Abstract
Our study focused on a Mesorhizobium sp. that is phylogenetically affiliated by 16S rRNA gene sequence to other marine and saline bacteria of this genus. Liquid chromatography-mass spectrometry investigations of the extract obtained from solid-phase extraction of cultures of this bacterium indicated the presence of several N-acyl homoserine lactones (AHLs), with chain lengths of C10 to C16. Chromatographic separation of the active bacterial extract yielded extraordinarily large amounts of two unprecedented acylated homoserine lactones, 5-cis-3-oxo-C12-homoserine lactone (5-cis-3-oxo-C12-HSL) (compound 1) and 5-cis-C12-HSL (compound 2). Quorum-sensing activity of compounds 1 and 2 was shown in two different biosensor systems [Escherichia coli MT102(pSB403) and Pseudomonas putida F117(pKR-C12)]. Furthermore, it was shown that both compounds can restore protease and pyoverdin production of an AHL-deficient Pseudomonas aeruginosa PAO1 lasI rhlI double mutant, suggesting that these signal molecules maybe used for intergenus signaling. In conclusion, these data indicate that the quorum-sensing activity of compounds 1 and 2 is modulated by the chain length and functional groups of the acyl moiety. Additionally, compound 1 showed antibacterial and cytotoxic activities.
Homoserine lactones (HSLs) possess, in addition to the lactone moiety, an acyl side chain. The chain length (C4 to C18), substitution at position 3′, and existence of a double bond in the acyl chain vary in naturally occurring HSLs.
Bacteria employ sophisticated cell-to-cell communication systems to coordinate their activities and to act in a concerted manner. Gram-negative bacteria produce N-acyl homoserine lactones (AHLs), signal molecules which allow the producer to monitor its own population density in a process known as quorum sensing (QS) (for reviews see references 3, 5, and 39). These regulatory systems typically rely on two proteins, an AHL synthase, which is usually a member of the LuxI family of proteins, and an AHL receptor protein belonging to the LuxR family of transcriptional regulators. At low population densities, cells produce a basal level of AHLs via the activity of the AHL synthase. As the cell density increases, AHLs accumulate in the growth medium. Upon reaching a critical threshold concentration, AHL molecules bind to their cognate receptor, which in turn activates or represses expression of target genes.
Compared to the case for bacteria from terrestrial environments, to date relatively little information is available on AHL-based QS systems in marine bacteria. Most notably, however the first organism for which AHL-dependent cell-to-cell signaling was demonstrated was the marine bacterium Vibrio fischeri. Moreover, recent work has shown that AHLs are of major importance for the transcriptional control in many marine Vibrio and Photobacterium species (4), and different Roseobacter strains isolated from marine snow were capable of AHL production (9). In the free-living marine bacterium Rhodobacter sphaeroides, a QS system was identified that relies on an AHL molecule with a C14 side chain and a double bond at C-7 [7-cis-N-(tetradecenoyl)HSL], the synthesis of which is directed by the AHL synthase CerI (21). Inactivation of the cerI gene resulted in increased exopolysaccharide production and the formation of large aggregates of cells in liquid culture.
Mesorhizobium species are present in marine habitats and were found during investigation of the bacterial population in sponge samples. They have been isolated from two Demospongiae (28) and were detected by DGGE analysis in the sponge Cymbastela concentrica (34). The biological nature of the sponge-bacterium association, however, is not known; bacteria may be either symbiotic, specific and permanently associated but not symbiotic, or merely commensally present.
The genus Mesorhizobium belongs to the class Alphaproteobacteria and to the order Rhizobiales, together with Sinorhizobium, Bradyrhizobium, and Rhizobium. Bacteria of these genera are well-investigated nitrogen-fixing organisms living in symbiosis with plants (6). Members of the genus Mesorhizobium were formerly classified in the genus Rhizobium; phylogenetic differences and specific phenotypic characteristics, however, led to their transfer into the separate genus Mesorhizobium (12). Mesorhizobium species are not homogenous but are highly variable, containing symbiotic and nonsymbiotic bacteria (23). These organisms are well known for their cell-cell communication by secretion and sensing of small diffusible signaling molecules (8). Thus, AHL production was shown to occur in the symbiotic plant bacterium Mesorhizobium huakuii (37, 42) but has not yet been observed for marine-derived Mesorhizobium species.
AHL production by marine bacteria isolated from sponge samples has to date been described solely for a Vibrio sp. and an alphaproteobacterium from the Roseobacter-Ruegeria subgroup (33). A Mesorhizobium sp. obtained from the marine sponge Phakellia ventilabrum thus attracted our attention due to the extremely high levels of AHLs detected in its culture medium. AHL molecules differ in the length and substitution pattern of the acyl side chain, structural motifs that seem to determine the specificity of the regulators’ response. In this context, the recently identified unusual long-chain AHLs in marine Alphaproteobacteria are of special interest (36). Here we report the isolation and structure of two unusual long-chain (C12) AHLs from the culture of a marine Mesorhizobium sp. Both compounds showed bioactivity in a range of assay systems for QS: Escherichia coli and Pseudomonas putida biosensor strains carrying plasmids with luxR-gfp and lasB-gfp transcriptional fusions, which are optimally responsive to AHLs with short (C6) and long (C12) chain lengths, and a lasI rhlI double mutant of Pseudomonas aeruginosa. Furthermore, compound 1 exhibited antimicrobial activity against Bacillus brevis and cytotoxic activity against two tumor cell lines, whereas compound 2 was inactive.
MATERIALS AND METHODS
Sponge collection.
The sponge Phakellia ventilabrum (class Demospongiae) was collected from the Korsfjord near Bergen, Norway, at a depth of 250 m. After sampling, the sponge was directly transferred into a container with fresh seawater and kept in the cold until arrival in the Berlin laboratory. The sponge was cut into approximately 0.5- by 1-cm pieces and transferred into a continuous-culture system, where the pieces of sponge tissue were fed with oligotrophic medium (0.1% MB [Difco], 3% Reef Crystals [Aquarium Systems, France], 5% silicate, 1 μM retinol [Sigma]) at 8°C in a cold room under aerobic conditions.
Isolation of bacteria.
After 53 days of cultivation in continuous culture, a piece of the sponge tissue was harvested and homogenized in sterile artificial seawater in a glass tube squeezer. The extract was serially diluted and distributed on agar plates with low nutrient concentrations (substrate concentrations were as mentioned above, with 1% MB and 1.5% Bacto agar). Subcultivation was always performed on MB agar plates. Incubation was performed at 8°C.
Phylogenetic characterization.
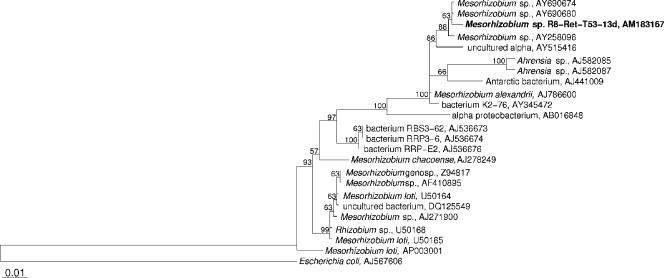
DNA was extracted from liquid cultures of the bacterium by the alkaline lysis method (0.25% sodium dodecyl sulfate, 50 mM NaOH) and incubation at 95°C for 15 min. Amplification of the 16S rRNA gene was performed with the universal bacterial primers 616V (5′-AGA GTT TGA TYM TGG CTC AG) and 1525R (5′-AAG GAG GTG WTC CAR CC). Cycling conditions were as follows: 1.5 min at 96°C; 35 cycles of 2 min at 96°C, 3 min at 57°C, and 1.5 min at 72°C; and a final extension step at 72°C for 10 min. The complete 16S rRNA gene sequence was determined by Services in Molecular Biology, Berlin. Sequence data were assembled with the ABI Prism autoassembler program, analyzed with the ARB software (14), and compared to the EMBL/GenBank nucleotide sequence database. The tree (see Fig. 1) was calculated with partial 16S rRNA gene sequences (1,420 bp; E. coli positions 42 to 1462) by using the maximum-parsimony and maximum-likelihood methods implemented in ARB.
FIG. 1.
Phylogenetic maximum-parsimony tree calculated with partial 16S rRNA gene sequences (1,420 bp). The isolated Mesorhizobium sp. strain R8-Ret-T53-13d is indicated in bold. Maximum-parsimony (1,000 resamplings) bootstrap values are provided for relevant groups. Only values of >50% are shown. The maximum-likelihood consensus tree shares the same topology. The scale bar indicates 1% sequence divergence. The tree was rooted using Escherichia coli as an outgroup.
Cultivation, extraction, and isolation.
The Mesorhizobium sp. (strain R8-Ret-T53-13d) was chosen for cultivation due to striking antimicrobial and cytotoxic activity found in a screening program for bioactive secondary metabolites. The strain was cultivated in 20 liters of LB2 medium (tryptone, 2.5 g/liter; yeast extract, 2.5 g/liter; glucose, 5 g/liter; NaCl, 10 g/liter; sea weed extract, 2.5 ml/liter; marine salts, 17 g/liter) with 1 kg Diaion HP21 resin (Mitsubishi Chemical Industry Ltd., Düsseldorf) at 22°C, 150 rpm, and 4.5 liters air/min in a fermentor (Biostat A-20; Braun, Melsungen) for 4 days. As the inoculum, 12 ml of a 3-day-old culture in LB2 was used. The cultures were harvested after 4 days by filtration over a Büchner funnel. The HP21 resin was stirred with 3 liters acetone and 100 ml water for 30 min. After filtration, the acetone was evaporated and the water phase extracted twice with ethyl acetate. Evaporation of the combined ethyl acetate phases yielded 250 to 440 mg crude extract per fermentor.
Isocratic fractionation (2 ml/min, acetonitrile-H2O gradient) with a reversed-phase C18 column (Macherey-Nagel Nucleodur 100; 250 mm by 10 mm, 5 μm) yielded 10 fractions. High-pressure liquid chromatography (HPLC) separation of fraction 8 yielded 3 mg of compound 1. Fraction 10 gave 25 mg of pure compound 2. Recultivation of the bacteria under identical conditions in 60 liters culture broth and reisolation gave 36 mg of compound 1 and 35 mg of compound 2. HPLC was carried out using a Waters system consisting of a 600 pump, a 996 photodiode array detector, a 717 plus autosampler, and a fraction collector.
Structure elucidation.
All nuclear magnetic resonance (NMR) spectra were recorded on Bruker Avance 500 DRX or 300 DPX spectrometer. Spectra were referenced to residual solvent signals with resonances at δH/C 7.26/77.0 (CDCl3-d1). UV and infrared (IR) spectra were obtained with Perkin-Elmer Lambda 40 and Perkin-Elmer Spectrum BX instruments, respectively. High-resolution electrospray ionization-Fourier transform-ion cyclotron resonance-mass spectrometry (HR-ESI-FT-ICR-MS) measurements were recorded on a Bruker Daltonics APEX III FT-ICR-MS spectrometer. Low-resolution (LR)-ESI-MS measurements were recorded with an API 2000 instrument (Applied Biosystems/MDS Sciex).
5-cis-3-oxo-C12-HSL (compound 1).
Data for compound 1 (36 mg, 0.6 mg/liter) are as follows: [α]25D −12° (c 0.94 MeOH); UV (MeOH) λmax (log ɛ) 223 (3.57), 286 (3.38) nm; IR (ATR) νmax 3316, 2925, 1776, 1710, 1658, 1537, 1172 cm−1; LR-ESI-MS m/z 296 [M + H+], 294 [M − H+]; HR-FT-ICP-MS m/z 294.17120 (calculated for [M − H+] C16H24NO4 294.17053). 1H and 13C NMR data are given in Table 1.
TABLE 1.
One- and two-dimensional NMR spectral data for compounds 1 and 2 in CDCl3
| Position | Compound 1 (5-cis-3-oxo-C12-HSL)
|
Compound 2 (5-cis-C12-HSL)
|
||||
|---|---|---|---|---|---|---|
| 13Ca,c δ (ppm) | 1Hb,c δ (ppm) (multiplicity, J [Hz]) | COSYb,d | HMBCb,e | 13Ca,c δ (ppm) | 1Ha,c δ (ppm) (multiplicity, J [Hz]) | |
| 2 | 174.7 (Cf) | 175.5 (C) | ||||
| 3 | 49.0 (CH) | 4.58 (m) | 4a/b | 49.3 (CH) | 4.57 (m) | |
| 4a/b | 29.9 (CH2) | 2.76 (m), 2.25 (m) | 3, 5a/b | 2, 3 | 30.7 (CH2) | 2.89 (m), 2.18 (m) |
| 5a/b | 66.0 (CH2) | 4.48 (t, 8.8), 4.28 (m) | 4a/b | 2, 3 | 66.1 (CH2) | 4.49 (t, 8.4), 4.31 (m) |
| 1′ | 166.2 (C) | 173.6 (C) | ||||
| 2′ | 47.3 (CH2) | 3.51 (s) | 1′, 3′ | 35.5 (CH2) | 2.28 (t, 7.3) | |
| 3′ | 204.5 (C) | 25.3 (CH2) | 1.73 (p, 7.3) | |||
| 4′ | 42.6 (CH2) | 3.28 (d, 7.0) | 5′ | 3′, 5′, 6′ | 26.5 (CH2) | 2.10 (q, 7.3) |
| 5′ | 119.0 (CH) | 5.51 (dt, 9.6, 7.0) | 4′, 6′, 7′ | 128.2 (CH) | 5.36 (m)g | |
| 6′ | 135.3 (CH) | 5.67 (dt, 9.6, 7.0) | 5′, 7′ | 131.3 (CH) | 5.45 (m)g | |
| 7′ | 27.6 (CH2) | 2.03 (q, 7.0) | 5′, 6′ | 5′, 6′, 8′ | 27.3 (CH2) | 2.02 (q, 6.6) |
| 8′ | 29.2 (CH2) | 1.29 (m) | 29.6 (CH2) | 1.29 (m) | ||
| 9′ | 28.9 (CH2) | 1.29 (m) | 29.0 (CH2) | 1.29 (m) | ||
| 10′ | 31.7 (CH2) | 1.29 (m) | 31.7 (CH2) | 1.29 (m) | ||
| 11′ | 22.6 (CH2) | 1.29 (m) | 12′ | 12′, 10′ | 22.6 (CH2) | 1.29 (m) |
| 12′ | 14.1 (CH3) | 0.88 (t, 7.0) | 11′ | 11′, 10′ | 14.1 (CH3) | 0.90 (t, 7.0) |
300/75.5 MHz.
500/125.7 MHz.
Assignments are based on extensive one- and two-dimensional NMR measurements (HMBC, HSQC, and COSY).
Numbers refer to proton resonances.
Numbers refer to carbon resonances.
Implied multiplicities determined by DEPT.
JH5′-H6′ = 10.6 Hz, determined with homonuclear decoupling NMR experiments.
5-cis-C12-HSL (compound 2).
Data for compound 2 (25 mg, 1.3 mg/liter) are as follows: [α]25D −7° (c 0.42 MeOH); UV (MeOH) λmax (log ɛ) 217 (3.44) nm; IR (ATR) νmax 3311, 2923, 1777, 1650, 1535, 1175, 1021 cm−1; LR-ESI-MS m/z 282 [M + H+], 280 [M − H+]; HR-FT-ICP-MS m/z 280.19190 (calculated for [M − H+] C16H26NO3 280.19182). 1H and 13C NMR data are given in Table 1.
Antimicrobial assays.
Antibacterial activities were tested with Bacillus brevis (ATCC 9999), Micrococcus luteus (ATCC 21415), Staphylococcus aureus (ATCC 11632), Acinetobacter calcoaceticus (DSM 30006), and Proteus vulgaris (DSM 30119), for antifungal activities Nematospora corylii (ATCC 10647), Mucor miehei (Tü 284), Fusarium solani (CBS 166.87), and Candida albicans (ATCC 10231) were used. Bacteria were grown on nutrient agar (Difco) at 37°C and fungi on YMG agar. Test plates were seeded with 5 × 105 cells or spores per ml. Filter disks (6-mm diameter) were loaded with the compounds to be tested (50 μg) and placed on the agar plates. The diameters of inhibition zones were monitored after 24 h.
Cytotoxic activities.
Cytotoxic activity was assayed as described previously (41) with slight modifications. Jurkat cells (ATCC TIB 152) were grown in RPMI 1640 medium (GIBCO, BRL), and HeLa S3 cells (ATCC CCL 2.2) were grown in Dulbecco modified Eagle medium (GIBCO, BRL), supplemented with 10% fetal calf serum (GIBCO, BRL), 65 μg/ml of penicillin G, and 100 μg/ml of streptomycin sulfate. The assay mixtures contained 1 × 105 cells/ml medium.
Detection of AHLs.
The AHL biosensor strains Escherichia coli MT102(pSB403) (40) and Pseudomonas putida F117(pKR-C12) (29) were grown overnight in Luria-Bertani (LB) medium (1) at 30°C, diluted fourfold in fresh medium, and grown for another hour. Aliquots (100 μl) of cultures were pipetted into the wells of microtiter plates (FluoroNunc; Polysorp). Compound 1, compound 2, or synthetic AHLs (3-oxo-C6-HSL and 3-oxo-C12-HSL) were added to the wells at final concentrations ranging from 0 to 1,000 nM. The microtiter plates were incubated at 30°C for 4 h before bioluminescence and green fluorescent protein fluorescence were measured with a Sirius HT reader (MWG Biotech). Data were processed with the KC4 software (Bio-Tek Instruments). The biosensor P. putida F117(pKR-C12) was also used to visualize compounds 1 and 2 on thin-layer chromatography (TLC) plates. To this end, 10-μl samples of compounds 1 and 2 and 5 μl of the AHL standard 3-oxo-C12-HSL were applied to C18 reversed-phase TLC plates (Merck no. 1.15389) and dried with a stream of cold air. Samples were separated using methanol (60%, vol/vol) in water as the mobile phase. For detection of AHLs, the TLC plate was overlaid with a thin film of 0.8% (wt/vol) LB agar (143 ml) seeded with 7 ml of an exponentially grown AHL biosensor and was then incubated at 30°C for 24 h. Green fluorescence was visualized by illuminating plates with blue light, using an HQ 480/40 filter (F44-001; AHF-Analysentechnik, Tübingen, Germany) in combination with a halogen lamp (Intralux 5000-1; Volpi, Schlieren, Switzerland) as a light source. Illumination took place in a darkbox equipped with a C2400-40 camera connected to a Pentax CCTV camera lens and an HQ 535/20 filter (F42-001; AHF-Analysentechnik).
Measurement of protease activity and pyoverdin production in Pseudomonas aeruginosa.
Wild-type P. aeruginosa PAO1 and the AHL-deficient lasI rhlI double mutant were grown overnight in LB medium in the absence and in the presence of 350 nM 3-oxo-C12-HSL and 700 nM of compounds 1 and 2. Proteolytic activity was measured as described previously (19). Briefly, 250 μl of 2% (wt/vol) azocasein (Sigma) in 50 mM Tris-HCl and 150 μl of the sterile filtered supernatants were incubated for 4 h at 4°C. After precipitation of undigested substrate with 1.2 ml of 10% (wt/vol) trichloroacetic acid for 15 min at room temperature, followed by 10 min of centrifugation at 15,000 rpm, 1.4 ml of 1 M NaOH was added to the supernatant. The A440 of the supernatant was measured. Pyoverdin production in the same cultures was monitored by absorbance of sterile filtered supernatants at 380 nm as described elsewhere (21).
Nucleotide sequence accession number.
The 16S rRNA gene sequence was deposited in the EMBL database under accession number AM183167.
RESULTS
Isolation and phylogenetic characterization.
By using a continuous-culture system with an oligotrophic medium and subsequent serial dilutions on low-nutrient agar plates, the bacterium R8-Ret-T53-13d (BOSMAN stock collection) was isolated from the marine sponge Phakellia ventilabrum. The sponge originated from 250 m of depth from the Korsfjord near Bergen in Norway. The isolated bacterium is phylogenetically closely related to the genus Mesorhizobium of the class Alphaproteobacteria. As shown in Fig. 1, the 16S rRNA gene of the isolated bacterium has more than 99% homology to two Mesorhizobium species (accession no. AY690674 and AY690680; unpublished) isolated from the rhizosphere soil of salt marshes from the southwestern coasts of Korea as well as to the Mesorhizobium sp. strain DG1023 (AY258096) (10) associated with the dinoflagellate Gymnodinium catenatum, which causes paralytic shellfish poisoning. Mesorhizobium alexandrii obtained from the toxic dinoflagellate Alexandrium minutum (AJ786600; unpublished), bacterium K2-76 from the Hawaiian Lake Kauhako (AY345472; unpublished), an Antarctic bacterium from microbial mats of Antarctic lakes (AJ441009) (35), and an uncultured alphaproteobacterium from the coastal region of the German Wadden Sea (AY515416) (30) show 98 to 99% homology to Mesorhizobium sp. strain R8-Ret-T53-13d. Furthermore, Mesorhizobium sp. strain R8-Ret-T53-13d shows 97 to 98% homology to the two Ahrensia spp. from the dinoflagellate Alexandrium lusitanicum (AJ582085 and AJ582087) (20). All mentioned nearest neighbors are from marine (i.e., saline) origins. Other phylogenetically affiliated bacteria that share less than 97% homology to our isolate are either soil bacteria or symbiotic plant bacteria.
Cultivation and structure elucidation.
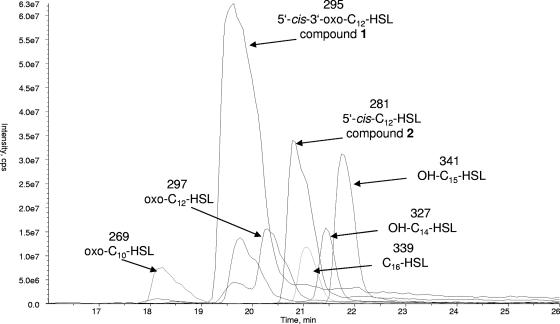
Mesorhizobium sp. strain R8-Ret-T53-13d was cultivated in the presence of an adsorber resin. Liquid chromatography-MS (LC-MS) investigations (Fig. 2) of the extract obtained from solid-phase extraction indicated the presence of several AHLs with chain lengths of C10 to C16. Chromatographic separation of the active bacterial extract using reversed-phase HPLC yielded unusually large amounts (0.6 mg/liter for compound 1 and 1.3 mg/liter for compound 2) of the two major acylated homoserine lactones (compounds 1 and 2) in this bacterium (Fig. 3).
FIG. 2.
LC-ESI(+)-MS fingerprint of raw extract, showing, in addition to compounds 1 and 2 (molecular masses of 295 and 281, respectively), several masses corresponding to other AHLs. The chromatogram represents overlaid mass chromatograms for the [M + H]+ ions extracted from the total ion chromatogram.
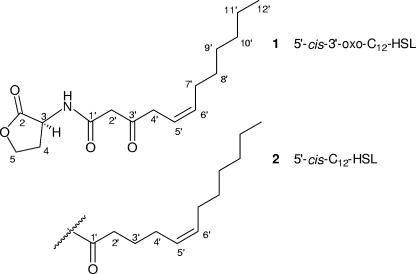
FIG. 3.
Acylated homoserine lactones isolated from the Mesorhizobium sp.
The molecular formula of compound 1 was deduced from accurate mass measurement (HR-FT-ICR-MS) to be C16H25NO4, implying five degrees of unsaturation. The 13C NMR spectrum contained 16 resonances resulting from one methyl, nine methylene, three methine, and three quaternary carbons (Table 1). The distortionless enhancement by polarization transfer (DEPT) spectrum revealed 24 protons attached to carbons. Therefore, the remaining proton has to be present as an OH/NH group. 1H-1H correlation spectroscopy (COSY) correlations between H-3, H2-4 and H2-5 as well as 1H-13C heteronuclear multiple-bond correlation (HMBC) cross peaks from H2-4 and H2-5 to C-2 (δC 174.7) indicated a γ-butyrolactone. The 13C and 1H NMR shift value of CH-3 (δH/C 4.58/49.0) determined this group to be connected to a nitrogen atom, resulting in a homoserine moiety. The remaining part of the molecule was identified as an acyl group. One of the methylene groups (δH/C 3.51/47.3) within this acyl moiety appeared as a singlet in 1H NMR spectra and showed 1H-13C HMBC cross peaks to the quaternary carbon δC 166.2 (C-1′) and to the keto function δC 204.5 (C-3′). Thus, it has to be located between C-1′ and C-3′. The chemical shift value of C-1′ is in agreement with a connection to the NH group forming an amide function. The residual protons belonged to an unsaturated alkyl chain with one CH=CH double bond (δH/C 5.51/119.0, δH/C 5.67/135.3), six methylene groups, and a terminal methyl group. Methylene group CH2-4′ (δH/C 3.28/42.6) showed a doublet multiplicity in 1H NMR spectra and 1H-13C HMBC correlations to both double-bond methine groups (C-5′/C-6′) and C-3′. Therefore, C-4′ was directly bonded to the carbon-carbon double bond Δ5′, 6′ and to the quaternary keto function C-3′. From 1H and 13C NMR spectra, 1H-1H COSY, and 1H-13C HMBC correlations, an n-hexyl chain (CH2-7′ to CH3-12′) was deduced, which clarified the other end of the double bond, i.e., C-7′ to C-12′. The planar structure of compound 1 (Fig. 3) was thus determined, leaving the configuration at C-3 and of the Δ5′, 6′ to be established. H-5′ showed 1H-1H coupling constants of 7.0 Hz to H2-4′ and of 9.6 Hz to H-6′. The value of the JH5′-H6′ coupling enabled us to assign the cis configuration for Δ5′, 6′.
All natural occurring HSLs are regarded as having the l configuration; however, even though a large number of literature reports deal with AHLs, hardly any proof for their absolute configuration has been presented. For purchasable enantiomerically pure l-homoserine lactone, optical rotation values are given by the supplier (Sigma-Aldrich) (l-HSL·HBr, [α]20D −21°; l-HSL·HCl, −28° [H2O]). We measured the specific optical rotation of l-3-oxo-hexanoyl-HSL (3-oxo-C6-HSL; Sigma K3007) as [α]25D −21° (MeOH). N-Octanoyl-l-homoserine lactone was recently synthesized (36) and had a specific optical rotation of [α]25D −24° (MeOH). The optical rotation of compound 1 was determined as [α]25D −12° (MeOH), and thus the configuration at C-3 was assigned to be l. The systematic IUPAC name for compound 1 is (Z)-3-oxo-N-[(3S)-2-oxotetrahydro-3-furanyl]-5-dodecenamide. According to literature precedents, we propose the half systematic name 5,6-cis-3-oxo-dodecenoyl-HSL (= 5-cis-3-oxo-C12-HSL).
HR-MS data for compound 2 showed the molecular formula of this molecule to be C16H27NO3. Comparison of NMR spectral data for compound 2 with those of compound 1 revealed close similarity (Table 1). From NMR and MS data it can be deduced that in compound 2 a carbonyl group is replaced by a methylene moiety. This was confirmed by the presence of additional NMR signals for a CH2 group at δH/C 1.73/25.3 showing 1H-1H COSY cross peaks to H2-2′ (δH/C 2.28/35.5) and H2-4′ (δH/C 2.10/26.5). A 13C NMR signal for the keto group C-3′ was missing. All other deductions from COSY, heteronuclear single-quantum coherence (HSQC), and HMBC NMR experiments were identical to those for compound 1, revealing that compound 2 possessed a methylene group at position 3′ instead of the keto function as deduced for compound 1. In 1H NMR spectra the methine protons of the double bond (Δ5′, 6′) appeared as multiplets. Homonuclear decoupling NMR experiments revealed JH5′-H6′ to be 10.6 Hz and endorsed the cis configuration. According to the optical rotation value of [α]25D −7° (MeOH), the configuration at C-3 was attributed to the l form. We propose the name 5,6-cis-dodecenoyl-HSL (= 5-cis-C12-HSL) for compound 2.
UV data for compound 1 revealed an unexpected maximum at 286 nm (MeOH), although on first sight the structure possesses no extended chromophore. In this context it is also noteworthy that NMR spectra of compound 1 when measured in deuterated methanol show no signals for CH2-2′, due to the occurrence of keto-enol tautomerism. Obviously, the β-diketo function connected to the homoserine moiety and its tautomeric forms provide an expanded π-electron system which is responsible for the UV maximum at 286 nm. Compound 2 is lacking a UV maximum beyond 217 nm, which is consistent with the missing keto functionality at position 3′.
According to LC-MS and 1H/13C NMR data, at least five additional and structurally different AHLs are produced by this bacterial strain. ESI MS data (Fig. 2) indicate molecular masses of 269 and 297, presumably derived from 3-oxo-C10-HSL and 3-oxo-C12-HSL, previously identified from Vibrio aguillarum (16) and Pseudomonas aeruginosa (18, 24), respectively. The molecular masses of 327 and 339 are assumed to correspond to hydroxy-C14-HSL and C16-HSL (15, 25), respectively, whereas 341 accounts for an AHL with an uneven number of carbon atoms, that is, hydroxy-C15-HSL.
Biological activity.
Compound 1 showed antibacterial activity against Bacillus brevis (8-mm inhibition zone at 50 μg/disk). No activity was found for the other tested organisms. Cytotoxic activity was found against Jurkat and HeLa S3 cells: at 10 μg/ml, proliferation of Jurkat cells was reduced by 90%; at 50 μg/ml, proliferation of HeLa S3 cells was decreased by 50%. Compound 2 appeared to be inactive in the antimicrobial as well as in the cytotoxic bioassays.
To test whether compounds 1 and 2 are QS functional, we employed two AHL biosensors with different specificities: Escherichia coli MT102(pSB403) (38) and Pseudomonas putida F117(pKR-C12) (29). The sensor plasmid pSB403 contains the Vibrio fischeri luxR gene together with the luxI promoter region as a transcriptional fusion to the bioluminescence genes luxCDABE of Photorhabdus luminescens. The QS system of V. fischeri relies on 3-oxo-hexanoyl-HSL (3-oxo-C6-HSL), and the sensor plasmid consequently exhibits the highest sensitivity for this AHL molecule. When the two compounds were tested with the aid of this sensor, activation was observed only at a concentration of at least 2 μM (data not shown). For comparison, the sensor is fully induced in the presence of 10 nM 3-oxo-C6-HSL.
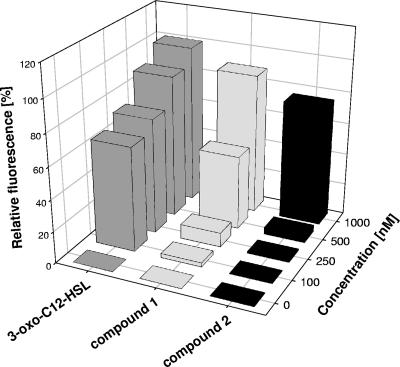
The second sensor plasmid, pKR-C12, contains a translational fusion of the lasB elastase gene of Pseudomonas aeruginosa to the gfp gene encoding the green fluorescent protein. The sensor also expresses LasR, the cognate 3-oxo-C12-HSL receptor protein. As expression of lasB is controlled by the las QS system, this biosensor is most sensitive for 3-oxo-C12-HSL and related long-chain AHLs and cannot be activated by AHLs with acyl chains shorter than C8. As expected from their structures, both compounds activated this biosensor (Fig. 4). Compounds 1 and 2 were approximately 20- and 100-fold less efficient in the bioassay than 3-oxo-C12-HSL, respectively. We also employed the biosensor to visualize the compounds on TLC plates. The compounds exhibited mobilities that were similar but not identical to that of 3-oxo-C12-HSL (data not shown).
FIG. 4.
Detection of Mesorhizobium sp. signal molecules by the AHL monitor strain P. putida F117(pKR-C12). The biosensor was grown in the presence of 3-oxo-C12-HSL or compound 1 or 2 in a concentration range of 0 to 1,000 nM. Green fluorescence of the cultures was recorded with a microtiter plate reader at 515 nm. The measured values were corrected for autofluorescence and plotted as a function of AHL concentration. Fluorescence of the sensor in the presence of 1,000 nM 3-oxo-C12-HSL was arbitrarily set to 100%.
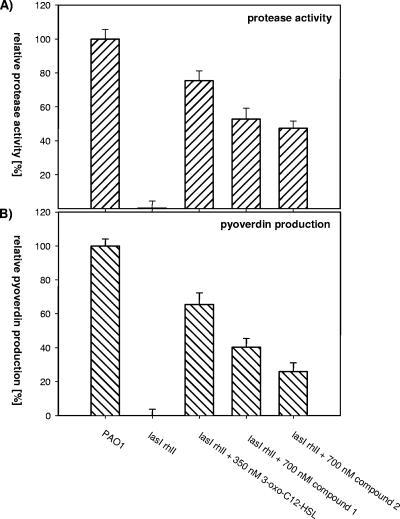
In the opportunistic human pathogen P. aeruginosa, production of extracellular proteases and the siderophore pyoverdine is dependent on the synthesis of 3-oxo-C12-HSL. When a las rhl double mutant, which is defective in both QS circuits operating in P. aeruginosa and thus produces greatly reduced levels of pyoverdin and proteases (17, 31), was grown in the presence of compounds 1 and 2 or 3-oxo-C12-HSL, the defects were at least partially rescued (Fig. 5). In conclusion, these data provide clear evidence that the two new compounds isolated from the Mesorhizobium sp. are biologically active and can stimulate QS systems present in other bacteria.
FIG. 5.
Measurements of protease activity (A) and pyoverdin production (B) in wild-type P. aeruginosa PAO1 and the AHL-deficient lasI rhlI double mutant grown in the absence or presence of 350 nM of 3-oxo-C12-HSL and 700 nM of compound 1 or 2. Sterile filtered supernatants were used for enzymatic measurements of proteolytic activities on azocasein and spectrophotometric quantification of pyoverdin. The activity of the wild type was arbitrarily set to 100%. The data represent mean values from three independent experiments. Error bars represent the standard errors of the means.
DISCUSSION
Bacteria of the genus Mesorhizobium have rarely been observed in connection with marine sponges. By employing molecular techniques, a range of sponges was analyzed in regard the microbial diversity associated with them, but this did not yield any data suggesting a widespread occurrence of Mesorhizobium spp. (11, 38). In one of our previous large-scale investigations of marine sponge-associated bacteria from the Sula Ridge (Norway), no Mesorhizobium species could be detected (2). Studies on two sponges from the Korsfjord led to more than 800 bacterial isolates (data not shown), only one of which was a Mesorhizobium species (strain R8-Ret-T53-13d). From these data we assume that there is no close and specific association between the isolated Mesorhizobium strain and the sponge. Nevertheless, to date, there are three further reported cases of such microorganisms obtained from Demospongiae (28, 34).
Our isolate affiliates in the vicinity of strains derived from marine (i.e., saline) origins (such as marine dinoflagellates), soil of salt marshes, and different lakes (Fig. 1). Mesorhizobium sp. strain R8-Ret-T53-13d produces extraordinarily large amounts of different AHLs (0.6 mg/liter for compound 1). Typically, AHLs are present at only nanomolar concentrations or less in culture media (36). LC-MS data suggest the presence of at least seven structurally different AHL compounds (Fig. 2) with chain lengths of C10 to C15. In some cases the acyl chain bears additional functional groups such as keto and hydroxyl functionalities, as well as carbon-carbon double bonds. Schripsema et al. (26) reported the isolation of a new unsaturated HSL from Rhizobium leguminosarum (7-cis-3-OH-C14-HSL), Puskas and coworkers (22) isolated 7-cis-C14-HSL from the photosynthetic bacterium Rhodobacter sphaeroides, and HSL molecules with side chains composed of 16 and 18 carbon atoms (15, 25), some with one or two double bonds (36), were also reported. AHLs with an uneven number of carbon atoms, e.g., C15, are extremely rare (36).
The two major AHLs of the Mesorhizobium sp. were isolated in pure form, and their structures were rigorously characterized by making use of spectroscopic data. The reference data provided are of prime importance, because despite the many reports on AHL signaling molecules, frequently physicochemical characterizations are incomplete and a definite structure is merely implied. This of course is not astonishing given that AHL molecules are usually produced in nanomolar concentrations. In the case of our Mesorhizobium sp., extraordinarily large amounts (i.e., in the milligram range) of AHLs could be retrieved. It is estimated on the basis of LC-MS data for HPLC fractions of the raw extract that overall, approximately one-quarter of the crude extract is composed of this class of substances. This amounts to a production of about 5 mg of HSLs per liter of cultivation medium.
As discussed by Wagner-Döbler et al. (36), the method of cultivation and extraction (e.g., addition of adsorber resin to the culture medium) may influence the amounts extracted and the type of HSLs. Our screening results from the cultivation of our Mesorhizobium sp. on four different media showed that the production of biologically active metabolites was greatly enhanced when the adsorber resin was added to the medium.
QS activity of compounds 1 and 2 was evaluated in three biosensor systems, i.e., E. coli and. P. putida biosensor strains responsive to AHLs with short (C6) and long (C12) chain lengths and a lasI rhlI double mutant of P. aeruginosa. Since the E. coli biosensor is best induced with 3-oxo-C6-HSL, the observed lower activity of compounds 1 and 2 is in accordance with the extended carbon chain (C12) and the presence of a carbon-carbon double bond in our compounds. These results are also in agreement with previous studies that showed that long-chain AHLs can be detected by the sensor, albeit with low sensitivity (7, 40). Compounds 1 and 2 were also approximately 20- and 100-fold less efficient in the bioassay employing the P. putida F117(pKR-C12) biosensor than was 3-oxo-C12-HSL. The diminished activity of our compounds (compounds 1 and 2) compared to the control may be explained by the presence of a double bond in a side chain, clearly modulating the bioactivity. In all QS bioassays, compound 1 was somewhat more efficient than compound 2, and both were less active than the indigenous AHL molecules 3-oxo-C6-HSL and 3-oxo-C12-HSL. The slightly enhanced bioactivity of 5-cis-3-oxo-C12-HSL (compound 1) compared to 5-cis-C12-HSL (compound 2) must be due to the additional keto function in compound 1.
In our assays compound 1 showed antibacterial activity and cytotoxicity towards cultured tumor cells. The relatively high concentration of AHLs detected in our bacterial strain may point towards additional biological functions of these molecules. For bacteriocin small (7-cis-3-OH-C14-HSL), antibacterial activity towards the producing strain was reported (26). Kaufmann et al. described additional functions of AHLs, particularly bactericidal activity against gram-positive bacteria (13). Cytotoxic activity of 3-oxo-C12-HSL was observed in neutrophils and monocytic cell lines (32). In order to judge the ecological role of AHLs produced by bacteria in the marine environment, the in situ production of these molecules must be analyzed. Recently, Schupp et al. (27) described the enhanced detection of AHLs in sponge material by using solid-phase extraction.
Acknowledgments
We thank H. T. Rapp for obtaining the sponge material from the Korsfjord and for his valuable help during our visit in Bergen. For expert technical assistance, we thank A. Spohn and M. Saul. We thank H. Hamacher, Bayer Analytics Leverkusen, Germany, for recording HR-MS spectra.
Financial support was provided by the Bundesministerium für Bildung und Forschung (BMBF), research program 03F0415A. In addition, we gratefully acknowledge the generous support of the Improving Human Potential Programme of the EU, contract no. HPRI-CT-1999-00056, Bergen Marine.
Footnotes
Published ahead of print on 30 March 2007.
REFERENCES
- 1.Bertani, G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62:293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dieckmann, R., I. Graeber, I. Kaesler, U. Szewzyk, and H. von Döhren. 2005. Rapid screening and dereplication of bacterial isolates from marine sponges of the Sula Ridge by intact-cell-MALDI-TOF mass spectrometry (ICM-MS). Appl. Microbiol. Biotechnol. 67:539-548. [DOI] [PubMed] [Google Scholar]
- 3.Eberl, L. 1999. N-Acyl homoserinelactone-mediated gene regulation in Gram-negative bacteria. Syst. Appl. Microbiol. 22:493-506. [DOI] [PubMed] [Google Scholar]
- 4.Flodgaard, L. R., P. Dalgaard, J. B. Andersen, K. F. Nielsen, M. Givskov, and L. Gram. 2005. Nonbioluminescent strains of Photobacterium phosphoreum produce the cell-to-cell communication signal N-(3-hydroxyoctanoyl)homoserine lactone. Appl. Environ. Microbiol. 71:2113-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuqua, C., S. C. Winans, and E. P. Greenberg. 1996. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 50:727-751. [DOI] [PubMed] [Google Scholar]
- 6.Gage, D. J. 2004. Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol. Mol. Biol. Rev. 68:280-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geisenberger, O., M. Givskov, K. Riedel, N. Hoiby, B. Tümmler, and L. Eberl. 2000. Production of N-acyl-l-homoserine lactones by P. aeruginosa isolates from chronic lung infections associated with cystic fibrosis. FEMS Microbiol. Lett. 184:273-278. [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez, J. E., and M. M. Marketon. 2003. Quorum sensing in nitrogen-fixing rhizobia. Microbiol. Mol. Biol. Rev. 67:574-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gram, L., H. P. Grossart, A. Schlingloff, and T. Kiorboe. 2002. Possible quorum sensing in marine snow bacteria: production of acylated homoserine lactones by Roseobacter strains isolated from marine snow. Appl. Environ. Microbiol. 68:4111-4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Green, D. H., L. E. Llewellyn, A. P. Negri, S. I. Blackbur, and C. J. S. Bolch. 2004. Phylogenetic and functional diversity of the cultivable bacterial community associated with the paralytic shellfish poisoning dinoflagellate Gymnodinium catenatum. FEMS Microbiol. Ecol. 47:345-357. [DOI] [PubMed] [Google Scholar]
- 11.Hentschel, U., J. Hopke, M. Horn, A. B. Friedrich, M. Wagner, J. Hacker, and B. S. Moore. 2002. Molecular evidence for a uniform microbial community in sponges from different oceans. Appl. Environ. Microbiol. 68:4431-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jarvis, B. D. W., P. van Berkum, W. X. Chen, S. M. Nour, M. P. Fernandez, J. C. Cleyet-Marel, and M. Gillis. 1997. Transfer of Rhizobium loti, Rhizobium huakuii, Rhizobium ciceri, Rhizobium mediterraneum, and Rhizobium tianshanense to Mesorhizobium gen. nov. Int. J. Syst. Bacteriol. 47:895-898. [Google Scholar]
- 13.Kaufmann, G. F., R. Sartorio, S.-H. Lee, C. J. Rogers, M. M. Meijler, J. A. Moss, B. Clapham, A. P. Brogan, T. J. Dickerson, and K. D. Janda. 2005. Revisiting quorum sensing: discovery of additional chemical and biological functions for 3-oxo-N-acylhomoserine lactones. Proc. Natl. Acad. Sci. USA 102:309-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marketon, M. M., M. R. Gronquist, A. Eberhard, and J. E. Gonzalez. 2002. Characterization of the Sinorhizobium meliloti sinR/sinI locus and the production of novel N-acyl homoserine lactones. J. Bacteriol. 184:5686-5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Milton, D. L., A. Hardman, M. Camara, S. R. Chhabra, B. W. Bycroft, G. S. Stewart, and P. Williams. 1997. Quorum sensing in Vibrio anguillarum: characterization of the vanI/vanR locus and identification of the autoinducer N-(3-oxodecanoyl)-l-homoserine lactone. J. Bacteriol. 179:3004-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Passador, L., J. M. Cook, M. J. Gambello, L. Rust, and B. H. Iglewski. 1993. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science 260:1127-1130. [DOI] [PubMed] [Google Scholar]
- 18.Pearson, J. P., K. M. Gray, L. Passador, K. D. Tucker, A. Eberhard, B. H. Iglewski, and E. P. Greenberg. 1994. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc. Natl. Acad. Sci. USA 91:197-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peters, L., G. M. König, A. D. Wright, R. Pukall, E. Stackebrand, L. Eberl, and K. Riedel. 2003. Secondary metabolites of Flustra foliacea and their influence on bacteria. Appl. Environ. Microbiol. 69:3469-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pradella, S., M. Allgaier, C. Hoch, O. Pauker, E. Stackebrandt, and I. Wagner-Döbler. 2004. Genome organization and localization of the pufLM genes of the photosynthesis reaction center in phylogenetically diverse marine Alphaproteobacteria. Appl. Environ. Microbiol. 70:3360-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prince, R. W., C. D. Cox, and M. L. Vasil. 1993. Coordinate regulation of siderophore and exotoxin A production: molecular cloning and sequencing of the Pseudomonas aeruginosa fur gene. J. Bacteriol. 175:2589-2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Puskas, A., E. P. Greenberg, S. Kaplan, and A. L. Schaefer. 1997. A quorum-sensing system in the free-living photosynthetic bacterium Rhodobacter sphaeroides. J. Bacteriol. 179:7530-7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sawada, H., L. D. Kuykendall, and J. M. Young. 2003. Changing concepts in the systematics of bacterial nitrogen-fixing legume symbionts. J. Gen. Appl. Microbiol. 49:155-179. [DOI] [PubMed] [Google Scholar]
- 24.Schaefer, A. L., B. L. Hanzelka, M. R. Parsek, and E. P. Greenberg. 2000. Detection, purification, and structural elucidation of the acylhomoserine lactone inducer of Vibrio fischeri luminescence and other related molecules. Methods Enzymol. 305:288-301. [DOI] [PubMed] [Google Scholar]
- 25.Schaefer, A. L., T. A. Taylor, J. T. Beatty, and E. P. Greenberg. 2002. Long-chain acyl-homoserine lactone quorum-sensing regulation of Rhodobacter capsulatus gene transfer agent production. J. Bacteriol. 184:6515-6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schripsema, J., K. E. de Rudder, T. B. van Vliet, P. P. Lankhorst, E. de Vroom, J. W. Kijne, and A. A. van Brussel. 1996. Bacteriocin small of Rhizobium leguminosarum belongs to the class of N-acyl-l-homoserine lactone molecules, known as autoinducers and as quorum-sensing cotranscription factors. J. Bacteriol. 178:366-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schupp, P., T. S. Charlton, M. W. Taylor, S. Kjelleberg, and P. D. Steinberg. 2005. Use of solid-phase extraction to enable enhanced detection of acyl homoserine lactones (AHLs) in environmental samples. Anal. Bioanal. Chem. 383:132-137. [DOI] [PubMed] [Google Scholar]
- 28.Sfanos, K., D. Harmody, P. Dang, A. Ledger, S. Pomponi, P. McCarthy, and J. Lopez. 2005. A molecular systematic survey of cultured microbial associates of deep-water marine invertebrates. Syst. Appl. Microbiol. 28:242-264. [DOI] [PubMed] [Google Scholar]
- 29.Steidle, A., K. Sigl, R. Schuhegger, A. Ihring, M. Schmid, S. Gantner, M. Stoffels, K. Riedel, M. Givskov, A. Hartmann, C. Langebartels, and L. Eberl. 2001. Visualization of N-acylhomoserine lactone-mediated cell-cell communication between bacteria colonizing the tomato rhizosphere. Appl. Environ. Microbiol. 67:5761-5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stevens, H., M. Stubner, M. Simon, and T. Brinkhoff. 2005. Phylogeny of Proteobacteria and Bacteroidetes from oxic habitats of a tidal flat ecosystem. FEMS Microbiol. Ecol. 54:351-365. [DOI] [PubMed] [Google Scholar]
- 31.Stintzi, A., K. Evans, J. M. Meyer, and K. Poole. 1998. Quorum-sensing and siderophore biosynthesis in Pseudomonas aeruginosa: lasR/lasI mutants exhibit reduced pyoverdine biosynthesis. FEMS Microbiol. Lett. 166:341-345. [DOI] [PubMed] [Google Scholar]
- 32.Tateda, K., Y. Ishii, M. Horikawa, T. Matsumoto, S. Miyairi, J. P. Pechere, T. J. Standiford, M. Ishiguro, and K. Yamaguchi. 2003. The Pseudomonas aeruginosa autoinducer N-3-oxododecanoyl homoserine lactone accelerates apoptosis in macrophages and neutrophils. Infect. Immun. 71:5785-5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor, M. W., P. J. Schupp, H. J. Baillie, T. S. Charlton, R. de Nys, S. Kjelleberg, and P. D. Steinberg. 2004. Evidence for acyl homoserine lactone signal production in bacteria associated with marine sponges. Appl. Environ. Microbiol. 70:4387-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor, M. W., P. J. Schupp, R. de Nys, S. Kjelleberg, and P. D. Steinberg. 2005. Biogeography of bacteria associated with the marine sponge Cymbastela concentrica. Environ. Microbiol. 7:419-433. [DOI] [PubMed] [Google Scholar]
- 35.Van Trappen, S., J. Mergaert, S. Van Eygen, P. Dawyndt, M. C. Cnockaert, and J. Swings. 2002. Diversity of 746 heterotrophic bacteria isolated from microbial mats from ten Antarctic lakes. Syst. Appl. Microbiol. 25:603-610. [DOI] [PubMed] [Google Scholar]
- 36.Wagner-Döbler, I., V. Thiel, L. Eberl, M. Allgaier, A. Bodor, S. Meyer, S. Ebner, A. Hennig, R. Pukall, and S. Schulz. 2005. Discovery of complex mixtures of novel long-chain quorum sensing signals in free-living and host-associated marine Alphaproteobacteria. Chem. Biochem. 6:2195-2206. [DOI] [PubMed] [Google Scholar]
- 37.Wang, H., Z. T. G. Zhong, T. Cai, S. P. Li, and J. Zhu. 2004. Heterologous overexpression of quorum-sensing regulators to study cell-density-dependent phenotypes in a symbiotic plant bacterium Mesorhizobium huakuii. Arch. Microbiol. 182:520-525. [DOI] [PubMed] [Google Scholar]
- 38.Webster, N. S., K. J. Wilson, L. L. Blackall, and R. T. Hill. 2001. Phylogenetic diversity of bacteria associated with the marine sponge Rhopaloeides odorabile. Appl. Environ. Microbiol. 67:434-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whitehead, N. A., A. M. Barnard, H. Slater, N. J. Simpson, and G. P. Salmond. 2001. Quorum-sensing in Gram-negative bacteria. FEMS Microbiol. Rev. 25:365-404. [DOI] [PubMed] [Google Scholar]
- 40.Winson, M. K., S. Swift, P. J. Hill, C. M. Sims, G. Griesmayr, B. W. Bycroft, P. Williams, and G. S. Stewart. 1998. Engineering the luxCDABE genes from Photorhabdus luminescens to provide a bioluminescent reporter for constitutive and promoter probe plasmids and mini-Tn5 constructs. FEMS Microbiol. Lett. 163:193-202. [DOI] [PubMed] [Google Scholar]
- 41.Zapf, S., M. Hossfeld, H. Anke, R. Velten, and W. Steglich. 1995. Darlucins A and B, new isocyanide antibiotics from Sphaerellopsis filum (Darluca filum). J. Antibiot. 48:36-41. [DOI] [PubMed] [Google Scholar]
- 42.Zhu, J., Y. R. Chai, Z. T. Zhong, S. P. Li, and S. C. Winans. 2003. Agrobacterium bioassay strain for ultrasensitive detection of N-acylhomoserine lactone-type quorum-sensing molecules: detection of autoinducers in Mesorhizobium huakuii. Appl. Environ. Microbiol. 69:6949-6953. [DOI] [PMC free article] [PubMed] [Google Scholar]