator DegU. Rok, CodY, and AbrB are repressors of the comK promoter. In addition, there is a specific adaptor protein, MecA, that targets ComK for degradation by the ClpCP system. This proteolytic control mechanism is regulated by the quorum-sensing pathways, in which, surprisingly, srfA also takes part. Even when conditions are optimal, only about 10 to 20% of cells in a B. subtilis 168 culture induce ComK. The mechanism of this bistable expression was recently solved (24, 41). The comK promoter has a low basal level of expression that fluctuates stochastically between individual cells. When in certain cells the concentration of ComK reaches a threshold level, the autostimulatory induction of comK starts to kick in, and only these cells will fully activate comK and become competent. All of the regulatory proteins listed above, aside from ComK itself, are there to keep the threshold level high to prevent premature activation of autostimulatory comK expression. The low levels of competence attained in most natural isolates of B. subtilis are likely due to ComK threshold levels that are kept too high in most cells to initiate comK autostimulation. Of course, it is also possible that the comK gene is mutated in some strains.
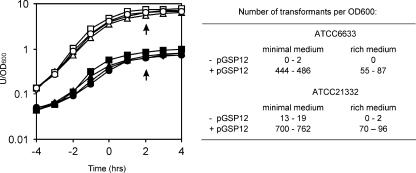
Several years ago, van Sinderen and Venema (48) showed that the introduction of a low-copy-number plasmid containing the comK gene (pGSP12) stimulates competence development and bypasses the normal medium requirements, resulting in competence development in rich medium. This observation can now be explained by assuming that the expression of the additional copies of comK lowers the comK activation threshold level substantially. We reasoned that plasmid pGSP12 could be used to stimulate genetic competence in natural isolates of B. subtilis. To test this, we introduced pGSP12 into B. subtilis strains ATCC 6633 and ATCC 21332 by means of conventional protoplast transformation. Competence was measured by transforming the different strains with chromosomal DNA from a B. subtilis strain containing a chloramphenicol resistance marker. The results are shown in Fig. 1. In minimal (competence) medium, the presence of pGSP12 increased transformation efficiencies almost 100-fold, and even in rich TY broth, which normally inhibits competence development, a substantial number of chloramphenicol-resistant transformants were obtained when pGSP12 was present.
FIG. 1.
pGSP12-stimulated transformation of B. subtilis ATCC 6633 and ATCC 21332. Growth curves for B. subtilis ATCC 6633, with (▴/▵) and without pGSP12 (▪/□), and B. subtilis ATCC 21332, with (•/○) and without (⧫/⋄) pGSP12, are presented in the graph. Closed symbols refer to growth in minimal medium, and open symbols refer to growth in rich medium. The time scale indicates hours before and after the transition from the exponential to the stationary growth phase, and arrows indicate the time of transformation. The numbers of transformants obtained are shown in the table.
srfA expression is low in B. subtilis ATCC 6633.
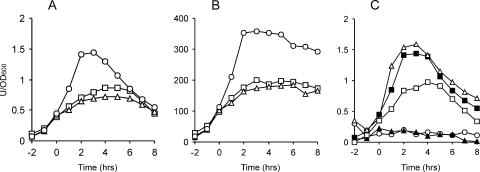
Once we had raised the competence of ATCC 6633 to convenient levels, it was possible to use classic B. subtilis genetic strategies to examine gene regulation in this strain (all ATCC 6633 derivatives were made using pGSP12, which is not mentioned further in the text). We first examined the expression of srfA. In B. subtilis 168, expression of srfA is induced at the end of the exponential growth phase and reaches the highest levels in minimal medium (49). To examine whether srfA shows similar medium- and growth phase-dependent expression in B. subtilis ATCC 6633, a srfA-lacZ transcriptional reporter gene fusion was constructed (BV12E13). As a control, we used B. subtilis strain 8G5, which is a derivative of strain 168 (BV12E15) (2). For clarity, we refer to this strain as B. subtilis 168-8G5. The activity of the promoter of srfA was measured in rich, minimal, and sporulation media at hourly intervals. Figure 2 shows that the expression of srfA in ATCC 6633 was about 200-fold lower than that in strain 168-8G5 (note the different scales). Despite this much lower expression level, the growth phase-related induction and the medium dependency of srfA expression were comparable in both strains.
FIG. 2.
srfA-lacZ expression in B. subtilis. The expression levels of srfA-lacZ in strains ATCC 6633 (A) and 168-8G5 (B), grown in rich medium (□), minimal medium (○), and sporulation medium (▵), are depicted. (C) Effects of various mutations on the expression of srfA-lacZ in B. subtilis ATCC 6633. The following cultures were grown in minimal medium: ▪, wild type; ○, ΔcomA mutant; ▵, ΔcodY mutant; ▴, Δspo0H mutant; and □, Δspo0K mutant. The time scales refer to hours before and after the transition from exponential to stationary-phase growth (defined as time zero). β-Galactosidase activities are presented in activity units per OD600 unit.
srfA induction is ComA but not CodY dependent in B. subtilis ATCC 6633.
To assess whether regulation of srfA in B. subtilis ATCC 6633 is governed by the same regulators as that in B. subtilis 168, we introduced several mutations into our srfA-lacZ reporter strain. The β-galactosidase activities of the different mutants were measured in minimal medium, and the results are depicted in Fig. 2C. First, the gene encoding the main activator of srfA in B. subtilis 168, comA, was deleted (BV12I11). From the resulting β-galactosidase levels, it is clear that also in the ATCC 6633 background, ComA is essential for srfA expression. The transcription factor CodY is a known repressor of srfA. However, in strain ATCC 6633, a codY mutation had almost no effect on srfA expression (BV12I38). We also introduced mutations into the σH-encoding gene spo0H and the oligopeptide permease gene spo0K (BV12E33 and BV12E32). Both genes modulate PhrC levels (22). Due to the stimulating effect of PhrC on ComA activity, it was not surprising that mutations in spo0H and spo0K also affected srfA expression in ATCC 6633. However, it should be mentioned that the negative effects of a spo0K mutant were not as dramatic as those published for B. subtilis 168 (13).
myc expression in B. subtilis ATCC 6633.
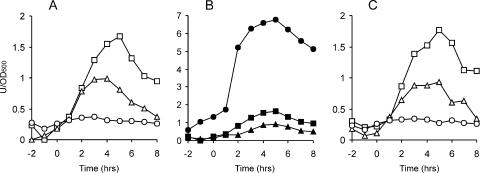
To study expression of the mycosubtilin synthetase operon, we constructed a myc-lacZ reporter gene fusion (BV12E12). Figure 3A shows the β-galactosidase activities of an ATCC 6633 strain containing this reporter fusion and grown in different media. The levels of myc expression were comparable to those found for srfA in ATCC 6633 and were also maximal in the stationary phase of growth. In contrast to the case for srfA, the expression of myc was highest in rich medium and lowest in minimal medium. Sporulation medium gave intermediate levels of expression.
FIG. 3.
myc-lacZ expression in B. subtilis. (A) Expression levels of myc-lacZ in strain ATCC 6633, grown in rich medium (□), minimal medium (○), and sporulation medium (▵). (B) Effects of various mutations on the expression of myc-lacZ in B. subtilis ATCC 6633. The following cultures were grown in rich medium: ▪, wild type; •, ΔabrB mutant; and ▴, Δspo0H mutant. (C) Expression profiles of a myc-lacZ reporter fusion in B. subtilis 168-8G5, grown in rich medium (□), minimal medium (○), and sporulation medium (▵). The time scales refer to hours before and after the transition from exponential to stationary-phase growth (defined as time zero). β-Galactosidase activities are presented in activity units per OD600 unit.
The different responses to medium compositions suggested that there are differences in the regulation of srfA and myc. A comA mutant (BV12E16) confirmed this, as the introduction of a comA mutation did not lead to reduced β-galactosidase levels (result not shown). We tested several other regulators involved in post-exponential-phase gene expression (abrB, codY, cssS [18], degU, sinR [10], spo0H, and spo0K), but only an abrB mutation (BV12E18) gave a strong response, resulting in a fivefold increase in myc induction (Fig. 3B [note the difference in scale]). Of the other regulators tested, the spo0H mutant gave a mild reduction in β-galactosidase activity (BV12E31). These data clearly show that the surfactin and mycosubtilin synthetase operons are regulated differently in B. subtilis ATCC 6633.
Construction of a myc promoter fusion by PEG ligation.
Since srfA expression was much stronger in 168-8G5 than in the ATCC 663 background, we were curious whether this would also be the case for the expression of myc. The difficulty is that B. subtilis 168 does not contain a myc operon. Instead, this strain contains, at the same locus, another lipopeptide synthetase operon, that for fengycin (8). Thus, the flanking regions of the myc operon in ATCC 6633 are exactly the same as the flanking regions of the fengycin synthetase operon in strain 168. From an evolutionary point of view, this exchange of antibiotic synthetases is fascinating, but for our purpose, the absence of myc sequences in 168 was a complicating factor. We decided to construct a myc promoter-lacZ fusion and to integrate this reporter fusion at the same locus as that in ATCC 6633, that is, downstream of dacC. The promoter of myc is likely to reside in the 960-bp intergenic region between dacC and fenF, with the latter being the first gene of the myc operon (8). This was confirmed by an RT-PCR that located a transcriptional start site within a 30-bp region 180 bp upstream of fenF (data not shown). Within this 30-bp region, there is a perfect −10 consensus sequence for a σA-dependent promoter, but no clear −35 consensus sequence is present. To be sure, the whole intergenic region between dacC and fenF was chosen for the lacZ reporter fusion. Unfortunately, cloning of the myc promoter into the cloning host E. coli appeared to be impossible. We therefore had to consider cloning the myc promoter-lacZ fusion directly into B. subtilis (see Materials and Methods for details). Campbell integrations in competent B. subtilis cells require the uptake of multimeric DNA (7). It is known that the presence of PEG during the ligation reaction inhibits the formation of closed circular monomeric DNA (32), thus resulting in a large percentage of large linear multimeric DNA molecules, the ideal substrate for Campbell-type integrations in B. subtilis. Moreover, macromolecular crowding caused by PEG strongly stimulates the ligation reaction itself. We found that the B. subtilis transformation efficiencies for DNA ligation products increased 30- to 60-fold when PEG 8000 was included in the ligation mixture (data not shown). This facilitated the direct integration of a myc promoter-lacZ fusion into the genome of B. subtilis 168-8G5 (BV12E27). As shown in Fig. 3C, the myc expression profiles for 168-8G5 were the same as those for ATCC 6633 (BV12E12). We also tested mutations in different regulators, with the same effect; also, in B. subtilis 168-8G5, the myc promoter was strongly induced, up to fivefold, when abrB was deleted (BV12E39), whereas a mutation in comA (BV12E35) had no effect (data not shown).
DISCUSSION
Here we describe two methods to facilitate genetic studies of B. subtilis. In the first method, we boost the expression of the competence transcription factor ComK by the introduction of plasmid pGSP12. There are several reasons why such an approach is of interest, including (i) pGSP12 can easily be artificially introduced into low-competence or noncompetent B. subtilis strains by means of protoplast transformation or electroporation; (ii) if necessary, the plasmid can be removed by plasmid curing; and (iii) the method will introduce a new comK gene in those strains that have lost an active copy of the gene. It is likely that the efficiency of homologous recombination depends on the measure of homology between DNA fragments. During the isolation and sequencing of the myc operon, we noticed sequence differences of up to 2% in certain genes compared with the sequence of B. subtilis 168. The differences in genome sequence between both strains might explain why we had difficulties deleting codY or degU from B. subtilis ATCC 6633 when using donor DNAs from strain 168 derivatives. Therefore, when chromosomal DNA from a different donor strain is used for transformation, good levels of competence are essential. Introduction of plasmid pGSP12 can help with this. The second method describes how ligation mixtures can be transformed into B. subtilis more efficiently when PEG is present in the reaction mix. In fact, the use of ligation products for Campbell-type integrations in B. subtilis renders the use of shuttle vectors redundant. We have used this method on several occasions to integrate antibiotic markers into the genome. Recently, a method for B. subtilis protoplast electroporation was also described, and it shows potential for transformation of natural B. subtilis isolates (36).
The two methods were applied to study the regulation of mycosubtilin synthesis. It appeared that the expression of the mycosubtilin synthetase is regulated differently from that of surfactin. We also tested whether the actual production of the lipopeptide antibiotics would trigger the induction of synthetases, but neither expression of srfA nor that of myc showed any change upon the presence of surfactin and/or mycosubtilin in the medium (data not shown).
The response regulator ComA is one of the main regulators of srfA expression. The regulation of srfA has been studied extensively because this large operon harbors a small gene, comS, which is essential for competence development. ComS blocks the degradation of ComK by the ClpCP protease complex (45). The low expression level of srfA, and therefore comS, in ATCC 6633 might explain the low level of competence of this strain. We do not know why srfA expression is much lower in ATCC 6633 than in the 168 strain. In B. subtilis 168, a mutation of the oligopeptide permease Spo0K has a detrimental effect on srfA expression because the small secreted signaling peptide PhrC cannot be taken up anymore, and therefore inhibition of ComA by ParC is not blocked (42). The genome of B. subtilis harbors another oligopeptide permease operon, namely, app. In strain 168, this gene is mutated, and the encoded permease is inactive (21). Presumably, this alternative permease is active in strain ATCC 6633, which would explain why a spo0K mutation shows only a mild effect on expression of srfA. Why a mutation in the regulator codY had no consequences for srfA expression in B. subtilis ATCC 6633 is unknown.
The ComA/ComP signal transduction pathway is the main quorum-sensing system in B. subtilis. It is therefore not surprising that this control mechanism directs the synthesis of antibiotics. In fact, the synthesis of the antibiotics bacilysin in B. subtilis and lichenysin A in Bacillus licheniformis is also dependent on ComA (26, 51). Nevertheless, the expression of mycosubtilin is governed by another regulatory cascade, in which AbrB forms the center. AbrB is a very pleiotropic regulator and is one of the main transition-state regulators in B. subtilis. In the case of the tyrocidin operon of Bacillus brevis, there is also evidence for AbrB-dependent control (26). Since σH is indirectly involved in the repression of abrB during the transition to stationary-phase growth (34), this would explain the reduced expression of myc in a spo0H background. According to Fig. 3A, an abrB mutant of ATCC 6633 still shows growth-phase-dependent induction of myc, so apparently there are more regulators involved in the regulation of this operon. With the new genetic tools described in this paper, it will now be easier to examine this in more detail.
Supplementary Material
Acknowledgments
Part of this work was supported by EU grant PL950176 and by a Wellcome Trust research career development fellowship awarded to L. W. Hamoen.
Footnotes
Published ahead of print on 6 April 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Besson, F., F. Peypoux, G. Michel, and L. Delcambe. 1979. Antifungal activity upon Saccharomyces cerevisiae of iturin A, mycosubtilin, bacillomycin L and of their derivatives; inhibition of this antifungal activity by lipid antagonists. J. Antibiot. (Tokyo) 32:828-833. [DOI] [PubMed] [Google Scholar]
- 2.Bron, S., and G. Venema. 1972. Ultraviolet inactivation and excision-repair in Bacillus subtilis. IV. Integration and repair of ultraviolet-inactivated transforming DNA. Mutat. Res. 15:395-409. [DOI] [PubMed] [Google Scholar]
- 3.Chang, S., and S. N. Cohen. 1979. High frequency transformation of Bacillus subtilis protoplasts by plasmid DNA. Mol. Gen. Genet. 168:111-115. [DOI] [PubMed] [Google Scholar]
- 4.Core, L., and M. Perego. 2003. TPR-mediated interaction of RapC with ComA inhibits response regulator-DNA binding for competence development in Bacillus subtilis. Mol. Microbiol. 49:1509-1522. [DOI] [PubMed] [Google Scholar]
- 5.Cosmina, P., F. Rodriguez, F. de Ferra, G. Grandi, M. Perego, G. Venema, and D. van Sinderen. 1993. Sequence and analysis of the genetic locus responsible for surfactin synthesis in Bacillus subtilis. Mol. Microbiol. 8:821-831. [DOI] [PubMed] [Google Scholar]
- 6.D'Souza, C., M. M. Nakano, and P. Zuber. 1994. Identification of comS, a gene of the srfA operon that regulates the establishment of genetic competence in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 91:9397-9401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dubnau, D. 1999. DNA uptake in bacteria. Annu. Rev. Microbiol. 53:217-244. [DOI] [PubMed] [Google Scholar]
- 8.Duitman, E. H., L. W. Hamoen, M. Rembold, G. Venema, H. Seitz, W. Saenger, F. Bernhard, R. Reinhardt, M. Schmidt, C. Ullrich, T. Stein, F. Leenders, and J. Vater. 1999. The mycosubtilin synthetase of Bacillus subtilis ATCC6633: a multifunctional hybrid between a peptide synthetase, an amino transferase, and a fatty acid synthase. Proc. Natl. Acad. Sci. USA 96:13294-13299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garrido, N., J. Becerra, C. Marticorena, E. Oehrens, M. Silva, and E. Horak. 1982. Antibiotic properties of ectomycorrhizae and saprophytic fungi growing on Pinus radiata D. Don I. Mycopathologia 77:93-98. [DOI] [PubMed] [Google Scholar]
- 10.Gaur, N. K., J. Oppenheim, and I. Smith. 1991. The Bacillus subtilis sin gene, a regulator of alternate developmental processes, codes for a DNA-binding protein. J. Bacteriol. 173:678-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grunewald, J., and M. A. Marahiel. 2006. Chemoenzymatic and template-directed synthesis of bioactive macrocyclic peptides. Microbiol. Mol. Biol. Rev. 70:121-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guillen, N., Y. Weinrauch, and D. A. Dubnau. 1989. Cloning and characterization of the regulatory Bacillus subtilis competence genes comA and comB. J. Bacteriol. 171:5354-5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hahn, J., and D. Dubnau. 1991. Growth stage signal transduction and the requirements for srfA induction in development of competence. J. Bacteriol. 173:7275-7282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamoen, L. W., H. Eshuis, J. Jongbloed, G. Venema, and D. van Sinderen. 1995. A small gene, designated comS, located within the coding region of the fourth amino acid-activation domain of srfA, is required for competence development in Bacillus subtilis. Mol. Microbiol. 15:55-63. [DOI] [PubMed] [Google Scholar]
- 15.Hamoen, L. W., W. K. Smits, A. de Jong, S. Holsappel, and O. P. Kuipers. 2002. Improving the predictive value of the competence transcription factor (ComK) binding site in Bacillus subtilis using a genomic approach. Nucleic Acids Res. 30:5517-5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamoen, L. W., G. Venema, and O. P. Kuipers. 2003. Controlling competence in Bacillus subtilis: shared use of regulators. Microbiology 149:9-17. [DOI] [PubMed] [Google Scholar]
- 17.Hayashi, K., T. Ohsawa, K. Kobayashi, N. Ogasawara, and M. Ogura. 2005. The H2O2 stress-responsive regulator PerR positively regulates srfA expression in Bacillus subtilis. J. Bacteriol. 187:6659-6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hyyrylainen, H. L., A. Bolhuis, E. Darmon, L. Muukkonen, P. Koski, M. Vitikainen, M. Sarvas, Z. Pragai, S. Bron, J. M. van Dijl, and V. P. Kontinen. 2001. A novel two-component regulatory system in Bacillus subtilis for the survival of severe secretion stress. Mol. Microbiol. 41:1159-1172. [DOI] [PubMed] [Google Scholar]
- 19.Jaacks, K. J., J. Healy, R. Losick, and A. D. Grossman. 1989. Identification and characterization of genes controlled by the sporulation-regulatory gene spo0H in Bacillus subtilis. J. Bacteriol. 171:4121-4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kearns, D. B., and R. Losick. 2003. Swarming motility in undomesticated Bacillus subtilis. Mol. Microbiol. 49:581-590. [DOI] [PubMed] [Google Scholar]
- 21.Koide, A., M. Perego, and J. A. Hoch. 1999. ScoC regulates peptide transport and sporulation initiation in Bacillus subtilis. J. Bacteriol. 181:4114-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lazazzera, B. A., I. G. Kurtser, R. S. McQuade, and A. D. Grossman. 1999. An autoregulatory circuit affecting peptide signaling in Bacillus subtilis. J. Bacteriol. 181:5193-5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leclere, V., M. Bechet, A. Adam, J. S. Guez, B. Wathelet, M. Ongena, P. Thonart, F. Gancel, M. Chollet-Imbert, and P. Jacques. 2005. Mycosubtilin overproduction by Bacillus subtilis BBG100 enhances the organism's antagonistic and biocontrol activities. Appl. Environ. Microbiol. 71:4577-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maamar, H., and D. Dubnau. 2005. Bistability in the Bacillus subtilis K-state (competence) system requires a positive feedback loop. Mol. Microbiol. 56:615-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maget-Dana, R., L. Thimon, F. Peypoux, and M. Ptak. 1992. Surfactin/iturin A interactions may explain the synergistic effect of surfactin on the biological properties of iturin A. Biochimie 74:1047-1051. [DOI] [PubMed] [Google Scholar]
- 26.Marahiel, M. A., M. M. Nakano, and P. Zuber. 1993. Regulation of peptide antibiotic production in Bacillus. Mol. Microbiol. 7:631-636. [DOI] [PubMed] [Google Scholar]
- 27.Mulligan, C. N., and B. F. Gibbs. 1989. Correlation of nitrogen metabolism with biosurfactant production by Pseudomonas aeruginosa. Appl. Environ. Microbiol. 55:3016-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakano, M. M., N. Corbell, J. Besson, and P. Zuber. 1992. Isolation and characterization of sfp: a gene that functions in the production of the lipopeptide biosurfactant, surfactin, in Bacillus subtilis. Mol. Gen. Genet. 232:313-321. [DOI] [PubMed] [Google Scholar]
- 29.Nakano, M. M., L. A. Xia, and P. Zuber. 1991. Transcription initiation region of the srfA operon, which is controlled by the comP-comA signal transduction system in Bacillus subtilis. J. Bacteriol. 173:5487-5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nissen, E., G. Pauli, J. Vater, and D. Vollenbroich. 1997. Application of surfactin for mycoplasma inactivation in virus stocks. In Vitro Cell Dev. Biol. Anim. 33:414-415. [DOI] [PubMed] [Google Scholar]
- 31.Perego, M., G. B. Spiegelman, and J. A. Hoch. 1988. Structure of the gene for the transition state regulator, abrB: regulator synthesis is controlled by the spo0A sporulation gene in Bacillus subtilis. Mol. Microbiol. 2:689-699. [DOI] [PubMed] [Google Scholar]
- 32.Pheiffer, B. H., and S. B. Zimmerman. 1983. Polymer-stimulated ligation: enhanced blunt- or cohesive-end ligation of DNA or deoxyribooligonucleotides by T4 DNA ligase in polymer solutions. Nucleic Acids Res. 11:7853-7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piazza, F., P. Tortosa, and D. Dubnau. 1999. Mutational analysis and membrane topology of ComP, a quorum-sensing histidine kinase of Bacillus subtilis controlling competence development. J. Bacteriol. 181:4540-4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Predich, M., G. Nair, and I. Smith. 1992. Bacillus subtilis early sporulation genes kinA, spo0F, and spo0A are transcribed by the RNA polymerase containing sigma H. J. Bacteriol. 174:2771-2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roggiani, M., and D. Dubnau. 1993. ComA, a phosphorylated response regulator protein of Bacillus subtilis, binds to the promoter region of srfA. J. Bacteriol. 175:3182-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Romero, D., A. Perez-Garcia, J. W. Veening, A. de Vicente, and O. P. Kuipers. 2006. Transformation of undomesticated strains of Bacillus subtilis by protoplast electroporation. J. Microbiol. Methods 66:556-559. [DOI] [PubMed] [Google Scholar]
- 37.Rudner, D. Z., J. R. LeDeaux, K. Ireton, and A. D. Grossman. 1991. The spo0K locus of Bacillus subtilis is homologous to the oligopeptide permease locus and is required for sporulation and competence. J. Bacteriol. 173:1388-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schaeffer, P., J. Millet, and J. P. Aubert. 1965. Catabolic repression of bacterial sporulation. Proc. Natl. Acad. Sci. USA 54:704-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Serror, P., and A. L. Sonenshein. 1996. CodY is required for nutritional repression of Bacillus subtilis genetic competence. J. Bacteriol. 178:5910-5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sieber, S. A., and M. A. Marahiel. 2003. Learning from nature's drug factories: nonribosomal synthesis of macrocyclic peptides. J. Bacteriol. 185:7036-7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smits, W. K., C. C. Eschevins, K. A. Susanna, S. Bron, O. P. Kuipers, and L. W. Hamoen. 2005. Stripping Bacillus: ComK auto-stimulation is responsible for the bistable response in competence development. Mol. Microbiol. 56:604-614. [DOI] [PubMed] [Google Scholar]
- 42.Solomon, J. M., B. A. Lazazzera, and A. D. Grossman. 1996. Purification and characterization of an extracellular peptide factor that affects two different developmental pathways in Bacillus subtilis. Genes Dev. 10:2014-2024. [DOI] [PubMed] [Google Scholar]
- 43.Spizizen, J. 1958. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc. Natl. Acad. Sci. USA 44:1072-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stein, T. 2005. Bacillus subtilis antibiotics: structures, syntheses and specific functions. Mol. Microbiol. 56:845-857. [DOI] [PubMed] [Google Scholar]
- 45.Turgay, K., L. W. Hamoen, G. Venema, and D. Dubnau. 1997. Biochemical characterization of a molecular switch involving the heat shock protein ClpC, which controls the activity of ComK, the competence transcription factor of Bacillus subtilis. Genes Dev. 11:119-128. [DOI] [PubMed] [Google Scholar]
- 46.van Sinderen, D., G. Galli, P. Cosmina, F. de Ferra, S. Withoff, G. Venema, and G. Grandi. 1993. Characterization of the srfA locus of Bacillus subtilis: only the valine-activating domain of srfA is involved in the establishment of genetic competence. Mol. Microbiol. 8:833-841. [DOI] [PubMed] [Google Scholar]
- 47.van Sinderen, D., A. Luttinger, L. Kong, D. Dubnau, G. Venema, and L. Hamoen. 1995. comK encodes the competence transcription factor, the key regulatory protein for competence development in Bacillus subtilis. Mol. Microbiol. 15:455-462. [DOI] [PubMed] [Google Scholar]
- 48.van Sinderen, D., and G. Venema. 1994. comK acts as an autoregulatory control switch in the signal transduction route to competence in Bacillus subtilis. J. Bacteriol. 176:5762-5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Sinderen, D., S. Withoff, H. Boels, and G. Venema. 1990. Isolation and characterization of comL, a transcription unit involved in competence development of Bacillus subtilis. Mol. Gen. Genet. 224:396-404. [DOI] [PubMed] [Google Scholar]
- 50.Venema, G., R. H. Pritchard, and T. Venema-Schroeder. 1965. Fate of transforming deoxyribonucleic acid in Bacillus subtilis. J. Bacteriol. 89:1250-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yazgan, A., G. Ozcengiz, and M. A. Marahiel. 2001. Tn10 insertional mutations of Bacillus subtilis that block the biosynthesis of bacilysin. Biochim. Biophys. Acta 1518:87-94. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.