Abstract
The endolithic environment, the pore space in rocks, is a ubiquitous microbial habitat. Photosynthesis-based endolithic communities inhabit the outer few millimeters to centimeters of rocks exposed to the surface. Such endolithic ecosystems have been proposed as simple, tractable models for understanding basic principles in microbial ecology. In order to test previously conceived hypotheses about endolithic ecosystems, we studied selected endolithic communities in the Rocky Mountain region of the United States with culture-independent molecular methods. Community compositions were determined by determining rRNA gene sequence contents, and communities were compared using statistical phylogenetic methods. The results indicate that endolithic ecosystems are seeded from a select, global metacommunity and form true ecological communities that are among the simplest microbial ecosystems known. Statistical analysis showed that biogeographical characteristics that control community composition, such as rock type, are more complex than predicted. Collectively, results of this study support the idea that patterns of microbial diversity found in endolithic communities are governed by principles similar to those observed in macroecological systems.
The endolithic environment, the pore space in rocks, is a ubiquitous habitat for microorganisms (16). Photosynthesis-based microbial communities inhabit the upper few millimeters to centimeters of rock exposed to the surface, forming unique microbial ecosystems. In extremely desiccated environments, such as deserts and alpine environments, endolithic ecosystems harbor much of the extant life (15). Endolithic life forms an important interface between biology and geology. Remnants of endoliths preserved in rocks can serve as biosignatures that provide clues about past life (38).
Endolithic ecosystems are among the simplest microbial ecosystems known and consequently provide a tractable model with which to test ecological principles, which remain largely untested in microbial ecology due to experimental limitations and the extraordinary diversity of microorganisms (27). Previous studies have suggested specific ecological hypotheses for endolithic communities (14, 15), but these hypotheses have been difficult to examine with traditional culture-based microbiological techniques because most environmental microbes are not cultured by standard techniques (5).
Therefore, we determined the compositions of selected endolithic communities with cultivation-independent, rRNA-based molecular phylogenetic methods. The results provided incisive identification of microbial constituents and specific DNA sequence information that can be compared universally to study microbial ecosystems. We used these data, along with data from a previous study of Antarctic endolithic communities (9), to test the following hypotheses: (i) endolithic communities are among the simplest microbial ecosystems known (15); (ii) endolithic communities are ecological communities that are characterized by consistent compositions of microorganisms that cooccur within a defined habitat (14); (iii) the endolithic environment is extreme from the human perspective and, as such, is expected to be seeded from a relatively small reservoir (metacommunity) of microorganisms highly adapted to the endolithic environment (14); and (iv) biogeographical characteristics, such as the physical and chemical properties of rock types, climate, and direction of exposure, influence the specific microbial composition of endolithic communities (15).
MATERIALS AND METHODS
Study sites.
The study sites are located in semiarid montane zones in Colorado and Wyoming. Unless stated otherwise, samples were collected from south- to southeast-facing, nearly vertical cliff faces. The Forest Quarry site (39.98472°N, 105.28833°W) is an historical sandstone quarry in Boulder, CO, and is a member of the Lyons formation sandstone unit. Exclamation point (39.56028°N, 107.31937°W) is a highly pure (>97.5%) limestone cliff near Glenwood Springs, CO, and is part of the Leadville limestone unit (3). Sinks Canyon (42.74175°N, 108.82486°W) is located near Lander, WY, and contains granite, sandstone, and limestone cliffs (8). Samples were collected from limestone, sandstone, and granite cliffs along the northwest canyon walls, which have southeast exposures to the sun. Samples of sandstone and limestone communities were collected from sites <10 m apart in a horizontally layered deposit. Owl Canyon (45.75100°N, 10.79800°W) is near Fort Collins, CO, and cuts through layers of uniformly sorted sandstone and nearly pure limestone.
Sample collection.
Samples up to a depth of ∼1 cm were collected with flame-sterilized chisels. Duplicate samples were collected ∼30 cm apart and frozen at ≤−80°C for transport and storage. All samples were collected in April 2001.
Genomic DNA extraction.
Genomic DNA was extracted from ∼0.5-g crushed rock samples by bead beating with phenol and sodium dodecyl sulfate (28). Rock samples (∼5 g) were crushed and homogenized with a flame-sterilized steel ore crusher (Fisher Scientific). DNA concentrations were determined with a PicoGreen double-stranded DNA quantitation kit (Molecular Probes) and a Turner Quantech fluorometer (Barnstead Thermolyne). The average yields of extracted DNA were ∼5 μg/g of crushed rock. Control extractions were performed with washed sand (Fisher Scientific) that was sterilized for 24 h at ∼300°C. Procedures were performed in a UV-sterilized AirClean 600 PCR workstation (AirClean Systems).
PCR amplification of rRNA genes.
Small-subunit rRNA genes were amplified by PCR performed with community DNA as the template and universal primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) or 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 1391R (5′-GACGGGCGGTGWGTRCA-3′) (21). Each 50-μl PCR mixture contained ∼50 ng template DNA, 1.25 U HotMaster Taq DNA polymerase (Eppendorf), 1× HotMaster buffer (2.5 mM Mg2+), 1 mg/ml bovine serum albumin, each deoxynucleoside triphosphate at a concentration of 50 μM, and each primer at a concentration of 0.2 μM. The PCR conditions were 94°C for 2 min, 30 cycles of 94°C for 15 s, 52°C for 15 s. and 65°C for 60 s, and a final 20-min extension at 65°C. PCR controls with no template or extraction controls as the template were negative.
Clone library construction.
Two to 13 libraries of 96 randomly selected rRNA gene clones were constructed from independent PCR and cloning reactions for each community DNA sample. PCR-amplified products from four independent PCRs were pooled to reduce the chances of PCR artifacts (20), purified by agarose gel electrophoresis with a Montage DNA gel extraction kit (Millipore), and cloned into the pCR4.0 vector with a TOPO TA cloning kit and TOP10 chemically competent cells (Invitrogen). Clones were grown for ∼16 h at 37°C in ∼1 ml of LB broth on an orbital shaker at ∼250 rpm. Plasmids were isolated by an alkaline lysis miniprep method (18) or by the “boiled biomass” method (a 1:4 dilution of a culture in 1× Tris-EDTA was heated in a thermal cycler to ∼85°C for 10 min and centrifuged at 3,000 relative centrifugal force for 5 min).
RFLP analysis.
Unique rRNA gene clones were identified by restriction fragment polymorphism (RFLP) analysis of PCR-amplified plasmids with the HinPI and MspI restriction enzymes (18).
DNA sequence determination.
rRNA gene clones were sequenced with a Licor Global 4200 (Licor) or MegaBACE 1000 (Amersham Biosciences) DNA sequencer.
DNA sequence analysis.
DNA sequence data were analyzed as previously described (28). Sequences were compared to known sequences in the GenBank database by BLAST. Sequences were aligned with the NAST sequence aligner (10). The phylogenetic analysis was performed with the Arb software package (26) and Mr. Bayes (30).
Statistical methods.
To estimate richness and coverage, sequences were collected into operational taxonomic units based on sequence identity at different fixed levels of identity from 95 to 100% in 1% increments, as previously described (28). Uncorrected distance matrices were calculated with Arb. Operational taxonomic unit richness and coverage estimators were calculated with the software program EstimateS, as previously described (28). Communities were compared with the UniFrac software package (24) with phylogenetic trees calculated in Arb.
Nucleotide sequence accession numbers.
The sequences of 588 rRNA gene clones determined have been deposited in the GenBank database under accession numbers EF522190 to EF522777.
RESULTS
A previous study showed that endolithic communities inhabit up to ∼90% of exposed rock formations at numerous sites across the Colorado Plateau, a physiographic region that spans ∼340,000 km2 of Utah, Colorado, Arizona, and New Mexico (5). We surveyed large, exposed outcrops of sandstone, limestone, and granite in the Rocky Mountain region and found abundant endolithic growth just beneath the surface of fractured rock samples. Based on this survey, four distinct study sites were chosen.
The microbial composition of Rocky Mountain endolithic communities was determined by rRNA gene census. For each community, two 96-clone libraries were constructed from duplicate community DNA samples extracted from rock samples collected ∼30 cm apart. Unique clones were identified by RFLP analysis (11). Clones representative of RFLP types were sequenced and compared to known sequences to determine their phylogenetic affiliations (28). Approximately 2,500 clones were analyzed by the RFLP method, and ∼1,000 sequences were determined. The results are summarized in Tables 1 and 2.
TABLE 1.
Phylogenetic compositions of Rocky Mountain endolithic communitiesa
| Taxon | % of rRNA clones in community
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Sandstone
|
Limestone
|
Sinks Canyon granite | Avg | |||||
| Forest Quarry | Owl Canyon | Sinks Canyon | Exclamation Point | Owl Canyon | Sinks Canyon | |||
| Bacteria | ||||||||
| Acidobacteria | 1 | 4 | 4 | 4 | 1.9 | |||
| Actinobacteria | 5 | 31 | 21 | 16 | 22 | 32 | 41 | 24.0 |
| Bacteroidetes | 1 | 21 | 2 | 2 | 9 | 7 | 1 | 6.1 |
| Chlorobi | 1 | 0.1 | ||||||
| Chloroflexi | 2 | 1 | 7 | 4 | 2 | 6 | 3.1 | |
| Cyanobacteria | 23 | 22 | 48 | 48 | 23 | 24 | 2 | 27.1 |
| Deinococcus | 1 | 8 | 1.3 | |||||
| Firmicutes | 2 | 2 | 1 | 1 | 2 | 9 | 2.4 | |
| Gemmimonas | 1 | 2 | 1 | 0.6 | ||||
| OP10 | 1 | 2 | 2 | 0.7 | ||||
| Planctomycetes | 1 | 1 | 0.3 | |||||
| Proteobacteria | 6 | 7 | 11 | 13 | 21 | 18 | 7 | 11.9 |
| Verrucomicrobia | 6 | 0.9 | ||||||
| Archaea | ||||||||
| Crenarchaeota | 19 | 2 | 8 | 25 | 7.7 | |||
| Eucarya | ||||||||
| Chlorophytes | 58 | 1 | 12 | 2 | 10 | 1 | 12.1 | |
| Total | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
The results represent combined RFLP and sequence data for replicate libraries.
TABLE 2.
Data for Rocky Mountain endolithic communitiesa
| Community | Avg BLAST identity (%)b | No. of libraries analyzed | No. of clones analyzed | No. of RFLP types | No. of clones sequenced | Chao1 richness | CACE coverage (%) | No. of operational taxonomic units (97% identity) |
|---|---|---|---|---|---|---|---|---|
| Sandstone | ||||||||
| Forest Quarry | 94 | 13 | 1,185 | 29 | 300 | 35 | 98 | 30 |
| Owl Canyon | 92 | 4 | 384 | 18 | 104 | 18 | 77 | 17 |
| Sinks Canyon | 94 | 2 | 192 | 57 | 91 | 93 | 87 | 48 |
| Limestone | ||||||||
| Exclamation Point | 91 | 6 | 576 | 18 | 107 | 28 | 96 | 14 |
| Owl Canyon | 92 | 4 | 384 | 40 | 142 | 38 | 84 | 45 |
| Sinks Canyon | 95 | 2 | 192 | 43 | 84 | 61 | 69 | 41 |
| Sinks Canyon granite | 94 | 2 | 192 | 38 | 165 | 46 | 87 | 21 |
| Avg or sum | 93 | 33 | 3,105 | 243 | 993 | 319 | 85 | 216 |
The results represent combined RFLP and sequence data for replicate libraries.
Average sequence identity of closest GenBank relative for each community.
Sample representativeness.
Microbial communities are complex, and gene surveys rarely sample the complete genetic diversity. Therefore, we assessed the richness and coverage of samples with the Chao1 and CACE estimators, as well as the autosimilarity of the duplicate samples (28). The results, shown in Table 2, indicate that the typical goal, to sample ≥50% of the estimated richness, was achieved for most samples. The autosimilarity of duplicate libraries was assessed with the Morisita-Horn similarity index (7) for RFLP data and with UniFrac for sequence data (24). The Morisita-Horn results ranged from 0.47 to 0.96 (1 indicates identity), with an average of 0.79, and the UniFrac results indicated no significant differences (P > 0.26) between replicate libraries. Collectively, these results suggest that the clone libraries are representative samples of the incidence and abundance of the rRNA genes present. The similarity observed between replicate community samples from the same site indicates that the communities are well-defined assemblages, at least on a scale of ∼30 cm. Thus, the results show that endolithic communities are not relatively random assemblages of organisms at any location.
Microbial composition.
Overall, the bacterial sequences were most abundant and diverse, representing ∼15 of the ∼75 known bacterial phyla (23). Eucaryal plastid and archaeal sequences also were common, constituting up to ∼25% of the clones in some communities. Comparison of the communities revealed significant similarities in overall composition at the broadest phylogenetic levels. Typically, >95% of sequences from a community belonged to just nine bacterial divisions. The actinobacterial, cyanobacterial, and proteobacterial sequences were the most common and accounted for the largest fractions of each community. Remarkably, most communities shared some rRNA genes with ≥97% identity, which corresponds approximately to species-level relationships. This shows that a few broadly related kinds of organisms are most common in Rocky Mountain endolithic communities and thus form the foundation of such communities.
Oxygenic phototrophs are considered the main primary producers of endolithic communities (16) and constituted a substantial fraction of the endolithic communities (Table 1). Heterotrophic organisms are known to inhabit endolithic communities and are considered mostly consumers of chemical energy derived from autotrophic primary producers (36). While it is convenient to attribute heterotrophy to organisms that are not oxygenic phototrophs, recent studies have revealed that many other kinds of autotrophic metabolism contribute substantially to primary productivity in microbial ecosystems This has blurred the traditional dichotomy between heterotrophy and autotrophy. Examples include lithotrophic crenarchaeaota and anoxygenic photosynthetic bacteria in the oceans (4, 19). While oxygenic photosynthesis is undoubtedly a driving energy source for endolithic communities, other kinds of autotrophic metabolism likely contribute to the overall energy budget, particularly among abundant organisms.
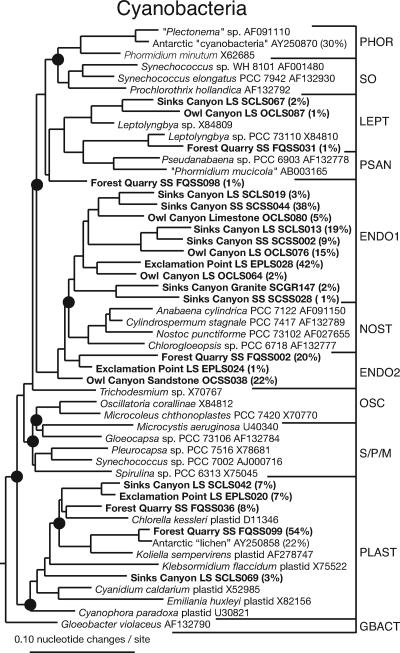
Phylogenetic analysis of sequences representative of oxygenic phototrophs showed that the majority belong to two novel cyanobacterial clades, as shown in Fig. 1. The average distances between these sequences were ∼11%, and all sequences were ≤97% identical. Comparison of cyanobacterial sequences from each community with UniFrac revealed significant differences between all communities (P ≥ 0.34), which implies that different cyanobacteria inhabit each community and that the organisms are considerably different phylogenetically from cultivated representatives. The Forest Quarry community was dominated by algal plastid sequences, the majority of which were most closely related (∼97%) to sequences from an Antarctic endolithic community (9). Nuclear rRNA genes representative of eucaryotic algae also were cloned in this study; however, the frequency was less (∼1% of plastid sequences).
FIG. 1.
Cyanobacteria: diagrammatic tree of Rocky Mountain endolithic cyanobacterial sequences and reference sequences representative of cyanobacterial diversity (35). Representative endolithic sequences are indicated by bold type. The percentages indicate the abundance of sequence types in the community. Solid circles indicate nodes with ≥70% bootstrap support as determined by neighbor joining with maximum likelihood rate corrections and Bayesian inference. The tree was rooted with Escherichia coli. The following sequence groups (35) are indicated by brackets on the right: Phormidium (PHOR), Synechococcus (SO), Leptolynbia (LEPT), Pseudoanabaena (PSAN), endolith-specific group 1 (ENDO1), Nostocales (NOST), endolith-specific group 2 (ENDO2) Oscillatoria (OSC), Synechocystis/Pleurocapsa/Microcystis (S/P/M), chloroplasts (PLAST), and Gloeobacter (GBACT). Rock types are indicated as follows: SS, sandstone; LS, limestone; and GR, granite.
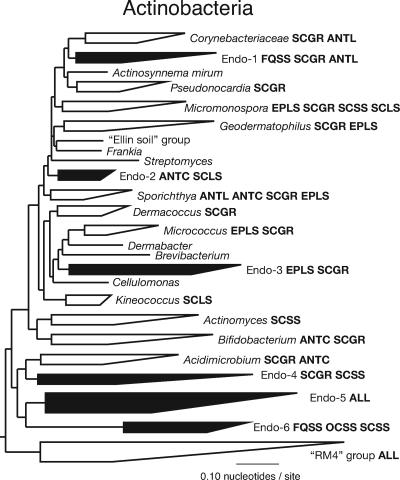
Sequences representative of organisms other than oxygenic phototrophs comprised a substantial portion of the endolithic communities (average, ∼60% of clones). These sequences were related to known sequences at an average level of only ∼93% (at least genus-level difference) and were associated with ∼16 bacterial phyla, as well as crenarchaeotes. Such organisms most likely contribute to both heterotrophic metabolism and autotrophic metabolism. Indeed, the high abundance of some sequence types perhaps indicates that there is unrecognized autotrophy in the phylogenetic group indicated by the sequences. The most conspicuous sequence groups included the groups representative of the Actinobacteria, Bacteroidetes, Chloroflexi, Firmicutes, Proteobacteria, and Crenarchaeota. Collectively, along with the oxygenic phototrophs, such organisms likely form the core of Rocky Mountain endolithic communities. Of these phyla, actinobacterial sequences were the most common, abundant (average, 24%), and diverse group, as shown in Fig. 2.
FIG. 2.
Actinobacteria: diagrammatic tree showing the broad distribution of endolithic rRNA gene sequences. The open wedges represent the main recognized groups within the Actinobacteria (32). The solid wedges represent new actinobacterial groups formed by endolithic sequences (Endo-1 to Endo-6). Communities with sequences that belong to a group are indicated by four-letter codes that indicate the study site (FQ, Forest Quarry; EP, Exclamation Point; OC, Owl Canyon; SC, Sinks Canyon) and the rock type (SS, sandstone; LS, limestone; GR, granite). The general topology of the tree is supported by neighbor-joining bootstrap and Bayesian analysis (not shown) and is consistent with previous phylogenetic analyses (32).
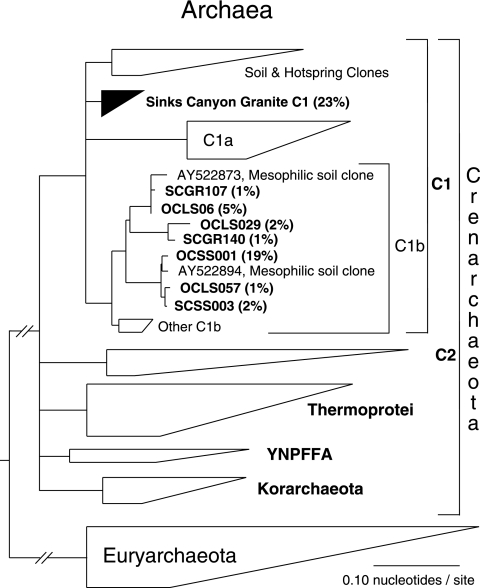
All Rocky Mountain communities had actinobacterial rRNA sequences most closely related (∼92 to 98% sequence identity) to a variety of actinobacterial rRNA sequences cloned from Antarctic endolithic communities (9). Archaea are increasingly recognized as important, global players in the biosphere (22) and constituted as much as ∼25% of the clones of Rocky Mountain endolithic communities. All archaeal sequences were representatives of the “C1” group of the Crenarchaeota (Fig. 3), which are among the most commonly encountered and abundant environmental archaeal sequences (20). A substantial portion, ∼25%, of the clones from the Sinks Canyon granite community were archaeal, and the majority of these clones represent a new group within the “C1” group of the Crenarchaeota (Fig. 3).
FIG. 3.
Archaea: diagrammatic tree showing the phylogenetic affiliations of representative Rocky Mountain endolithic sequences (bold type) determined in the context of a recent comprehensive archaeal phylogeny (29). The tree was rooted with bacterial sequences. Representative clones and their fractions (expressed as percentages) in the community are shown in bold type. The communities were Sinks Canyon granite (SCGR), Sinks Canyon sandstone (SCSS), and Owl Canyon limestone (OCLS).
Community comparisons.
In order to compare the phylogenetic compositions of Rocky Mountain endolithic communities and to test the hypothesis that rock type or other site-specific characteristics influence the phylogenetic composition of endolithic communities, we statistically compared sequence sets representative of each community with UniFrac (24). Sequence sets representative of two distinct Antarctic sandstone communities (24) were included in this analysis in order to compare their phylogenetic composition to that of Rocky Mountain communities. We also included a sequence set that represents a high-temperature (∼70°C) hot spring community (31) as an outgroup community.
The results indicate that the overall phylogenetic compositions of most communities differ significantly (P < 0.05) at the finest phylogenetic scales, approximately equivalent to the species level and finer. However, lineage-specific analysis that compared sequences at higher taxonomic levels (UniFrac G test) (25) revealed that all communities shared significantly similar lineages (P > 0.05) at taxonomic levels of approximately genus and higher. One notable exception of the species-level comparisons is that of the Forest Quarry sandstone and Antarctic “lichen-dominated” communities, where UniFrac analysis indicated a strong probability that the sequences were drawn from a similar phylogenetic population (P = 0.887). These results suggest that site-specific characteristics other than rock type alone likely influence the finer-scale phylogenetic structure of these communities, but that on broader phylogenetic scales the communities have significant similarities in their compositions.
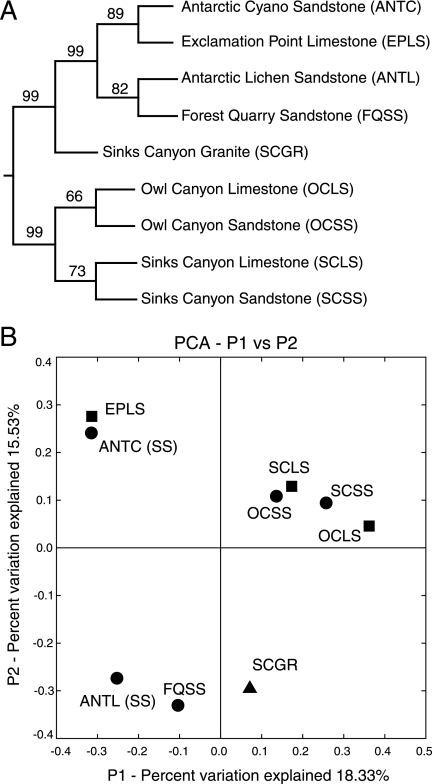
To explore further the relationships between the phylogenetic compositions of the communities, we compared a matrix of UniFrac distances from each pairwise comparison of representative sequence sets with the unweighted pair group method with arithmetic mean (UPGMA) cluster algorithm (24) and principal coordinate analysis (25), as shown in Fig. 4. The results show a pattern of relationships more complicated than expected. For example, most communities do not cluster by rock type. Instead, sandstone and limestone communities from Owl Canyon and Sinks Canyon cluster by site, which suggests that other site-specific characteristics or geography may have a stronger influence on community composition than rock type. However, this does not explain the relatively distant relationship of the Sinks Canyon granite community, which appears as an outlier. Remarkably, the Antarctic communities are more closely related to Rocky Mountain communities than to each other. Manual and statistical comparison of sequences representative of clustered communities offered possible explanations for this.
FIG. 4.
UniFrac statistical comparison of Rocky Mountain endolithic communities based on phylogenetic compositions: pairwise UniFrac distances calculated from an Arb phylogenetic tree. (A) UPGMA tree of UniFrac distances showing the overall phylogenetic relationships of the communities. The values indicate the percent Jackknife support at each node based on 1,000 random samplings. The tree was rooted with a Yellowstone hot spring community (31). (B) Principal coordinate analysis (PCA) of UniFrac distances resulted in a clustering pattern similar to that observed by UPGMA. The rock types are sandstone (•), limestone (▪), and granite (▴).
The Forest Quarry community and the Antarctic “lichen-dominated” community are the only communities with significantly similar phylogenetic structures of rRNA gene sequences, which indicates that the microbial compositions of these communities are similar. Both communities were dominated by nearly identical sequences (∼99% sequence identity with GID AY250858) representative of eucaryotic algae. Analysis of these sequences indicated that their closest known relative, cultured or uncultured, is only ∼90% identical (Fig. 1). This shows that unique, closely related algal species dominate endolithic communities in these distant environments. Other closely related sequences shared by these communities included representatives of the α-proteobacterial order Rhizobiales (∼94% identity between the communities) and the genus Acetobacter (95 to 97% identity [genus level]). These communities also are distinguished by a relatively low abundance of actinobacterial sequences.
Cyanobacterial sequences prevailed in the Forest Quarry community and were most closely related to other Rocky Mountain sequences. The putatively lichen-dominated Antarctic communities have proven to be rare elsewhere, and the original observation that such communities were devoid of cyanobacteria was disproved previously (14). In the present study no evidence of fungi was obtained by rRNA sequence analysis of the Forest Quarry community, and microscopic analysis (37) showed the algae occur free in the rock pore spaces. Thus, the term “lichen” as sometimes applied to these communities is a misnomer.
The Exclamation Point and Antarctic “cyanobacterium-dominated” communities are characterized by low rRNA gene diversity relative to other communities in this study. Comparison of the communities showed a remarkable similarity in rRNA gene sequences in four phylogenetic groups, which were ∼95 to 98% identical. This shows that organisms related at the genus and species levels inhabit these two communities in different environments in different substrate rocks (sandstone and limestone) separated by thousands of kilometers. The closely related sequences shared by these two communities included representatives of the genus Rhodobacter (∼95% identical, α-Proteobacteria phylum), the genus Cytophaga (∼96%, Bacteroidetes phylum), and two actinobacterial genera, Micrococcus (∼97%) and Frankia (∼93 to 96%). In contrast, cyanobacterial sequences in the different communities were related at levels of identity of only ∼83 to 90%, which showed only remote relationships. This difference is the most significant difference between the communities as measured by UniFrac analysis (P < 0.001).
A previous study of Antarctic endolithic communities suggested that anoxygenic bacterial photosynthesis may be a significant metabolism in these communities based on an abundance of sequences related to known bacterial phototrophs, such as Rhodobacter spp. (24). Rocky Mountain communities harbored similar sequences. However, attempts to use PCR to amplify the bacterial photosystem genes pufL and pufM (1) were unsuccessful (37). Furthermore, pigment analyses failed to detect pigments associated with anoxygenic photosynthesis (e.g., bacteriochlorophylls) (37). Nonetheless, the abundance of the sequences suggests that these organisms are important to the ecology of endolithic communities.
Sandstone and limestone communities from Owl Canyon and Sinks Canyon were among the most diverse and complex communities examined. Relatively few sequences in these communities were related at levels of identity of >95%; the exceptions were a few cyanobacterial sequences. Nonetheless, sequences from these four communities tended to group together to the exclusion of sequences from the other communities. Furthermore, the sandstone and limestone communities from the same study site shared more distinct sequence groups. These associations help explain the clustering patterns observed in the UniFrac analysis (Fig. 4). The four communities clustered due to related cyanobacterial, actinobacterial, and proteobacterial sequences, which accounted for the majority of the sequences from these communities. Communities from the same study site further associated because they shared unique groups of Bacteroidetes and crenarchael sequences (Owl Canyon) and acidobacterial, Firmicutes, and Chloroflexi sequences (Sinks Canyon).
DISCUSSION
Studies of endolithic ecosystems have relied primarily on culture and microscopy to identify microorganisms. Most environmental microbes (>99%) are not cultured with standard methods (2, 27), and morphological identification is often unreliable, even for morphologically conspicuous organisms, such as cyanobacteria (9, 17). Culture-independent molecular methods for the identification of microbes transformed our perception of the natural microbial world, and increasingly large data sets aim to resolve the extent, structure, and ecological implications of microbial diversity. Although we acknowledge current technological limitations, such as small sample size and potential biases due to experimental artifacts (28), the results provide a basis for evaluating the ecological hypotheses articulated above (see the Introduction).
Simplicity of endolithic communities.
The observed and predicted sequence diversities of Rocky Mountain communities are consistent and support the hypothesis that endolithic communities are among the simplest microbial ecosystems (14). We estimate that the diversity of Rocky Mountain and Antarctic endolithic communities inspected so far by molecular methods is on the order of 102 species-level clades. In contrast, recent diversity estimates for well-studied soil and seawater environments range from 105 to 109 species cm−3; the large range is due to limited sample size and uncertainty in community structure (12). Our assessments of sample representativeness suggest that clone libraries from this study indeed were representative samples of the abundant rRNA genes.
Ecological communities.
The endolithic communities studied had relatively consistent and uniform compositions at a scale of ∼30 cm. A related study at the Forest Quarry site revealed no significant variation in community composition at spatial scales of up to ∼20 m and temporal scales of up to ∼1.5 years (37). These findings indicate that endolithic ecosystems form true ecological communities, characterized by consistent compositions that cooccur within a defined habitat (14).
Global metacommunity.
Significant broad-scale similarities among the endolithic communities studied show that they are comprised of organisms from a relatively limited reservoir of phylogenetic diversity. This supports the idea that endolithic communities are seeded from a distinct global metacommunity uniquely adapted to the endolithic niche (14). For example, all communities shared sequences that were >97 to 98% identical, which indicates that similar species inhabit different communities. In one case, the Forest Quarry and Antarctic “lichen-dominated” communities were not significantly different, and ∼50% of the sequences were ∼97 to 98% identical, a remarkable concordance seldom seen in molecular community studies. Thus, endolithic ecosystems may be viewed as “extreme,” in the sense that relatively few kinds microorganisms appear to be adapted to the endolithic environment due to its specific requirements.
Biogeographical controls.
The biogeographical characteristics that control endolithic community composition are more complex than predicted. Cluster analysis revealed unexpected associations between endolithic communities. Associations of sandstone and limestone communities at the Sinks Canyon and Owl Canyon sites suggest that a site-specific characteristic, such as local climate or water chemistry, has a stronger influence than rock type alone. The two sites have morphologically similar banded deposits of fine-grained sandstone and almost pure limestone. Although the exact nature of the correlation between the canyon sites is unknown, this correlation suggests a possible biogeographical explanation for the composition of the communities. Further study is required to discern the biogeographical characteristics that control community composition.
Ecological implications.
Endolithic communities are considered true ecosystems because they function as isolated systems within the endolithic environment (14). This is especially true in desiccated environments, where little other life exists. Ecosystems often are studied using large-scale measures, such as energy fluxes, nutrient cycles, and biodiversity patterns. Such characteristics are emergent properties of ecosystems that reflect complex dynamics between organisms and their environments. A classic example from macroecology correlates biodiversity and ecosystem productivity (34). From this standpoint, the endolithic communities are relatively low in diversity and therefore are predicted to have relatively low productivity.
The distribution patterns of microbial diversity are poorly known. While these patterns often are predicted to resemble patterns observed in macroecological systems, few studies support these predictions. In fact, a general lack of discernible patterns advanced the idea that free-living microbes are globally dispersed, and high invasion rates discourage speciation and high microbial diversity (13). Some workers have argued that the microbial world is too dynamic for communities to have characteristic diversity (6). However, the results of this study and other studies (40) support the hypothesis that microbial diversity exhibits distinct patterns that reflect ecosystem function.
Collectively, results of this study support a more complicated pattern of community composition between endolithic communities than expected. Nevertheless, the broad-scale similarity and low diversity of endolithic communities suggest that they may be seeded from a cosmopolitan metacommunity of organisms adapted to the endolithic environment, likely disbursed by wind (14). The desiccation resistance of endolithic organisms (5, 14) and the patterns of colonization of building materials (39) are consistent with wind dispersal. The fine-scale phylogenetic differences among the communities indicate an undetermined biogeographical influence on community composition or a stochastic component to community establishment. Probably both factors pertain.
Regardless of the mechanism that selects the particular community composition, spatial and temporal studies of Rocky Mountain endolithic communities support the idea that endolithic communities are highly stable ecosystems (14) that may persist on geological time scales (∼104 years) at some locations (33). This apparent stability is an exception to the diversity-stability hypothesis, which predicts greater stability for more diverse ecosystems. We propose that the stability of these simple yet stable endolithic systems results from a lack of resource competition due to the unusual requirements for endolithic life. The stability of endolithic ecosystems also may be explained by the oligotrophic nature of the endolithic environment, low productivity, and low turnover rates (5, 14). Further study of endolithic ecosystems should shed light on these and other basic questions in microbial ecology.
Acknowledgments
We thank J. Spear for assistance with sample collection, C. Robertson and K. Harris for help with phylogeny, and all Pace lab members for their support and input.
This work was supported by NSF grant DEB-0085490 to N.R.P.
Footnotes
Published ahead of print on 6 April 2007.
REFERENCES
- 1.Achenbach, L. A., J. Carey, and M. T. Madigan. 2001. Photosynthetic and phylogenetic primers for detection of anoxygenic phototrophs in natural environments. Appl. Environ. Microbiol. 67:2922-2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barton, H. A., M. R. Taylor, and N. R. Pace. 2004. Molecular phylogenetic analysis of a bacterial community in an oligotrophic cave environment. Geomicrobiol. J. 21:11-20. [Google Scholar]
- 4.Beja, O., E. N. Spudich, J. L. Spudich, M. Leclerc, and E. F. DeLong. 2001. Proteorhodopsin phototrophy in the ocean. Nature 411:786-789. [DOI] [PubMed] [Google Scholar]
- 5.Bell, R. A. 1993. Cryptoendolithic algae of hot semiarid lands and deserts. J. Phycol. 29:133-139. [Google Scholar]
- 6.Brock, T. D. 1987. The study of microorganisms in situ: progress and problems. Symp. Soc. Gen. Microbiol. 41:1-17. [Google Scholar]
- 7.Colwell, R. R. 1997. Microbial diversity: the importance of exploration and conservation. J. Ind. Microbiol. Biotechnol. 18:302-307. [DOI] [PubMed] [Google Scholar]
- 8.Dahms, D. E. 2004. Relative and numeric age data for pleistocene glacial deposits and diamictons in and near Sinks Canyon, Wind River Range, Wyoming, USA. Arct. Antarct. Alp. Res. 36:59-77. [Google Scholar]
- 9.de la Torre, J. R., B. M. Goebel, E. I. Friedmann, and N. R. Pace. 2003. Microbial diversity of cryptoendolithic communities from the McMurdo Dry Valleys, Antarctica. Appl. Environ. Microbiol. 69:3858-3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeSantis, T. Z., P. Hugenholtz, K. Keller, E. L. Brodie, N. Larsen, Y. M. Piceno, et al. 2006. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 34:W394-W399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dojka, M. A., P. Hugenholtz, S. K. Haack, and N. R. Pace. 1998. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 64:3869-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunbar, J., S. M. Barns, L. O. Ticknor, and C. R. Kuske. 2002. Empirical and theoretical bacterial diversity in four Arizona soils. Appl. Environ. Microbiol. 68:3035-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finlay, B. J. 2002. Global dispersal of free-living microbial eukaryote species. Science 296:1061-1063. [DOI] [PubMed] [Google Scholar]
- 14.Friedmann, E. I., and R. Ocampo-Friedmann. 1984. Endolithic microorganisms in extreme dry environments: analysis of a lithobiontic microbial habitat, p. 177-185. In M. J. Klug and C. A. Reddy (ed.), Current perspectives in microbial ecology. American Society for Microbiology, Washington, DC.
- 15.Friedmann, E. I. 1982. Endolithic microorganisms in the Antarctic cold desert. Science 215:1045-1053. [DOI] [PubMed] [Google Scholar]
- 16.Friedmann, E. I., and R. Ocampo. 1976. Endolithic blue-green-algae in dry valleys—primary producers in Antarctic desert ecosystem. Science 193:1247-1249. [DOI] [PubMed] [Google Scholar]
- 17.Giovannoni, S. J., S. Turner, G. J. Olsen, S. Barns, D. J. Lane, and N. R. Pace. 1988. Evolutionary relationships among cyanobacteria and green chloroplasts. J. Bacteriol. 170:3584-3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hugenholtz, P., C. Pitulle, K. L. Hershberger, and N. R. Pace. 1998. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180:366-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ingalls, A. E., S. R. Shah, R. L. Hansman, L. I. Aluwihare, G. M. Santos, E. R. Druffel, et al. 2006. Quantifying archaeal community autotrophy in the mesopelagic ocean using natural radiocarbon. Proc. Natl. Acad. Sci. USA 103:6442-6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanagawa, T. 2003. Bias and artifacts in multitemplate polymerase chain reactions (PCR). J. Biosci. Bioeng. 96:317-323. [DOI] [PubMed] [Google Scholar]
- 21.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, New York, NY.
- 22.Leininger, S., T. Urich, M. Schloter, L. Schwark, J. Qi, G. W. Nicol, et al. 2006. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806-809. [DOI] [PubMed] [Google Scholar]
- 23.Ley, R. E., J. K. Harris, J. Wilcox, J. R. Spear, S. R. Miller, B. M. Bebout, et al. 2006. Unexpected diversity and complexity of the Guerrero Negro hypersaline microbial mat. Appl. Environ. Microbiol. 72:3685-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lozupone, C., and R. Knight. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228-8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lozupone, C., M. Hamady, and R. Knight. 2006. UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7:371-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, et al. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pace, N. R. 1997. A molecular view of microbial diversity and the biosphere. Science 276:734-740. [DOI] [PubMed] [Google Scholar]
- 28.Papineau, D., J. J. Walker, S. J. Mojzsis, and N. R. Pace. 2005. Composition and structure of microbial communities from stromatolites of Hamelin Pool in Shark Bay, Western Australia. Appl. Environ. Microbiol. 71:4822-4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robertson, C. E., J. K. Harris, J. R. Spear, and N. R. Pace. 2005. Phylogenetic diversity and ecology of environmental Archaea. Curr. Opin. Microbiol. 8:638-642. [DOI] [PubMed] [Google Scholar]
- 30.Ronquist, F., and J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 31.Spear, J. R., J. J. Walker, T. M. McCollom, and N. R. Pace. 2005. Hydrogen and bioenergetics in the Yellowstone geothermal ecosystem. Proc. Natl. Acad. Sci. USA 102:2555-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stackebrandt, E., F. A. Rainey, and N. L. Ward-Rainey. 1997. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int. J. Syst. Bacteriol. 47:479-491. [Google Scholar]
- 33.Sun, H. J., and E. I. Friedmann. 1999. Growth on geological time scales in the Antarctic cryptoendolithic microbial community. Geomicrobiol. J. 16:193-202. [Google Scholar]
- 34.Tilman, D., D. Wedin, and J. Knops. 1996. Productivity and sustainability influenced by biodiversity in grassland ecosystems. Nature 379:718-720. [Google Scholar]
- 35.Turner, S. 1997. Molecular systematics of oxygenic photosynthetic bacteria. Plant Syst. Evol. Suppl. 11:13-52. [Google Scholar]
- 36.Vestal, J. R. 1988. Carbon metabolism of the cryptoendolithic microbiota from the Antarctic Desert. Appl. Environ. Microbiol. 54:960-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walker, J. J. 2005. Ph.D. thesis. University of Colorado, Boulder.
- 38.Walker, J. J., J. R. Spear, and N. R. Pace. 2005. Geobiology of a microbial endolithic community in the Yellowstone geothermal environment. Nature 434:1011-1014. [DOI] [PubMed] [Google Scholar]
- 39.Warscheid, T., and J. Braams. 2000. Biodeterioration of stone: a review. Int. Biodeterior. Biodegrad. 46:343-368. [Google Scholar]
- 40.Whitaker, R. J., D. W. Grogan, and J. W. Taylor. 2003. Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science 301:976-978. [DOI] [PubMed] [Google Scholar]