Abstract
Lactobacillus salivarius DPC6005, a porcine intestinal isolate, produces a two-component bacteriocin, salivaricin P, with homology to ABP-118 produced by a human probiotic L. salivarius strain. Indeed, molecular characterization revealed that while the peptides Sln1 and ABP-118α are identical, their companion peptides (Sln2 and ABP-118β, respectively) differ by two amino acids. This observation suggests that two-component bacteriocins may be a common feature of intestinal L. salivarius strains.
A large number of lactic acid bacteria produce bacteriocins, which are ribosomally synthesized and secreted peptides which kill other microorganisms. Indeed, intensive research over the last 20 years has identified several novel bacteriocins, many of which are produced by intestinal and/or probiotic bacteria (13). The production of bacteriocins by Lactobacillus salivarius strains has been reported previously (1, 12, 14, 17), and ABP-118, a two-component class II bacteriocin produced by a human intestinal probiotic strain, L. salivarius subsp. salivarius UCC118, has been fully characterized at the molecular level (6). Indeed, recent genome sequencing of the producing strain revealed that this bacteriocin is encoded on a 242-kb megaplasmid (4). In this paper, we describe the characterization of a two-component bacteriocin which is remarkably similar to ABP-118 and is produced by a porcine intestinal isolate, L. salivarius DPC6005, which was included in a five-strain probiotic mixture which contributed to reducing the numbers of Enterobacteriaceae by up to 98% in a porcine model without affecting the total fecal Lactobacillus population (7). Furthermore, we describe detection of a number of other intestinal strains with the ability to produce salivaricin P, a finding which suggests that the production of two-component antilisterial bacteriocins is prevalent among intestinal members of L. salivarius.
All bacterial strains used in this study are shown in Table 1, and all strains were stored at −80°C in 40% (vol/vol) glycerol. L. salivarius subsp. salivarius UCC118 was isolated previously from the human gastrointestinal tract and was obtained from University College, Cork, Ireland, under a restricted-material transfer agreement. Lactobacilli were grown anaerobically at 37°C in MRS (Oxoid Ltd., Hampshire, England). Anaerobic conditions were maintained in Anaerocult anaerobic jars (Merck, Darmstadt, Germany), using Anaerocult A gas packs (Merck). Lactococcal strains were propagated at 30°C in M17 (Difco Laboratories, Detroit, MI) supplemented with 0.5% (wt/vol) lactose and grown aerobically. Listeria innocua DPC3572 and enterococcal strains were propagated at 37°C in M17 supplemented with 0.5% (wt/vol) glucose and grown aerobically. Pediococci and Bacillus subtilis were propagated at 30°C in M17 supplemented with 0.5% (wt/vol) glucose and grown aerobically. Leuconostoc sp. was grown anaerobically at 30°C in MRS. Solid agar media were prepared by addition of 1% (wt/vol) agar (Difco) to the broth media. The bacteriocin-producing capabilities of strains were determined using cell-free supernatants in a modified agar well diffusion assay described previously (11), while the activity of purified bacteriocins was also examined using this method. A zone of inhibition between wells showed the synergistic activity between peptides. L. innocua DPC3572 was used as the indicator strain in all cases unless stated otherwise.
TABLE 1.
Comparison of the inhibition spectrum of the culture supernatant of L. salivarius DPC6005 with the inhibition spectra of strains DPC6027, DPC6189, DPC6196, M7.2, 7.3, and L. salivarius subsp. salivarius UCC118
| Strain | Sourcea | Inhibition byb:
|
||||||
|---|---|---|---|---|---|---|---|---|
| DPC6005 | DPC6027 | DPC6189 | DPC6196 | M7.2 | 7.3 | UCC118 | ||
| Bacillus subtilis BD630 | DPC3344 | − | − | − | − | − | − | − |
| Enterococcus faecalis DPC1142 | Cornell University 10C1(1) | ++ | − | ++ | − | ++ | ++ | ++ |
| Enterococcus faecalis DPC1143 | Cornell University 10C1(2) | ++ | − | ++ | − | ++ | ++ | ++ |
| Enterococcus faecium DPC1146 | NCDO942 | + | − | − | − | + | + | + |
| Lactobacillus casei DPC3539 | ATCC 339 | ++ | ++ | ++ | − | ++ | ++ | − |
| Lactobacillus fermentum DPC3320 | NCDO1133 | +++ | ++ | +++ | − | +++ | +++ | +++ |
| Lactobacillus helveticus DPC3510 | DPC | ++ | + | ++ | − | ++ | ++ | ++ |
| Lactobacillus delbrueckii subsp. bulgaricus | ATCC 11842 | +++ | ++ | +++ | − | +++ | +++ | +++ |
| Lactobacillus salivarius DPC6005 | DPC | NA | − | − | − | − | − | − |
| Lactobacillus salivarius DPC6027 | DPC | − | NA | − | − | − | − | − |
| Lactobacillus salivarius DPC6189 | DPC | − | − | NA | − | − | − | − |
| Lactobacillus salivarius DPC6196 | DPC | − | − | − | NA | − | − | − |
| Lactobacillus salivarius UCC118 | University College Cork | − | − | − | − | − | − | NA |
| Lactococcus cremoris HP | NZDRI | − | − | − | − | − | − | + |
| Lactococcus lactis DPC3147 | DPC | − | − | − | − | − | − | − |
| Leuconostoc sp. strain DPC2276 | NCDO942 | ++ | ++ | ++ | − | ++ | ++ | ++ |
| Listeria innocua DPC3572 | DPC | ++ | − | ++ | − | ++ | ++ | ++ |
| Pediococcus pentosaceus DPC2445 | NCDO992 | + | − | + | − | + | + | + |
| Pediococcus pentosaceus FBB63 | DPC 3541 | ++ | − | + | − | ++ | ++ | ++ |
| Staphylococcus aureus DPC5245 | DPC | − | − | − | − | − | − | − |
| Staphylococcus aureus DPC5246 | DPC | − | − | − | − | − | − | − |
| Streptococcus thermophilus DPC | DPC | ++ | − | ++ | − | ++ | ++ | − |
DPC, Dairy Products Research Centre, Teagasc, Moorepark, Ireland; NCDO, National Collection of Dairy Organisms; ATCC, American Type Culture Collection.
The diameters of the zones of inhibition, including the diameter of the well, which was approximately 6 mm, are indicated as follows: +++, >15 mm; ++, 10 to 15 mm; +, 6 to 10 mm; −, no zone of inhibition. NA, not applicable.
Genomic DNA was isolated from L. salivarius by a previously described method (9). A region of the genetic locus of ABP-118 from L. salivarius UCC 118, consisting of the abp118α and abp118β genes and a section of the abp118IM gene, was amplified using the following primers: 118αF (5′ATG ATG AAG GAA TTT ACA G 3′) and 118imR (5′CCA CGC TCT CAC ATA AC 3′). DNA products of intestinal L. salivarius strains were also amplified using these primers and an Eppendorf Mastercycler gradient (Eppendorf, Hamburg, Germany).
PCRs were performed by standard procedures (15). The PCR cycles were preceded by an initial denaturation step of 94°C for 5 min. DNA was amplified for 30 cycles, with each cycle involving a denaturation step of 30 s at 94°C and an annealing step of 50°C for 30 s, followed by an elongation step at 72°C for 30 s. A final single elongation step consisting of 72°C for 7 min completed the amplification procedure.
Bacteriocins were purified from L. salivarius strains that were grown overnight at 37°C in MRS broth by a modified method described previously (10). The modification involved applying the bacteriocin-containing eluate to a reverse-phase high-performance liquid chromatography (RP-HPLC) column prior to lyophilization. The 100-ml bacteriocin-containing eluate was concentrated to a volume of ∼4 ml by removal of propan-2-ol by rotary evaporation. Aliquots (approximately 2 ml) were then applied to a Phenomenex (Cheshire, United Kingdom) C18 RP-HPLC column (Primesphere 10μ C18-MC 30; 250 by 10.0 mm; 10 μm) previously equilibrated with 30% (vol/vol) propan-2-ol containing 0.1% (vol/vol) trifluoroacetic acid (TFA). The column was subsequently developed in a gradient from 30% (vol/vol) propan-2-ol containing 0.1% (vol/vol) TFA to 70% (vol/vol) propan-2-ol containing 0.1% (vol/vol) TFA from 5 to 50 min at a flow rate of 2.5 ml/min. Absorbance was monitored at a wavelength of 214 nm. Individual fractions of interest were further purified by reapplying them to the RP-HPLC column under the conditions described above.
For pulsed-field gel electrophoresis, high-molecular-mass chromosomal DNA was isolated from stationary-phase L. salivarius cultures by a previously described method (16). The only modification to the procedure involved growing strains in MRS supplemented with 20 mM dl-threonine (Sigma-Aldrich, Dublin, Ireland).
Analysis of the DNA sequence was performed by Lark Technologies, United Kingdom. Sequencing was performed for both strands of DNA. Specific oligonucleotide primers 118αF and 118imR, ∼700 bp apart, were designed using the known sequence (GenBank accession number AF408405). The sequence analysis was performed using the DNAStar software (DNAStar, Madison, WI).
Peptide fractions exhibiting antibacterial activity were fractionated by using the RP-HPLC conditions described above, and then the fractions which inhibited the growth of the indicator strain, L. innocua DPC3572, were collected and the peptide molecular masses were analyzed by mass spectrometry (MS) as previously described (5). The amino acid sequences of the antimicrobial peptides were determined after Edman degradation by loading approximately 1 pmol onto an Applied Biosystems Procise 494 cLC cartridge. The phenylthiohydantoin amino acids produced were identified online by capillary HPLC using UV absorbance detection at 269 nm as recommended by the manufacturer.
Protease sensitivity assays indicated that the antimicrobial agent, which was found to be stable over a range of temperatures from 20 to 100°C (data not shown), was probably a bacteriocin since the inhibitory effect was eliminated by proteinase K (data not shown). Furthermore, the bacteriocin could be classified as having a medium inhibition spectrum (Table 1). Interestingly, the inhibition spectrum of the bacteriocin of L. salivarius DPC6005 was found to be very similar to that of the ABP-118 bacteriocin produced by L. salivarius subsp. salivarius UCC118 (Table 1), and cross-sensitivity assays involving culture supernatants of the two strains demonstrated that they are cross-immune. Following incubation in MRS broth for 18 h, 2,560 and 1,280 activity units against L. innocua were detected in the culture supernatants of L. salivarius DPC6005 and L. salivarius subsp. salivarius UCC118, respectively.
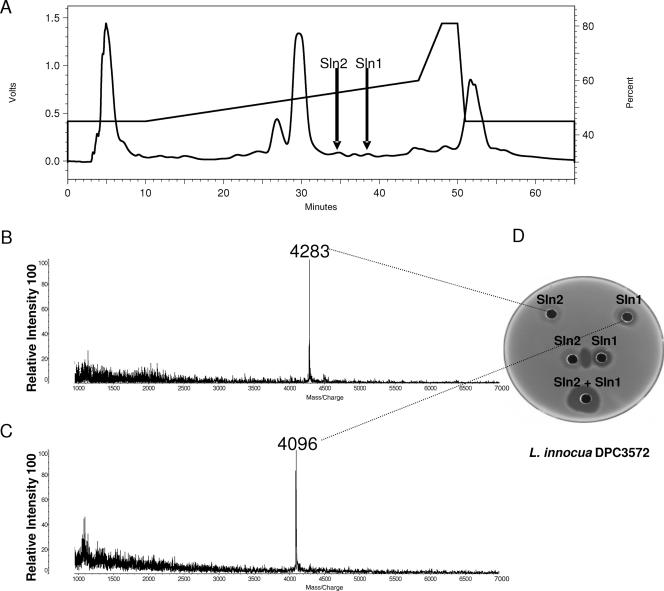
The bacteriocin produced by L. salivarius DPC6005 was purified and separated by adsorption onto a C18 silica column, followed by RP-HPLC, resulting in the chromatogram shown in Fig. 1. Agar well diffusion assays of the individually eluted fractions demonstrated that two peptides, designated Sln1 and Sln2, exhibited antimicrobial activity against the sensitive indicator strain L. innocua and that the activity was greatly enhanced when they were combined. Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) MS analysis of Sln1 and Sln2 revealed that their molecular masses are 4,096 and 4,283 Da, respectively (Fig. 1), while the molecular masses of the component peptides of the previously well-characterized bacteriocin ABP-118, ABP-118α and ABP-118β, were found to be 4,096 and 4,332 Da, respectively (data not shown). Edman degradation N-terminal amino acid sequencing of Sln1 revealed that the first 10 amino acids were identical to those of the α-peptide of ABP-118 (6) (data not shown).
FIG. 1.
(A) RP-HPLC profile of crude salivaricin P. Aliquots were applied to a Phenomenex C18 RP-HPLC column (Primesphere 10μ C18-MC 30; 250 by 10.0 mm; 10 μm. The column was subsequently developed in a gradient from 30% (vol/vol) propan-2-ol containing 0.1% (vol/vol) TFA to 70% (vol/vol) propan-2-ol containing 0.1% (vol/vol) TFA from 5 to 50 min at a flow rate of 2.5 ml/min. Absorbance was monitored at a wavelength of 214 nm. (B) MALDI-TOF MS data for Sln2. (C) MALDI-TOF MS data for Sln1. (D) Antilisterial activity of the Sln1 and Sln2 peptides, individually and together, against L. innocua DPC3572. The peptides were fractionated by RP-HPLC, and 50-μl portions of the fractions were added to wells.
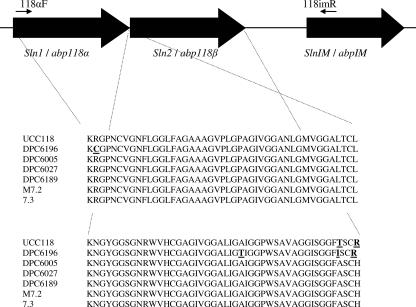
Due to the similarities between the bacteriocin produced by L. salivarius DPC6005 and ABP-118 (as determined by MALDI-TOF MS analysis, amino acid sequence analysis of the Sln1 peptide, cross-immunity analysis, and the similar inhibition spectra), specific oligonucleotide primers designed for the ABP-118 genetic locus were used to successfully amplify a fragment from the L. salivarius DPC6005 genome (Fig. 2). Analysis of the sequence of the DPC6005 gene coding for Sln1 production revealed two nucleotide differences between the Sln1 and ABP-118α genes. These differences, however, do not result in any amino acid change, and so the predicted peptides are identical (in agreement with the MALDI-TOF MS analysis [Fig. 1]). Sequencing of the genetic determinants responsible for production of the companion peptides, however, revealed six nucleotide differences between the Sln2 and ABP-118β genes, two of which result in differences in the predicted peptide sequences (Thr-43 and Arg-46 of ABP-118β are replaced by Ala and His in Sln2 [Fig 2]). This finding is supported by the observation that the actual mass of Sln2 differs from that of the ABP-118β peptide by 49.07 Da (as determined by MALDI-TOF MS analysis [Fig. 1]). DNA sequencing was performed for the ABP-118α and ABP-118β genes as positive controls, and the sequences were found to be identical to those reported previously (6) (accession number AF408405).
FIG. 2.
Schematic diagram of the structural genes involved in bacteriocin production and immunity in L. salivarius strains and the predicted peptide sequences of L. salivarius DPC6189, UCC118, DPC6005, DPC6196, DPC6027, M7.2, and 7.3. Amino acid differences are underlined.
The ability of two genetically distinct L. salivarius strains, UCC118 and DPC6005, to produce closely related two-component antilisterial bacteriocins led us to speculate that this ability may be a common feature of intestinal L. salivarius strains. Therefore, a further nine L. salivarius strains obtained from the Dairy Products Research Centre culture collection, of both human and porcine origin, were examined for the presence of the genes coding for the production of two-component antilisterial bacteriocins. Analysis of five genetically distinct strains (pulsed-field genomic data are not shown for four porcine isolates and one human isolate) generated a 702-bp PCR product corresponding to the genes required for the production of the two components of the antilisterial bacteriocins and a region of the immunity peptide.
Sequence analysis of four porcine L. salivarius isolates, M7.2 and 7.3, which are very closely related to DPC6005 (3), and the closely related isolates L. salivarius DPC6189 and DPC6027, revealed DNA sequences with 100% homology to the Sln1 and Sln2 genes of L. salivarius DPC6005 (data not shown). Sequencing of the genes coding for bacteriocin production in the human intestinal isolate L. salivarius DPC6196 revealed a 1-bp difference compared to the ABP-118α gene and a 2-bp difference compared to the Sln1 gene, while further sequence analysis revealed 2- and 8-bp differences compared with the ABP-118β and Sln2 genes, respectively. The differences in the DNA sequence of L. salivarius DPC6196 result in a single amino acid change in the predicted peptide sequence, from Arg-2 to Cys, compared with the ABP-118α and Sln1 peptides (Fig. 2), while the predicted peptide sequence of the companion peptide differs by two amino acids compared with the peptide sequence of ABP-118β and by three amino acids compared with the predicted peptide sequence of Sln2 (Fig. 2).
The porcine isolates L. salivarius DPC6189, DPC6027, M7.2, and 7.3 all produced salivaricin P, as determined by MALDI-TOF MS of their cell-free culture supernatants (data not shown); however, their inhibition spectra differed, and the zones of inhibition produced by the culture supernatants of DPC6005, DPC6189, M7.2, 7.3, and UCC118 with indicator strains were far larger than the zones produced by DPC6027 (Table 1). The human isolate L. salivarius DPC6196 did not exhibit any antimicrobial activity in a well diffusion assay despite possessing the genes for bacteriocin production, nor was a salivaricin P-like bacteriocin detected by MALDI-TOF MS (data not shown). Interestingly, all seven L. salivarius strains described in this study displayed cross-immunity.
It appears that the different inhibition spectra exhibited by the cell-free culture supernatants of L. salivarius DPC6005 and L. salivarius subsp. salivarius UCC118 may be due to the two substitutions in the companion peptides of salivaricin P and ABP-118 (Thr-43 and Arg-46 of ABP-118β are replaced by Ala and His in Sln2). The culture supernatant of DPC6005 inhibited Lactobacillus casei DPC3539 and Streptococcus thermophilus DPC1147 in a well diffusion assay, while the cell-free culture supernatant of UCC118 did not (Table 1). Conversely, the culture supernatant of L. salivarius subsp. salivarius UCC118 inhibited Lactococcus cremoris HP, while the culture supernatant of L. salivarius DPC6005 did not inhibit L. cremoris HP.
Interestingly, not only were the similarities between salivaricin P and ABP-118 highlighted, but it appears that salivaricin P production may be a common phenomenon among porcine intestinal L. salivarius strains. Clearly, while the genetic determinants for salivaricin P production are present in a number of porcine L. salivarius strains, the levels of production appear to differ. For example, the cell-free supernatant of L. salivarius DPC6027 inhibits Lactobacillus delbrueckii subsp. bulgaricus ATCC 11842 but not L. innocua DPC3572 in a well diffusion assay, despite the presence of salivaricin P in the culture supernatant. Furthermore, the human isolate L. salivarius DPC6196 possesses the genes for the production of a bacteriocin similar to both salivaricin P and ABP-118; however, the culture supernatant of this strain did not exhibit any antimicrobial activity against any of the indicator strains tested, despite the fact that the strain displays immunity to both salivaricin P and ABP-118. In addition to possessing the genes for bacteriocin production, the five L. salivarius strains described in this study all exhibited cross-immunity, suggesting that a similar immunity system operates in all five strains.
The results suggest that production of members of a family of two-component salivaricin P-like bacteriocins may be a common feature among intestinal L. salivarius strains, similar to mutacin production by Streptococcus mutans strains. Like mutacin-producing S. mutans strains, the L. salivarius strains described in this study were isolated from environments with diverse microbial populations, suggesting that bacteriocin production may confer an ecological advantage upon these strains in bacterial communities (2). Indeed, it is tempting to suggest that bacteriocin production by L. salivarius strains may help these strains predominate in the Lactobacillus population in the gut microflora. Interestingly, strong similarities in primary structure have also been reported to exist among a number of mutacins (2, 8), and it has been suggested that the production of similar bacteriocins by genetically unrelated S. mutans strains may be due to horizontal gene transfer (2). Perhaps similar events occurred in the genetically distinct porcine intestinal isolates which possess identical genetic determinants for salivaricin P production. A possible mechanism is plasmid transfer; indeed, a recent study identified the genes for ABP-118 bacteriocin production, as well as a number of suspected conjugation genes, on a megaplasmid, pMP118, in L. salivarius subsp salivarius UCC118 (4). In addition, pMP118-related megaplasmids of various sizes have been detected in a number of L. salivarius strains from various sources (4).
We concluded that the porcine cecal isolate L. salivarius DPC6005 produces a novel bacteriocin, salivaricin P, and that the production of such two-component bacteriocins may be a common feature among mammalian intestinal L. salivarius strains.
Acknowledgments
This work was funded by the Irish Government under National Development Plan 2000-2006, by the European Research and Development Fund, by EU project QLK1-2002-02362, and by Science Foundation Ireland. M. Hayes received a Teagasc Walsh Fellowship.
Footnotes
Published ahead of print on 6 April 2007.
REFERENCES
- 1.Arihara, K., S. Ogihara, T. Mukai, M. Itoh, and Y. Kondo. 1996. Salivacin 140, a novel bacteriocin from Lactobacillus salivarius subsp. salicinius T140 active against pathogenic bacteria. Lett. Appl. Microbiol. 22:420-424. [DOI] [PubMed] [Google Scholar]
- 2.Balakrishnan, M., R. S. Simmonds, M. Kilian, and J. R. Tagg. 2002. Different bacteriocin activities of Streptococcus mutans reflect distinct phylogenetic lineages. J. Med. Microbiol. 51:941-948. [DOI] [PubMed] [Google Scholar]
- 3.Casey, P. G., G. D. Casey, G. E. Gardiner, M. Tangney, C. Stanton, R. P. Ross, C. Hill, and G. F. Fitzgerald. 2004. Isolation and characterization of anti-Salmonella lactic acid bacteria from the porcine gastrointestinal tract. Lett. Appl. Microbiol. 39:431-438. [DOI] [PubMed] [Google Scholar]
- 4.Claesson, M. J., Y. Li, S. Leahy, C. Canchaya, J. P. van Pijkeren, A. M. Cerdeno-Tarraga, J. Parkhill, S. Flynn, G. C. O'Sullivan, J. K. Collins, D. Higgins, F. Shanahan, G. F. Fitzgerald, D. van Sinderen, and P. W. O'Toole. 2006. Multireplicon genome architecture of Lactobacillus salivarius. Proc. Natl. Acad. Sci. USA 103:6718-6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cotter, P. D., L. H. Deegan, E. M. Lawton, L. A. Draper, P. M. O'Connor, C. Hill, and R. P. Ross. 2006. Complete alanine scanning of the two-component lantibiotic lacticin 3147: generating a blueprint for rational drug design. Mol. Microbiol. 62:735-747. [DOI] [PubMed] [Google Scholar]
- 6.Flynn, S., D. van Sinderen, G. M. Thornton, H. Holo, I. F. Nes, and J. K. Collins. 2002. Characterization of the genetic locus responsible for the production of ABP-118, a novel bacteriocin produced by the probiotic bacterium Lactobacillus salivarius subsp. salivarius UCC118. Microbiology 148:973-984. [DOI] [PubMed] [Google Scholar]
- 7.Gardiner, G. E., P. G. Casey, G. Casey, P. B. Lynch, P. G. Lawlor, C. Hill, G. F. Fitzgerald, C. Stanton, and R. P. Ross. 2004. Relative ability of orally administered Lactobacillus murinus to predominate and persist in the porcine gastrointestinal tract. Appl. Environ. Microbiol. 70:1895-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hillman, J. D., J. Novak, E. Sagura, J. A. Gutierrez, T. A. Brooks, P. J. Crowley, M. Hess, A. Azizi, K. Leung, D. Cvitkovitch, and A. S. Bleiweis. 1998. Genetic and biochemical analysis of mutacin 1140, a lantibiotic from Streptococcus mutans. Infect. Immun. 66:2743-2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmann, C. S., and F. Winston. 1987. A ten minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57:267-272. [DOI] [PubMed] [Google Scholar]
- 10.Martin, N. I., T. Sprules, M. R. Carpenter, P. D. Cotter, C. Hill, R. P. Ross, and J. C. Vederas. 2004. Structural characterization of lacticin 3147, a two-peptide lantibiotic with synergistic activity. Biochemistry 43:3049-3056. [DOI] [PubMed] [Google Scholar]
- 11.McAuliffe, O., M. P. Ryan, R. P. Ross, C. Hill, P. Breeuwer, and T. Abee. 1998. Lacticin 3147, a broad-spectrum bacteriocin which selectively dissipates the membrane potential. Appl. Environ. Microbiol. 64:439-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ocana, V. S., A. A. Pesce De Ruiz Holgado, and M. E. Nader-Macias. 1999. Characterization of a bacteriocin-like substance produced by a vaginal Lactobacillus salivarius strain. Appl. Environ. Microbiol. 65:5631-5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Connor, E. B., E. Barrett, G. F. Fitzgerald, C. Hill, C. Stanton, and R. P. Ross. 2005. Production of vitamins, exopolysaccharides and bacteriocins by probiotic bacteria, p. 167-194. In A. Y. Tamime (ed.), Probiotic dairy products. Blackwell Publishing, Oxford, United Kingdom.
- 14.Robredo, B., and C. Torres. 2000. Bacteriocin production by Lactobacillus salivarius of animal origin. J. Clin. Microbiol. 38:3908-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed., vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 16.Simpson, P. J., C. Stanton, G. F. Fitzgerald, and R. P. Ross. 2002. Genomic diversity within the genus Pediococcus as revealed by randomly amplified polymorphic DNA PCR and pulsed-field gel electrophoresis. Appl. Environ. Microbiol. 68:765-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.ten Brink, B., M. Minekus, J. M. van der Vossen, R. J. Leer, and J. H. Huis in't Veld. 1994. Antimicrobial activity of lactobacilli: preliminary characterization and optimization of production of acidocin B, a novel bacteriocin produced by Lactobacillus acidophilus M46. J. Appl. Bacteriol. 77:140-148. [DOI] [PubMed] [Google Scholar]