Abstract
We analyzed the contributions of different heterotrophic bacterial groups to the uptake of several low-molecular weight compounds during a seasonal cycle on the northwestern Mediterranean coast (Blanes Bay Microbial Observatory). The bacterial assemblage structure had been shown to change substantially year-round for this site, but whether changes in the activities of the different bacterial groups also occurred on the seasonal scale was unknown. Microautoradiography combined with catalyzed reporter deposition fluorescence in situ hybridization was used to analyze the patterns of glucose, amino acid, and ATP uptake by different bacterial groups. Gammaproteobacteria and Bacteroidetes were not very active in the uptake of glucose at any time of the year (<10% of cells were active) compared to Alphaproteobacteria (generally >20% of cells were active). Dissolved free amino acids were taken up considerably by Alphaproteobacteria and Gammaproteobacteria but not by Bacteroidetes. Relatively high percentages of cells of the three broad phylogenetic groups actively took up ATP, which could be related to the important phosphorous limitation of bacterial production during most of the year in Blanes Bay. The contribution of SAR11 to the uptake of the monomers was variable year-round, generally with fewer than 30% of the cells being active. By contrast, Roseobacter were highly overrepresented in the uptake of all the substrates throughout all the year, with more than 50% of cells being active in all the samples and for all substrates. Our results suggest that substantial changes in the activity of some phylogenetic groups of bacteria occur throughout the year.
The heterotrophic activity of marine bacteria and their role in carbon cycling have been extensively studied in the ocean through bulk or global measurements. Consequently, current carbon models implicitly assume that all heterotrophic bacteria perform equally in carbon processing (19). This view has been challenged by the application of single-cell techniques, which have shown that activity is not homogeneously distributed throughout the bacterial assemblage (43) and have pointed to the need of studying the ecological roles of different bacterial populations in the ocean. One approach to that objective is the isolation of relevant oceanic microbes and the subsequent study of their metabolic properties in culture (37) or of their potential ecological functions through genome sequencing analysis (22). However, most cultured phylotypes are not representative of the true oligotrophic marine bacteria (21). The combination of single-cell and molecular techniques, such as microautoradiography combined with fluorescence in situ hybridization (MAR-FISH) constitutes a powerful alternative to study the metabolic strategies of natural communities.
To date and for the ocean, MAR-FISH has been used to show different uptake patterns of simple, low-molecular-weight (LMW) compounds in several studies (11, 34, 44). Although some MAR-FISH studies have focused on changes in bacterial use of organic compounds along spatial gradients (13, 18, 29), very little attention has been paid to temporal changes, and these studies have usually been limited to the comparison of two different periods (13, 44). Thus, it is difficult to know whether patterns of uptake of specific compounds by the bacterial groups can be generalized along temporal scales.
The activities of different bacterial groups could change with time, driven by variations in the availability of organic compounds or the need for specific nutrients in resource-limited situations. Indeed, a well-known response of organisms to nutrient depletion is an increase in the capacity for uptake of the limiting nutrient (e.g., reference 25). In this study we describe the bacterial uptake of three kinds of organic compounds (glucose, dissolved free amino acids, and ATP), which were chosen in order to represent the supply of carbon alone (glucose), carbon with extra nitrogen (amino acids), and carbon with extra nitrogen and phosphorous (ATP). The study was conducted across a seasonal range of trophic conditions, from nutrient-depleted (summer) to nutrient-replete (winter) periods in an oligotrophic coastal Mediterranean site.
Substantial year-round changes in bacterial assemblage structure have been described for Blanes Bay (5, 41). However, the identities of the active bacterial groups and whether the activities of specific bacterial groups change seasonally, were unknown. We specifically wanted to test whether the uptake activities of the different bacterial groups were a permanent feature throughout the year, despite differences in environmental conditions, or whether changes occurred. Ultimately, we wanted to know which members of the bacterioplankton community can be important in terms of carbon processing in this coastal site. To our knowledge, this is the first study to show annual dynamics in the single-cell activity of bacterial populations.
MATERIALS AND METHODS
Basic data.
Samples were taken from Blanes Bay (The Blanes Bay Microbial Observatory, northwestern Mediterranean Sea) at several dates along a seasonal cycle (March 2003 to February 2004) (Fig. 1). Surface water temperature was measured in situ with a mercury thermometer. For determination of chlorophyll a (Chl a) concentration, 150 ml of seawater was filtered on GF/F filters (Whatman) and subsequently extracted in acetone (90%, vol/vol) in the dark at 4°C for 24 h. Fluorescence was measured with a Turner Designs fluorometer. For analyses of dissolved nutrient concentrations, seawater samples were filtered through 0.2-μm-pore-size, 47-mm-diameter polycarbonate filter (Supor-200; Gelman Sciences) using a polycarbonate filtration device (Millipore). Dissolved inorganic nutrient concentrations were determined spectrophotometrically with an Alliance Evolution II autoanalyzer, following standard procedures (23). Phosphate concentrations were determined manually, using a 10-cm cuvette to increase the detection limit, and have been presented elsewhere (36).
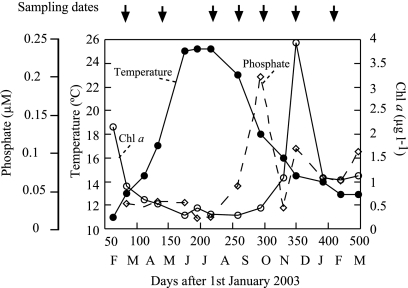
FIG. 1.
Temperature, Chl a, and phosphate concentrations throughout the seasonal study at Blanes Bay. The arrows indicate the dates at which samples were taken for MAR-CARD-FISH analyses.
Abundance of photosynthetic picoplankton.
Synechococcus and Prochlorococcus cells were enumerated by flow cytometry and distinguished by their different sizes and pigment properties in unstained samples following common procedures (41).
Bulk uptake of amino acids, glucose, and ATP.
Bulk uptake of the substrates (glucose, amino acids, and ATP) was determined monthly by the measurement of the radioactivity incorporated in subsamples withdrawn from the incubations used for MAR-catalyzed-reporter-deposition (CARD)-FISH analysis. As explained below, the three substrates were added at 0.5 nM, and the incubations lasted 4 h and were done in an incubator adjusted to the in situ temperature. For each sample (20 ml), four aliquots (1.2 ml) were taken in Eppendorf tubes, and 120 μl of cold 50% trichloroacetic acid was added to stop the incorporation. For every sample and compound, aliquots (5 ml) of cells killed with formalin before the isotope addition were used as controls. Samples were kept frozen at −20°C until processing, which was carried out by the centrifugation method (42). Finally, 1 ml of scintillation cocktail (Optimal HiSafe) was added to each Eppendorf tube, and radioactivity was counted on a Beckman scintillation counter after 24 h.
CARD-FISH.
CARD-FISH was carried out following the protocol described by Pernthaler et al. (35). Samples were fixed overnight with formaldehyde (1.8%) at 4°C and gently filtered on 0.2-μm polycarbonate filters (Millipore GTTP; 25-mm diameter). Filters were permeabilized with lysozyme (37°C, 1 h), and hybridizations were carried out overnight at 35°C. Several horseradish peroxidase-labeled probes were used to characterize the composition of the microbial communities in the samples: EUB 338-II and -III (target most Eubacteria) (6, 14), ALF968 (targets most Alphaproteobacteria) (33), GAM42a (targets most Gammaproteobacteria) (31), CF319 (targets many groups belonging to the Bacteroidetes group) (30), ROS537 (targets members of the Roseobacter-Sulfitobacter-Silicibacter group) (17), and SAR11-441R (targets the SAR11 cluster) (32). The EUB antisense probe NON338 (45) was used as a negative control. All probes were purchased from biomers.net (Ulm, Germany). Specific hybridization conditions were established by addition of formamide to the hybridization buffers (20% formamide for the NON338 probe, 45% formamide for the ALF968 and SAR11-441R probes, and 55% for the other probes). Counterstaining of CARD-FISH preparations was done with 4,6-diamidino-2-phenylindole (DAPI) (1 μg ml−1). Between 500 and 1,000 DAPI-positive cells were counted manually in a minimum of 10 fields.
MAR-CARD-FISH.
MAR-CARD-FISH was performed on seven occasions during the seasonal sampling. We followed the protocol described by Alonso and Pernthaler (2). Briefly, samples (20 ml) were incubated for 4 h in an incubator adjusted to the in situ temperature with the following tritiated substrates (0.5 nM final concentration): [3H]glucose (Amersham TRK85), a mixture of 3H-amino acids (Amersham TRK440), and [3H]ATP (Amersham TRK747). One replicate (for each compound and treatment) was killed with formaldehyde before the addition of the tritiated compounds and was used as a control. After the incubation, the samples were fixed overnight with formaldehyde (1.8%) at 4°C and gently filtered on 0.2-μm polycarbonate filters (Millipore GTTP; 25-mm diameter). The filters were then hybridized following the CARD-FISH protocol and subsequently glued onto glass slides with an epoxy adhesive (UHU plus; UHU GmbH, Bühl, Germany). For microautoradiography, the slides were embedded in a 46°C tempered photographic emulsion (KODAK NTB-2) containing 0.1% agarose (gel strength, 1%; >1 kg cm2) in a dark room. The slides were placed on an ice-cold metal bar for about 5 min to allow the emulsion to solidify and subsequently were placed inside black boxes at 4°C until development. The optimal exposure time was determined for each experiment and compound and was between 13 and 21 days depending on the compound and time of the season. For development, we submerged the exposed slides for 3 min in the developer (KODAK D19), followed by 30 s of rinsing with distilled water, 3 min in the fixer (KODAK Tmax), and 5 min of washing with tap water. The slides were then dried in a desiccator overnight and stained with DAPI (1 μg ml−1), and cells were counted in an Olympus BX61 epifluorescence microscope. Replicates or triplicates were counted for ca. 20% of the filters, generally in sections from different replicate filters and in a few cases in sections coming from the same filter. In any case, replicates were always processed separately, in different hybridizations and dipping procedures.
RESULTS
Seven samples were chosen in order to cover a full year throughout the period from March 2003 to February 2004 (Fig. 1). These samples are representative of the different bacterial assemblages that develop in Blanes Bay year-round, as assessed by monthly denaturing gradient gel electrophoresis and CARD-FISH analyses (5). Three samples were taken during winter, when the temperature was relatively low (12 to 13°C) and the concentrations of nutrients were relatively high (Fig. 1). One of the winter samplings coincided with a Chl a peak (December 2003), and the other two were taken in postbloom situations (March 2003 and February 2004). The summer period was characterized by high temperatures (over 22°C) and low Chl a and nutrient concentrations (Fig. 1). In this period, MAR-CARD-FISH analyses were carried out in August and September. Two additional samples were analyzed in spring (May) and autumn (October), the last coincident with a stormy period. The stormy events produced a peak in inorganic nutrient concentrations (Fig. 1) and the only measured instance of C instead of P limitation of bacterial production (36).
Bulk incorporation of glucose, amino acids, and ATP.
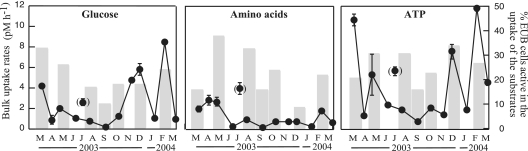
The bulk uptake rates of glucose, amino acids, and ATP (added at trace concentrations [0.5 nM]) ranged between <1 and 12 pM h−1 throughout the year (Fig. 2). In general, higher uptake rates were found in winter and spring, concomitant with higher concentrations of Chl a, while lower uptake rates were detected for the three compounds during the summer (Fig. 2). The highest incorporation rates were found for ATP (range, 1 to 12 pM h−1), with peaks in spring and winter. High glucose uptake rates were found in the autumn and winter periods (>4 pM h−1), while the highest amino acid uptake rates were found in spring (around 3 pM h−1) (Fig. 2). Cell-specific uptake rates followed the same trend and ranged between 2.5 × 10−7 and 9.9 × 10−6 fmol glucose bacterium−1 h−1, 2.5 × 10−7 and 4.9 × 10−6 fmol amino acids bacterium−1 h−1, and 9.5 × 10−7 and 1.4 × 10−5 fmol ATP bacterium−1 h−1 (details not shown).
FIG. 2.
Year-round rates of bulk substrate uptake in Blanes Bay and percentage of EUB+ cells actively incorporating each of the substrates (bars). Samples taken in July are presented as outliers, since the bacterial assemblage structure and carbon dynamics drastically changed in this sampling (5).
Single-cell activities of bacterial phylogenetic groups in the uptake of glucose, amino acids, and ATP throughout the year.
On average, 73% of total cell counts hybridized with the EUB338-II and -III probes (EUB+ cells) in Blanes Bay year-round (5). Alphaproteobacteria, Gammaproteobacteria, and Bacteroidetes together comprised, on average, 75% of the EUB+ cells (5), and Alphaproteobacteria were the dominant group year-round (average of 30% of cell counts). SAR11 cells were very abundant (average of 22% of cell counts) and peaked in the spring-summer period, while Roseobacter were always below 10% of cell counts and was more abundant in spring and the autumn-winter period. Bacteroidetes were rather constant (average of 11% of cell counts), and Gammaproteobacteria increased during the summer with a peak in August (8% of cell counts) (5).
The percentage of EUB+ cells that took up the different compounds was in the range of 9 to 38% (Fig. 2), and substantial changes were found seasonally, especially for amino acids and glucose. The percentage of EUB+ cells active in glucose uptake was high in early spring, decreased in summer, and increased again in autumn and winter. The percentage of EUB+ cells active in the uptake of amino acids was also high in spring but tended to decrease towards the summer and winter periods, with a minimum in December (9%) (Fig. 2). The percentage of EUB+ cells active in ATP uptake was more uniform, with a minimum in September (16% of active EUB+ cells) and a maximum in December (34%) (Fig. 2).
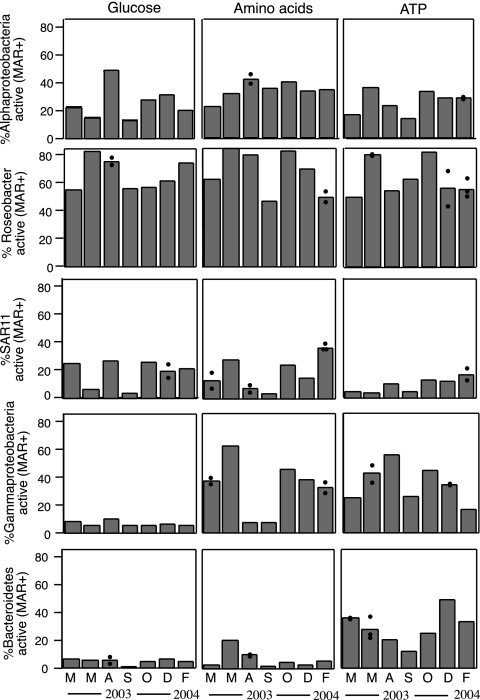
The uptake activities of the specific bacterial groups studied differed greatly depending on the compound and the time of the year (Fig. 3). Alphaproteobacteria showed substantial uptake activity for the three compounds throughout the year. Generally, more than 20% of the alphaproteobacterial cells were active (i.e., MAR+) (Fig. 3). By contrast, Gammaproteobacteria and Bacteroidetes were relatively inactive (<10% of the cells were MAR+) in the uptake of glucose and of glucose and amino acids, respectively. However, these two broad phylogenetic groups were active in the uptake of ATP.
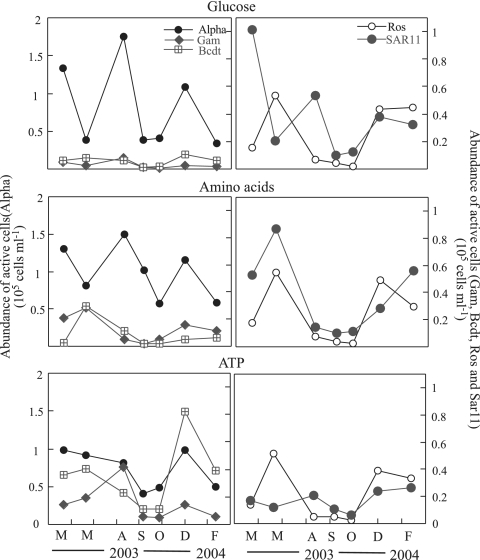
FIG. 3.
Seasonal dynamics of the percentages of probe-positive cells (Alphaproteobacteria, Roseobacter, SAR11, Gammaproteobacteria, and Bacteroidetes) active in the uptake of the three substrates (glucose, amino acids, and ATP). Individual data points are shown for those samples for which replicates or triplicates were counted.
Rather uniform percentages of alphaproteobacterial cells were active in the uptake of amino acids throughout the year (around 30%), but higher variability was found in the uptake of glucose (with a peak in August of 49% active cells) and ATP. When the alphaproteobacterial groups SAR11 and Roseobacter were studied, large differences were found between the two populations. Roseobacter were the most active group in the uptake of all substrates throughout the year (generally over 50% of the Roseobacter cells were MAR+), and the percentage of active cells varied, at most, twofold. The percentage of active SAR11 cells rarely exceeded 20%, and SAR11 cells were more variable in the uptake of the monomers throughout the year (up to sevenfold variation in glucose and amino acid uptake), although statistically significant differences between samples taken in different seasons could not be shown. The lowest percentages of SAR11 cells active in the uptake of substrates were always found in September.
The percentage of Gammaproteobacteria active in amino acid uptake was significantly higher in winter (March 2003, December 2003, and February 2004) than in summer (August and September 2003) (Student's t test, P < 0.05). By contrast, their ATP uptake activity tended to increase towards the summer (August). Bacteroidetes showed an trend opposite to that of Gammaproteobacteria in ATP uptake, with significantly lower uptake activity during the summer than during the winter (Student's t test, P < 0.05).
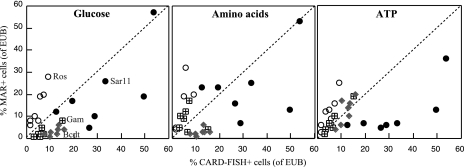
Figure 4 shows the percentages of cells (of the different groups) active in the uptake of glucose, amino acids, and ATP, plotted against their percent contributions to total abundance in the samples. Data points on the 1:1 line indicate groups that are participating in the uptake of the substrates proportionally to their in situ contributions to assemblage structure. Roseobacter cells were overrepresented in the uptake of all substrates compared to their in situ abundance. SAR11 cells were underrepresented or close to the 1:1 line when the uptake of glucose or amino acids was analyzed. However, this group was always underrepresented in the uptake of ATP. Gammaproteobacteria were rather close to the 1:1 line in the uptake of ATP but were usually overrepresented in the uptake of amino acids and underrepresented in the uptake of glucose. Finally, Bacteroidetes were close to the 1:1 line in the uptake of ATP but underrepresented in the uptake of glucose and amino acids.
FIG. 4.
Contributions of various phylogenetic groups (SAR11, Roseobacter [Ros], Gammaproteobacteria [Gam], and Bacteroidetes [Bcdt]) to assemblage structure, presented against their contributions to the uptake of glucose, amino acids, and ATP in Blanes Bay. Percentages were calculated relative to eubacterial cells (probes EUB338-II and -III).
Abundances of cells active in the uptake of glucose, amino acids, and ATP.
The abundance of cells of each phylogenetic group that were active in the uptake of substrates is shown in Fig. 5. The abundance of Alphaproteobacteria active in the uptake of glucose and amino acids exhibited a similar trend throughout the year, with peaks in March, August and December. By contrast, the abundance of Alphaproteobacteria active in ATP uptake decreased from March to September but also exhibited a peak in December. Given the high activity of Roseobacter in the uptake of all substrates, the abundance of active cells closely parallel their in situ total abundance trend (5), with peaks in May and winter. The year-round trend of active SAR11 cells differed more between the three substrates. For example, the maximum of SAR11 cells active in amino acid uptake was in May, coincident with a drastic decrease in cells active in glucose uptake. The abundance of Alphaproteobacteria taking up glucose was positively correlated with the abundance of Synechococcus (Spearman's rho = 0.75; P = 0.05), and the abundance of SAR11 cells taking up amino acids was inversely correlated with the abundance of Prochlorococcus (Spearman's rho = −0.89; P < 0.01).
FIG. 5.
Abundances of cells of the phylogenetic groups Alphaproteobacteria (Alpha), Gammaproteobacteria (Gam), Bacteroidetes (Bcdt), Roseobacter (Ros), and SAR11 that were active in the uptake of glucose, amino acids, and ATP.
The abundances of Bacteroidetes and Gammaproteobacteria cells active in the uptake of glucose and amino acids were generally low, except for a peak in the uptake of amino acids in May. These groups, though, showed different trends in ATP uptake. Gammaproteobacteria exhibited a peak in cells active in ATP uptake in August, and Bacteroidetes did so in December. The abundance of Bacteroidetes cells active in ATP uptake was positively correlated with the concentration of Chl a (Spearman's rho = 0.82; P < 0.03). The abundance of Gammaproteobacteria active in glucose uptake was inversely correlated with phosphate concentration (Spearman's rho = −0.85; P < 0.02), as was the abundance of Gammaproteobacteria active in ATP uptake (Spearman's rho = −0.79; P < 0.04).
Contributions of the different bacterial groups to total cells active in the uptake of glucose, amino acids, and ATP.
Most of the cells active in the uptake of the compounds could be assigned to the specific groups of bacteria studied, but in some cases, such as in the sample taken in February, a large fraction of eubacteria active in the uptake of the three compounds was unknown (i.e., not detected with the probes used) (Fig. 6).
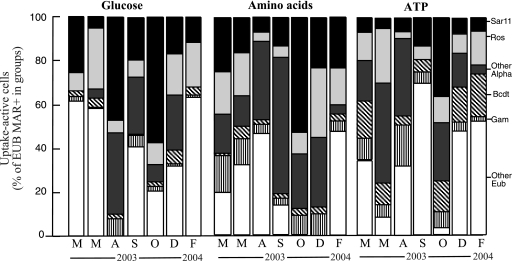
FIG. 6.
Seasonal contribution of each of the analyzed phylogenetic groups (SAR11, Roseobacter [Ros], other Alphaproteobacteria [Alpha], Bacteroidetes [Bcdt], and Gammaproteobacteria [Gam]) to total active bacteria in the uptake of glucose, amino acids, and ATP in Blanes Bay. Percentages were calculated relative to eubacterial cells (probes EUB338-II and -III).
Most glucose-incorporating bacteria belonged to the SAR11 group in two samples (August and October), but lower contributions of this group were found during the rest of the year. In spring (May), most glucose-incorporating bacteria were members of Roseobacter, and substantial contributions of this group were also found in winter (December and February). Alphaproteobacteria not detected with the probes for SAR11 or Roseobacter contributed significant proportions of glucose-incorporating cells during December and the summer period. Gammaproteobacteria also contributed more importantly to glucose incorporation during summer.
The majority of amino acid-incorporating cells were members of the SAR11 group in October, but more uniform contributions of this group to amino acid uptake were found for the rest of the year (around 25% of total active bacteria). Roseobacter contributed very importantly to the amino acid-incorporating cells in March, May, and winter. Similarly to the case for glucose, unknown Alphaproteobacteria were found to contribute significantly to the cells active in amino acid uptake during the summer and in December. Gammaproteobacteria contributed substantially to the amino acid uptake (around 10% of total active bacteria), except for the summer period and February.
SAR11 contributed substantially to the ATP-incorporating cell pool only in October, with relatively low contributions during the rest of the year. Roseobacter contributed the most to cells taking up ATP in May and the least during the summer. Gammaproteobacteria contributed importantly to bacteria active in ATP uptake during August but at lower proportions throughout the year. In contrast to their uptake of other compounds, Bacteroidetes showed relatively high contributions to ATP-incorporating cells except in the summer period, being the dominant analyzed group in December.
DISCUSSION
Comparison of uptake patterns with those from previous MAR-FISH studies.
Variability in the patterns of carbon use among phylogenetic groups of bacteria could have implications for carbon cycling. Whether the proportion of active cells differs between the bacterial groups and whether the activities of the bacterial groups are proportional to their in situ abundances are key issues to determine the partition of carbon flow for modeling or prediction purposes. Such questions have been assessed in different studies with the MAR-FISH technique, analyzing the carbon processed by different bacterial groups from the perspective of in situ uptake of representative substrates (at trace concentrations) (11) or biomass production (using leucine and/or thymidine uptake at saturating concentrations) (12).
Longnecker et al. (27) found a parallel increase or decrease in the average abundance of biosynthetically active cells (in leucine uptake) in the whole prokaryotic community along a trophic gradient in the California Current System, suggesting that no shifts in the activity of specific groups occurred. However, a great variability in the percentage of active cells within each phylogenetic group was found in each of the regions compared in their study (shelf, slope, and basin). Cottrell and Kirchman (12, 13) also found substantial changes in the percentage of active cells (in leucine and thymidine uptake) within different bacterial groups across a gradient in the Delaware estuary. These authors, by correlating the relative abundance and activity of these groups within the assemblage, concluded that only 50% of the contribution of the bacterial groups to biomass production could be explained by their relative proportions in the assemblage.
Studies analyzing the patterns of uptake of compounds at trace concentrations have also suggested that uptake activities are not uniformly distributed among groups (11, 18). Since LMW compounds can be easily transported across cell membranes, differences in the uptake patterns for monomers such as glucose or amino acids should be explained by the affinities of the uptake systems of each bacterial group. In this respect, Alonso and Pernthaler (3) carried out concentration-dependent experiments and showed that the contribution of a specific group of bacteria, such as SAR86 or the DE cluster 2 of Bacteroidetes, to the turnover of particular substrates was dependent on the concentrations at which these substrates were present in the waters.
In our study, we followed an annual cycle characterized by different trophic situations. We analyzed the patterns of uptake of three monomers that are common components of dissolved organic matter (DOM) in marine waters and are rapidly turned over by bacterioplankton: glucose (39, 40), amino acids (9), and ATP (8, 24). The rationale for using these three substrates was to study the supply of only carbon (glucose) or of carbon plus nitrogen (amino acids) and phosphorous (ATP) across a trophic range from nutrient-depleted (in summer) to nutrient-replete (winter) periods.
The higher rates of bulk uptake of ATP compared to glucose and amino acids throughout the year could be related to the phosphorous requirement of bacteria because of the strong year-round phosphorous limitation of bacterial production in Blanes Bay (36). Higher rates of bulk uptake of amino acids compared to ATP have been observed in several samples from the subtropical northeastern Atlantic Ocean, where bacteria showed carbon (instead of phosphorous) limitation (L. Alonso-Sáez and J. M. Gasol, unpublished data.). Other than phosphorous, ATP constitutes a source of carbon for bacteria. Several studies have reported that the best stimulation of growth is obtained when a combination of nutrients, and not a single nutrient, is supplied (16). This effect was also found in Blanes Bay, where the simultaneous addition of carbon and phosphorous stimulated bacterial growth (compared to addition of only phosphorous) during most of the year (36).
The rates of turnover of ATP have been shown to closely parallel those of glucose or amino acids (8, 24), and its assimilation is widespread in pure cultures of marine bacteria (8). These findings are in agreement with the results of our single-cell approach, in which ATP was taken up by all the bacterial groups studied, even if large differences were found for the other compounds (Fig. 3). The uptake of glucose and dissolved free amino acids has been more extensively studied, and apparently they can support a large fraction of bacterial growth in marine waters (8, 39, 40). Although it has been suggested that it is energetically advantageous to use preformed compounds such as amino acids compared to glucose, the energetic cost of transporting amino acids across the membranes can greatly offset this advantage (15). This seems to be the case in our system, where bulk uptake of amino acids was generally lower than that of glucose (Fig. 2). This could be related to the fact that nitrogen was not limiting bacterial production in Blanes Bay throughout the year (36), or it might simply reflect differences in the in situ availabilities of these substrates.
The MAR-CARD-FISH approach allowed the determination of the specific bacterial groups that were more successful in the uptake of these compounds year-round. Several reports have provided evidence that the ability to grow with a wide range of substrates is a typical adaptation of bacteria under nutrient-limited conditions (16). However, we found that some groups, such as Gammaproteobacteria and Bacteroidetes, did not substantially take up substrates as common as glucose at low concentrations. Furthermore, the seasonal measurement of the uptake activities allowed us to confirm some patterns in the uptake of LMW compounds shown in previous studies, such as the low activity of Bacteroidetes in amino acid uptake at trace concentrations (11), which appears to be a permanent feature year-round. In their study, Cottrell and Kirchman (11) proposed that this group shows higher affinity for high-molecular-weight compounds. However, we observed that Bacteroidetes could also competitively take up other LMW compounds such as ATP, probably forced by phosphorous limitation. It should be noted, though, that the incorporation of nucleotides requires previous extracellular hydrolysis (7), which could be regarded as a digestion process equivalent to that of particulate organic matter use.
The consistently low uptake of glucose by Bacteroidetes and Gammaproteobacteria throughout the year is also in agreement with the results of Alonso and Pernthaler (4). These authors showed that the majority of glucose-incorporating cells at the lower concentrations of the substrate were members of the Roseobacter and SAR11 lineages in the North Sea, whereas the proportion of Bacteroidetes significantly increased at higher levels of available substrate. Similar to our results, Gammaproteobacteria also showed low percentages of active cells incorporating glucose in Delaware Bay (18) and much higher uptake of amino acids (11), suggesting the preference of this group for amino acids rather than glucose as a carbon source.
Seasonal changes in single-cell activity of the bacterial groups.
The most novel contribution of this work is that we showed year-round changes in the proportions of active cells of specific groups and also in their contribution to the uptake of the compounds. Although seasonal changes in bulk bacterial production and respiration have been shown (26, 38), changes at the single-cell level of bacterial groups have not been systematically explored on the temporal scale.
Different seasonal patterns were found for distinct phylogenetic groups of bacteria. For example, low proportions of Gammaproteobacteria and Bacteroidetes took up amino acids and ATP, respectively, during the nutrient-limited season (summer), while higher percentages of cells (up to 60% of Gammaproteobacteria) took them up in spring and winter. In contrast, Roseobacter always showed high percentages of cells active in the uptake of the substrates throughout the year. These results could be related to the high ability of this group to take up substrates at all concentration ranges (3), and therefore, bacteria of the Roseobacter group seem well prepared to cope with different trophic conditions throughout the year. Their year-round low contributions to assemblage structure despite their high activity suggests that other factors, such as grazing, are controlling the in situ abundances of this group.
The study of the other alphaproteobacterial group, SAR11, is very relevant given their numerical dominance in marine regions (32), including our system, Blanes Bay (5). SAR11 was very abundant year-round, but its activity was quite variable throughout the year and much lower than that of Roseobacter. Substantial uptake of glucose and amino acids by this group has also been found in other studies (28). Because of the overrepresentation of SAR11 in the uptake of glucose and amino acids in their samples, those authors concluded that this group was a major contributor of bacterial biomass production and carbon flux and outcompeted other bacterial groups for carbon in the Sargasso Sea (28). However, the percentage of active SAR11 cells found in the Atlantic Ocean (over 80% active in amino acid uptake) was significantly higher than in our study (where it rarely exceeded 20%) and in other studies carried out in coastal areas such as the North Sea (4) (10 to 30%) or the Delaware Bay (18) (about 15%). Indeed, we found that this group contributed equally, or less than expected based on their abundance, to the total uptake of amino acids or glucose (Fig. 4), as was found by Elifantz et al. (18) in the Delaware Bay. Those authors suggested that these differences could be due to the eutrophic characteristics of the Delaware Bay compared to the Atlantic Ocean and to the selection of different phylotypes with diverse metabolism. However, even though Blanes Bay is a coastal site and not directly comparable to the open ocean, it is quite oligotrophic (yearly average Chl a is below 0.5 μg liter−1), and therefore our results suggest that SAR11 is not always a very active component of the carbon flux under nutrient-depleted conditions. We suggest that the contribution of this group to LMW DOM uptake can be highly variable year-round, at least in coastal ecosystems, although it can be high in some situations (Fig. 6).
Taxon substitutions within the bacterial groups could explain the seasonal changes in the uptake activities observed. In a fine-scale phylogenetic analysis, Acinas et al. (1) found four microdiverse clusters of the SAR11 lineage by grouping sequences at a 99% similarity level, which could represent different ecological and functional groups (i.e., ecotypes). However, in an exhaustive study on the bacterial assemblage structure in Blanes Bay (5), including five clone libraries, no marked seasonality was found for SAR11 sequences. This group showed substantial microdiversity (as in other studies [10, 20]) but high similarity between the sequences from spring (May), summer (August), autumn (October), and winter (February 2003, not sampled for MAR-FISH). Nevertheless, it will be very interesting to look at whether differences at a finer phylogenetic level can reveal distinct patterns of uptake activities of other bacterial groups.
In summary, our results confirm that (i) the activity of pelagic bacteria is substantially more dynamic than their population sizes and (ii) different groups of bacteria show very distinct DOM uptake patterns. Furthermore, we show that the activities of specific groups can vary year-round, particularly during the nutrient-limited season (summer). This could be due to different affinities of their uptake systems or phylotype substitution with different metabolisms. Further research on the seasonal activities of the phylogenetic groups of bacteria will be needed in order to better describe their individual contributions to total marine carbon heterotrophic processing.
Acknowledgments
This work was supported by the Spanish projects MicroDiff (REN2001-2110/MAR) and MODIVUS (CTM2005-04975/MAR) and the EU project BASICS (EVK3-CT-2002-00078) (to J.M.G.). It is also a contribution to the NoEs MARBEF and Eur-Oceans. Financial support was provided by a Ph.D. fellowship from the Spanish government to L. Alonso-Sáez.
We thank R. Malmstrom, M. Cottrell, D. Kirchman, C. Alonso, and J. Pernthaler for kindly teaching us the MAR-FISH technique. Special thanks go to M. Vila-Costa for her collaboration in setting the technique up in our lab, to D. Rubio and F. Auger for their help with the microscope analyses, to V. Balagué and C. Cardelús for organizing the Blanes Bay samplings, and to C. Pedrós-Alió for insightful comments.
Footnotes
Published ahead of print on 30 March 2007.
REFERENCES
- 1.Acinas, S. G., V. Klepac-Ceraj, D. E. Hunt, C. Pharino, I. Ceraj, D. L. Distel, and M. F. Polz. 2004. Fine-scale phylogenetic architecture of a complex bacterial community. Nature 430:551-554. [DOI] [PubMed] [Google Scholar]
- 2.Alonso, C., and J. Pernthaler. 2005. Incorporation of glucose under anoxic conditions by bacterioplankton from coastal North Sea surface waters. Appl. Environ. Microbiol. 71:1709-1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alonso, C., and J. Pernthaler. 2006. Concentration-dependent patterns of leucine incorporation by coastal picoplankton. Appl. Environ. Microbiol. 72:2141-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alonso, C., and J. Pernthaler. 2006. Roseobacter and SAR11 dominate glucose uptake in coastal North Sea waters. Environ. Microbiol. 8:2022-2030. [DOI] [PubMed] [Google Scholar]
- 5.Alonso-Sáez, L., V. Balagué, E. Sà, O. Sánchez, J. M. González, J. Pinhassi, R. Massana, J. Pernthaler, C. Pedrós-Alió, and J. M. Gasol. 2007. Seasonality in bacterial diversity in NW Mediterranean coastal waters: assessment through clone libraries, fingerprinting and fluorescence in situ hybridization. FEMS Microbiol. Ecol. 60:98-112. [DOI] [PubMed] [Google Scholar]
- 6.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ammerman, J. W., and F. Azam. 1985. Bacterial 5′-nucleotidase in aquatic ecosystems: a novel mechanism of phosphorous regeneration. Science 227:1338-1340. [DOI] [PubMed] [Google Scholar]
- 8.Azam, F., and R. E. Hodson. 1977. Dissolved ATP in the sea and its utilization by marine bacteria. Nature 267:696-697. [DOI] [PubMed] [Google Scholar]
- 9.Billen, G., and A. Fontigny. 1987. Dynamics of a Phaeocystis-dominated spring bloom in Belgian coastal waters. II. Bacteriplankton dynamics. Mar. Ecol. Prog. Ser. 37:249-257. [Google Scholar]
- 10.Brown, M. V., and J. A. Fuhrman. 2005. Marine bacterial microdiversity as revealed by internal transcribed spacer analysis. Aquat. Microb. Ecol. 41:15-23. [Google Scholar]
- 11.Cottrell, M. T., and D. L. Kirchman. 2000. Natural assemblages of marine Proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl. Environ. Microbiol. 66:1692-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cottrell, M. T., and D. L. Kirchman. 2003. Contribution of major bacterial groups to bacterial biomass production (thymidine and leucine incorporation) in the Delaware estuary. Limnol. Oceanogr. 48:168-178. [Google Scholar]
- 13.Cottrell, M. T., and D. L. Kirchman. 2004. Single-cell analysis of bacterial growth, cell size, and community structure in the Delaware estuary. Aquat. Microb. Ecol. 34:139-149. [Google Scholar]
- 14.Daims, H., A. Brühl, R. Amann, K. Schleifer, and M. Wagner. 1999. The domain-specific probe eub338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 15.del Giorgio, P. A., and J. J. Cole. 1998. Bacterial growth efficiency in natural aquatic systems. Annu. Rev. Ecol. Syst. 29:503-541. [Google Scholar]
- 16.Egli, T. 1995. The ecological and physiological significance of the growth of heterotrophic microorganisms with mixtures of substrates. Adv. Microb. Ecol. 14:305-386. [Google Scholar]
- 17.Eilers, H., J. Pernthaler, F. O. Glöckner, and R. Amann. 2000. Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl. Environ. Microbiol. 66:3044-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elifantz, H., R. R. Malmstrom, M. T. Cottrell, and D. L. Kirchman. 2005. Assimilation of polysaccharides and glucose by major bacterial groups in the Delaware Estuary. Appl. Environ. Microbiol. 71:7799-7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fasham, M. J. R., P. W. Boyd, and G. Savidge. 1999. Modeling the relative contributions of autotrophs and heterotrophs to carbon flow at a Lagrangian JGOFS station in the Northeast Atlantic: the importance of DOC. Limnol. Oceanogr. 44:80-94. [Google Scholar]
- 20.García-Martínez, J., and F. Rodríguez-Valera. 2000. Microdiversity of uncultured marine prokaryotes: the SAR11 cluster and the marine Archaea of Group I. Mol. Ecol. 9:935-948. [DOI] [PubMed] [Google Scholar]
- 21.Giovanonni, S. J., and M. Rappé. 2000. Evolution, diversity and molecular ecology of marine prokaryotes, p. 47-84. In D. L. Kirchman (ed.), Microbial ecology of the oceans. Wiley-Liss, New York, NY.
- 22.Giovanonni, S. J., H. J. Tripp, S. Givan, M. Podar, K. L. Vergin, D. Baptista, L. Bibbs, J. Eads, T. H. Richardson, M. Noordewier, M. S. Rappé, J. M. Short, J. C. Carrington, and E. J. Mathur. 2005. Genome streamlining in a cosmopolitan oceanic bacterium. Science 309:1242-1245. [DOI] [PubMed] [Google Scholar]
- 23.Grasshoff, K., M. Ehrhardt, and K. Kremling. 1983. Methods on seawater analysis, 2nd ed. Verlag Chemie, Weinheim, Germany.
- 24.Hodson, R. E., F. Azam, J. Fuhrman, A. F.Carlucci, D. M. Karl, and O. Holm-Hansen. 1981. Microbial utilization of dissolved organic matter in McMurdo Sound, Antarctica. Mar. Biol. 61:89-94. [Google Scholar]
- 25.Lean, D. R. S., and F. R. Pick. 1981. Photosynthetic response of lake plankton to nutrient enrichment: a test for nutrient limitation. Limnol. Oceanogr. 26:1001-1019. [Google Scholar]
- 26.Lemée, R., E. Rochelle-Newall, F. Van Wambeke, M.-D. Pizay, P. Rinaldi, and J.-P. Gattuso. 2002. Seasonal variation of bacterial production, respiration and growth efficiency in the open NW Mediterranean Sea. Aquat. Microb. Ecol. 29:227-237. [Google Scholar]
- 27.Longnecker, K., D. S. Homen, E. B. Sherr, and B. F. Sherr. 2006. Similar community structure of biosynthetically active prokaryotes across a range of ecosystem trophic states. Aquat. Microb. Ecol. 42:265-276. [Google Scholar]
- 28.Malmstrom, R. R., M. T. Cottrell, H. Elifantz, and D. L. Kirchman. 2005. Biomass production and assimilation of dissolved organic matter by SAR11 bacteria in the Northwest Atlantic Ocean. Appl. Environ. Microbiol. 71:2979-2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malmstrom, R. R., R. P. Kiene, M. T. Cottrell, and D. L. Kirchman. 2004. Contribution of SAR11 bacteria to dissolved dimethylsulfoniopropionate and amino acid uptake in the North Atlantic ocean. Appl. Environ. Microbiol. 70:4129-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manz, W., R. Amann, W. Ludwig, M. Vancanneyt, and H. Schleifer. 1996. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga-Flavobacter-Bacteroides in the natural environment. Microbiology 142:1097-1106. [DOI] [PubMed] [Google Scholar]
- 31.Manz, W., R. Amann, W. Ludwig, M. Wagner, and H. Schleifer. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593-600. [Google Scholar]
- 32.Morris, R. M., M. S. Rappé, S. A. Connon, K. L. Vergin, W. A. Slebold, C. A. Carlson, and S. J. Giovanonni. 2002. SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420:806-810. [DOI] [PubMed] [Google Scholar]
- 33.Neef, A. 1997. Anwendung der in situ-Einzelzell-Identifizierung von bakterin zur populationsanalyse in komplexen mikrobiellen Biozönosen. Ph.D. thesis. Technische Universität München, Munich, Germany.
- 34.Ouverney, C. C., and J. A. Fuhrman. 1999. Combined microautoradiography-16S rRNA probe technique for determination of radioisotope uptake by specific microbial cell types in situ. Appl. Environ. Microbiol. 65:1746-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pernthaler, A., J. Pernthaler, and R. Amann. 2004. Sensitive multi-color fluorescence in situ hybridization for the identification of environmental microorganisms, p. 711-726. In G. A. Kowalchuk, F. J. De Bruin, I. M. Head, A. D. L. Akkermans, and J. D. Van Elsas (ed.), Molecular microbial ecology manual, 2nd ed. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 36.Pinhassi, J., L. Gomez-Consarnau, L. Alonso-Sáez, M. M. Sala, M. Vidal, C. Pedrós-Alió, and J. M. Gasol. 2006. Seasonal changes in bacterioplankton nutrient limitation and their effects on bacterial community composition in the NW Mediterranean Sea. Aquat. Microb. Ecol. 44:241-252. [Google Scholar]
- 37.Rappé, M. S., S. A. Connon, K. L. Vergin, and S. J. Giovanonni. 2002. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418:630-633. [DOI] [PubMed] [Google Scholar]
- 38.Reinthaler, T., and G. J. Herndl. 2005. Seasonal dynamics of bacterial growth efficiencies in relation to phytoplankton in the southern North Sea. Aquat. Microb. Ecol. 39:7-16. [Google Scholar]
- 39.Rich, J. H., H. W. Ducklow, and D. L. Kirchman. 1996. Concentration and uptake of neutral monosaccharides along 140 degrees W in the Equatorial Pacific: contribution of glucose to heterotrophic bacterial activity and the DOM flux. Limnol. Oceanogr. 41:595-604. [Google Scholar]
- 40.Rich, J. H., M. Gosselin, E. Sherr, B. Sherr, and D. L. Kirchman. 1997. High bacterial production, uptake and concentrations of disolved organic matter in the central Arctic Ocean. Deep-Sea Res. II 44:1645-1663. [Google Scholar]
- 41.Schauer, M., V. Balagué, C. Pedrós-Alió, and R. Massana. 2003. Seasonal changes in the taxonomic composition of bacterioplankton in a coastal oligotrophic system. Aquat. Microb. Ecol. 31:163-174. [Google Scholar]
- 42.Smith, D. C., and F. Azam. 1992. A simple, economical method for measuring bacteria protein synthesis rates in seawater using 3H-leucine. Mar. Microbiol. Food Webs 6:107-114. [Google Scholar]
- 43.Smith, E. M., and P. A. del Giorgio. 2003. Low fractions of active bacteria in natural aquatic communities? Aquat. Microb. Ecol. 31:203-220. [Google Scholar]
- 44.Vila, M., R. Simó, R. P. Kiene, J. Pinhassi, J. A. Gonzalez, M. A. Moran, and C. Pedrós-Alió. 2004. Use of microautoradiography combined with fluorescence in situ hybridization to determine dimethylsulfoniopropionate incorporation by marine bacterioplankton taxa. Appl. Environ. Microbiol. 70:4648-4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wallner, G., R. Amann, and W. Beisker. 1993. Optimzing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 14:136-143. [DOI] [PubMed] [Google Scholar]