Abstract
The composition of ammonia-oxidizing bacteria from the β-Proteobacteria subclass (βAOB) was studied in the surface and upper-oxycline oxic waters (2- to 50-m depth, ∼200 to 44 μM O2) and within the oxygen minimum zone (OMZ) suboxic waters (50- to 400-m depth, ≤10 μM O2) of the eastern South Pacific off northern Chile. This study was carried out through cloning and sequencing of genes coding for 16S rRNA and the ammonia monooxygenase enzyme active subunit (amoA). Sequences affiliated with Nitrosospira-like cluster 1 dominated the 16S rRNA gene clone libraries constructed from both oxic and suboxic waters. Cluster 1 consists exclusively of yet-uncultivated βAOB from marine environments. However, a single clone, out of 224 obtained from the OMZ, was found to belong to Nitrosospira lineage cluster 0. To our knowledge, cluster 0 sequences have been derived from βAOB isolated only from sand, soil, and freshwater environments. Sequences in clone libraries of the amoA gene from the surface and upper oxycline could be grouped in a marine subcluster, also containing no cultured representatives. In contrast, all 74 amoA sequences originating from the OMZ were either closely affiliated with cultured Nitrosospira spp. from clusters 0 and 2 or with other yet-uncultured βAOB from soil and an aerated-anoxic Orbal process waste treatment plant. Our results reveal the presence of Nitrosospira-like βAOB in both oxic and suboxic waters associated with the OMZ but with a clear community shift at the functional level (amoA) along the strong oxygen gradient.
Oxygen minimum zones (OMZs) are permanent areas of the ocean with dissolved O2 concentrations of ≤22 μM at intermediate depths (15, 18) caused by low rates of ventilation and high biological O2 demand. The main zones are found in the eastern tropical Pacific and Arabian Sea. Associated with a sharp decrease of dissolved O2 concentration with depth is an intense nitrogen cycle that generates characteristic vertical distributions of nitrogen compounds within the OMZ: i.e., high NO3− deficit, a secondary nitrite maximum, and low NH4+ concentrations are found within the core, as well as N2O maxima towards the OMZ boundaries (8, 26, 30, 41). Such strong chemical gradients produced by OMZs could be considered as gene flow barriers and their oxygen-deficient waters as suitable environments for innovation, where organisms develop specific adaptations to exploit these unique conditions (9, 30, 42). In fact, novel and yet-uncultivated groups of denitrifiers have been found in the OMZ of the Arabian Sea (17a) and the eastern South Pacific (6a) by cloning and sequencing of the nirS genes. However, the diversity of most of the OMZ microbial world remains unexplored.
Ammonia-oxidizing bacteria (AOB) are considered to play a key role in the nitrogen cycling of these OMZs, contributing to NH4+ removal and NO2− and N2O production (26, 33, 52). AOB consist of a group of chemolithoautotrophic microorganisms able to oxidize aerobically NH4+ to NO2−, via NH2OH (hydroxylamine), a dissimilative metabolic process that also produces N2O as a by-product (20). Known AOB are found in three genera of the Proteobacteria phylum: Nitrosomonas and Nitrosospira, in the β class (βAOB), and Nitrosococcus, in the γ class (γAOB) (40, 50). In marine systems, the study of genes encoding the active site-containing subunit of ammonia monooxygenase (amoA), which is involved in the oxidation of NH4+ to NH2OH, has indicated that βAOB are predominant over γAOB (35). In addition, within the β subdivision, 16S rRNA genes libraries of AOB indicate that in planktonic marine samples sequences from Nitrosospira-like organisms are more frequent than those from Nitrosomonas-like bacteria, which in turn are mainly found associated with particles (3, 13, 16, 37, 39). However, in the OMZ off Peru, Ward et al. (52) found high chemoautotrophic activity related to Nitrosomonas sp. (marine) and Nitrosococcus oceanus by combining autoradiography with immunofluorescence methods. Thus, what the main AOB in OMZs are and what their contribution to the nitrogen cycle is remain unknown, since other NH4+ oxidizers could also be influencing the NH4+ removal in OMZs, such as the anammox bacteria (23, 49) and perhaps even the recently-reported ammonia-oxidizing archaea (11, 56).
Herein, we address the first question by exploring the community structure of βΑΟΒ in an O2 gradient associated with the OMZ off northern Chile (∼20°S) using phylogenetic analysis of the genes coding for the 16S rRNA and the ammonia monooxygenase enzyme active subunit (amoA).
MATERIALS AND METHODS
Sample collection and chemical analyses.
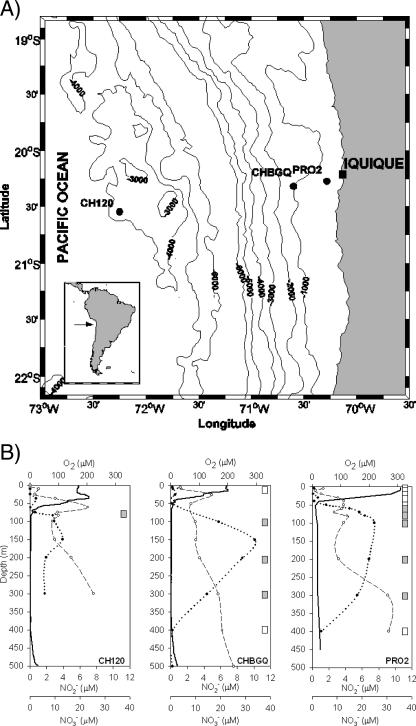
The study area (Fig. 1) was visited during March 2003 (stations CH120 and CHBGQ, CHUPS cruise onboard R/V Abate Molina) and July 2004 (station PRO2, PRODEPLOY cruise onboard R/V Carlos Porter). The physical and chemical characteristics of the water column were measured using a conductivity-temperature-depth profiler with an O2 sensor (Seabird). Profiles of dissolved O2 concentrations were examined to select the depths for collecting water samples for DNA extraction. The O2 gradient sampled was between ∼2 and 250 μM O2. Seawater (∼10 liters) was collected for environmental DNA isolation at each station using Niskin bottles attached to a rosette at the following nominal depths: CH120 (82 m), CHBGQ (10, 100, 200, 300, and 400 m), and PRO2 (2, 10, 20, 40, 50, 60, 80, 100, 200, 300, and 450 m).
FIG. 1.
(A) Map of the study area showing the sampling stations off the northern Chile coast (∼20°S, Iquique): CH120 and CHBGQ (CHUPS cruise, March 2003) and PRO2 (PRODEPLOY cruise, July 2004). (B) Distribution of dissolved oxygen (solid line), nitrite (dotted line with black circles), and nitrate (dashed line with white circles) and the sampling depths from which DNA was extracted. White and gray boxes indicate the samples derived from the more-oxygenated and oxygen-deficient waters, respectively.
Samples were prefiltered serially through a 20-μm mesh and a 5-μm filter and then filtered onto a 47-mm-diameter 0.22-μm membrane Nuclepore filter under a gentle vacuum flow. After filtration, each filter was soaked with 1 ml of DNA buffer (50 mM Tris-HCl [pH 9.0], 0.75 M sucrose, 400 mM NaCl) and stored in liquid nitrogen during transport and then at −80°C until DNA extraction in the laboratory.
In addition, discrete water samples were collected for determination of N2O, NH4+, NO3−, and NO2− concentrations. To avoid oxygenation, the headspace of the Niskin bottles was filled with N2 under atmospheric pressure. Samples for N2O analyses were processed according to Castro-González and Farías (7). NH4+ (40 ml, triplicate) was determined by a fluorometric method (17) using a Turner Designs 10-AU fluorometer. Samples for NO3− and NO2− determination were filtered (GF/F; Whatman) and stored frozen until later analysis with an automatic analyzer (ALPKEM, Flow Solution IV).
Hydrographic data associated with the CHUPS cruise have been published by Castro-González and Farías (7) and have been submitted for publication by V. Molina and L. Farías.
DNA extraction.
The procedure used to extract environmental DNA was modified from that described by West and Scanlan (55). Briefly, filters were cut into small pieces and placed in 3 ml of lysis solution (45 mM glucose, 23 mM Tris [pH 8.0], 59 mM EDTA) containing lysozyme at a concentration of 1 mg ml−1. This mixture was incubated for 1 h at 37°C, frozen at −20°C (15 min), and then thawed at 50°C. DNA was extracted by adding 350 μl of 10% sodium dodecyl sulfate and 33.5 μl of a proteinase K solution (10 mg ml−1), followed by incubation at 37°C (30 min) and at 55°C (10 min). This solution was mixed gently with the pipette, frozen at −20°C (15 min), and then thawed at 50°C. The thawed mixture was extracted with water-saturated phenol (neutralized with 0.5 M Tris-HCl buffer [pH 7.8 to 8.5])-chloroform-isoamyl alcohol (25:24:1) and centrifuged at 4,000 rpm (5 min). After centrifugation, the aqueous phase was carefully removed and again the DNA was extracted with chloroform-isoamyl alcohol (24:1) and centrifuged at 4,000 rpm (5 min). DNA was precipitated by adding 0.4 volume of sodium acetate (7.5 M) and 2 volumes of ice-cold ethanol (95%). The mixture was kept at −80°C for 20 min and then centrifuged at 13,000 rpm (10 min). The DNA pellet was washed with 70% ethanol, left to dry for 10 to 15 min, resuspended in 100 μl of TE (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]), and stored at −80°C. DNA was examined in an agarose gel. For PRODEPLOY cruise samples, the DNA extraction procedure was modified by the introduction of a physical breakage step according to Stevens et al. (48).
PCR.
The primers used to amplify 16S rRNA genes (CHUPS and PRODEPLOY cruise samples) and amoA genes (PRODEPLOY cruise samples), as well as their target groups, positions, particular annealing temperatures, and references are shown in Table 1. The forward primer amoA34f was designed to target a position close to the 5′ region of the amoA gene, in order to retrieve a longer amoA fragment than if the amoA1f primer was used. This new primer was generated by visual inspection of an alignment of 27 full-length amoA gene sequences from 14 AOB species of the following genera: Nitrosomonas (7 species), Nitrosospira (5 species, including Nitrosolobus multiformis and Nitrosovibrio tenuis), and Nitrosococcus (2 species). The primer targets a conserved region in βAOB and does not match γAOB. PCR amplification using amoA34f in combination with reverse primers, such as amoA2r (44), produces a fragment of approximately 800 bp. The primer specificity was evaluated experimentally with several cultures of known βAOB (e.g., Nitrosomonas eutropha, Nitrosomonas europaea, Nitrosospira briensis, Nitrosospira multiformis, and Nitrosovibrio tenuis) and also non-βAOB available in the laboratory (e.g., Mesorhizobium loti and Rhizobium tropici).
TABLE 1.
Primers used to amplify 16S RNA and ammonia monooxygenase active subunit (amoA) genes, specific target groups, nucleotide positions, and particular annealing temperature for PCR
| Primer set | Base sequence (5′ to 3′) | Nucleotide position | Target group | Annealing temp (°C) | Reference |
|---|---|---|---|---|---|
| EUB 1 | GAGTTTGATCCTGGCTCAG | 9a | General eubacteria | 40 | 25 |
| EUB 2 | AGAAAGGAGGTGATCCAGCC | 1542a | |||
| NIT A | CTTAAGTGGGGAATAACGCATCG | 137a | βAOB | 57 | 51 |
| NIT B | TTACGTGTGAAGCCCTACCCA | 1234a | |||
| βAMO f | TGGGGRATAACGCAYCGAAAG | 141a | βAOB | 57 | 28 |
| βAMO r | AGACTCCGATCCGGACTACG | 1320a | |||
| amoC305fd | GTGGTTTGGAACRGICARAGCAAA | 1012c | amoC | 57 | 36 |
| amoA1f | GGGGTTTCTACTGGTGGT | 332b | amoA | 50 | 44 |
| amoA2r | CCCCTCKGSAAAGCCTTCTTC | 822b | |||
| amoA34fd | GCGGCRAAAATGCCGCCGGAAGCG | 28b | amoA | 57 | This study |
E. coli 16S rRNA genes.
Nitrosomona europaea ammonia monooxygenase gene (amoA).
Nitrosospira sp. strain NpAV.
Combined with amoA2r.
Direct and nested PCRs, using genomic DNA and PCR-EUB 1-2 and amoC-amoA products as templates, respectively, were used in this study, due to the low abundance of AOB in environmental samples (53, 54). PCRs used to obtain 16S rRNA genes from known βAOB were conducted employing the nested PCR approach, using EUB 1-2 PCR products as template (with NIT A-B and βAMO f-r primer sets) and directly using genomic DNA as template (NIT A-B primers). For all of the 16S rRNA gene amplifications, a 50-μl PCR was carried out using a master mix containing 1 μl template DNA (∼10 ng) or PCR product (in the case of nested PCR) and a final concentration of 1× PCR buffer (50 mM KCl, 10 mM Tris-HCl, 1% Triton X-100, 1.25 mM MgCl2; Roche), 200 μM each deoxynucleotide triphosphate, 0.1 μM each primer (forward and reverse), and 1 U of Taq DNA polymerase (Roche). For the 16S rRNA gene amplification, a thermocycler program consisting of the following steps was used: 5 min at 94°C and then 35 cycles of 30 s at 94°C, 45 s at annealing temperature according to the primer (see Table 1), and 1.5 min at 72°C. PCRs used to obtain the amoA gene from known βAOB were done by a direct (amoA1f-2r primers) and nested (amoA1f-2r and amoA34f-2r) approach using amoC305f-amoA2r PCR products as a template (36, 44). For all of the amoA gene amplifications, a 25-μl PCR mixture containing 1 μl template DNA (∼10 ng) or PCR product (in the case of nested PCR) and a final concentration of 1× PCR buffer (50 mM KCl, 10 mM Tris-HCl; Roche), 4 mM MgCl2, 200 μM each deoxynucleotide triphosphate, 0.1 μM each primer (forward and reverse), 100 mg/ml bovine serum albumin (Sigma), 50% formamide, and 1 U of Taq DNA polymerase (Roche) was used. A thermocycler program consisting of 5 min at 94°C, then 35 cycles of 1 min at 94°C, with 1 min at annealing temperature according to the primer (see Table 1) and 2 min at 72°C, and with a final extension of 7 min at 72°C was used for all amo amplifications. All PCRs were carried out with positive (Nitrosomonas europaea) and blank controls. PCR products were run together with appropriate ladders (1,000 or 100 bp) and visualized on agarose gels using standard electrophoresis procedures.
Clone library construction, sequencing analyses, and phylogenetic affiliation within βAOB.
The DNA samples obtained during the CHUPS cruise were used to characterize βAOB through 16S rRNA gene analyses. In addition, DNA samples from a second cruise (PRODEPLOY) were used to study the amoA genes and to corroborate 16S rRNA genes results from the CHUPS cruise. βΑΜΟ f-r products were chosen at the beginning of this study because they are less specific than NIT A-B, thus reducing the risk of missing closely related sequences of βAOB (50) and avoiding potential biases against Nitrosospira-like sequences with NIT A-B (19). NIT A-B products were chosen during the second survey because a higher abundance of βAOB-like sequences were obtained in the NIT A-B clone libraries than with βΑΜΟ f-r, and the sequences that originated with NIT A-B and βΑΜΟ f-r were almost identical. The 16S rRNA gene clone libraries constructed during this study and specific labels are listed in Table 2. amoA gene clone libraries were constructed using a nested approach as follows. amoC305f-amoA2r products were used as templates (36, 44), and the following inner primers were used: amoA34f-amoA2r primers were used for samples PRO2-40 m, PRO2-80 m, and PRO2-300 m, and amoA1f-2r primers were used for samples PRO2-2 m, PRO2-10 m, PRO2-40 m, PRO2-80 m, and PRO2-300 m. A single clone library was also obtained using amoA1f-2r products originating directly from the genomic DNA obtained from sample PRO2-40 m. All of these products were gel purified by using QIAquick PCR purification kit (QIAGEN).
TABLE 2.
Number of βAOB-like sequences in the 16S rRNA gene clone libraries generated during this study
| Cruise | Depth (m)a | Primer or template used for library construction and PCR amplification
|
Library size (no. of clones) | No. of βAOB-like clones | |
|---|---|---|---|---|---|
| Primer | Template | ||||
| CH120 | 82* | AMO f-r | EUB 1-2 | 130 | 37 |
| NIT A-B | Genomic DNA | 36 | 36 | ||
| CHBGQ | 10 | βAMO f-r | EUB 1-2 | 46 | 12 |
| NIT A-B | Genomic DNA | 44 | 44 | ||
| 100* | βAMO f-r | EUB 1-2 | 91 | 1 | |
| PRO2 | 40 | NIT A-B | Genomic DNA | 76 | 76 |
| 80* | NIT A-B | Genomic DNA | 54 | 18 | |
| NIT A-B | EUB 1-2 | 30 | 0 | ||
| 300* | NIT A-B | EUB 1-2 | 46 | 0 | |
*, depth within the OMZ according to the presence of the secondary nitrite maximum (Fig. 1).
Cloning was performed using the TOPO TA cloning kit (Invitrogen) following the manufacturer's instructions. Briefly, the PCR products were ligated into a plasmid vector (pCR4-TOPO vector with genes coding for kanamycin resistance). Competent Escherichia coli cells (provided by the manufacturer) were transformed with the ligate and incubated at 37°C (∼24 to 48 h) on agar plates containing LB medium with kanamycin (50 μg ml−1) and spread with 40 μl (40 mg ml−1) of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). White or light-blue colonies were picked, and inserts of the correct size were evaluated by PCRs using M13 f-r vector-specific primers and agarose electrophoresis. For sequencing, M13 f-r PCR products were purified using multiscreen plates (Millipore). All cycle sequencing reactions were carried out using the Big Dye Terminator v3.1 cycle sequencing kit (ABI PRISM) and analyzed using a 3100 genetic analyzer (ABI, Applied Biosystems). 16S rRNA clone libraries were prescreened by sequencing using primer 341 f 16S rRNA genes of E. coli (32). The sequences were analyzed by generating multiple alignments and by comparison with the NCBI database using BLAST. Based on this screening, clones were selected for complete sequencing. Full-length sequencing was performed using five additional primers: M13 f, 926 f, M13 r, 534 r, and 907 r (24, 32). For amoA gene products, all clones were sequenced with M13 f and M13 r. Forward and reverse sequences were manually inspected using the Autoassembler (ABI Biosystems) and SeqMan (DNASTAR-Lasergene v6) software packages, and consensus sequences were constructed. These sequences were checked for chimeras (using CHECK-CHIMERA at the Ribosomal Database Project II and Bellerophon chimera detection programs, available from http://35.8.164.52/cgis/chimera.cgi?su = SSU). Consensus sequences constructed by full overlap of forward and reverse sequences were used for further analyses. The phylogenetic analyses were conducted using the Phylip software package (10), available from http://evolution.genetics.washington.edu/phylip/software.html, and the MEGA version 3.0 software package (22), available from http://www.megasoftware.net/. The phylogenetic analyses were based on maximum likelihood (ML) and distance matrix methods (neighbor joining [NJ]). Trees were drawn using the program Bosque for Linux (http://www.profc.udec.cl/bosque). Nucleotide and amino-acid sequences were used for phylogenetic analyses of the amoA gene. Amino acid sequences were translated from nucleotide sequences using MEGA and corroborated by the translate tool in ExPASy (http://www.expasy.org/tools/dna.html). The different phylogenetic analyses were compared visually, and a consensus tree was created based on ML topology. Multifurcations were generated when inconsistencies between ML and NJ results were found according to Ludwig et al. (27) and Purkhold et al. (40).
Nucleotide sequence accession numbers.
The full sequences of the 16S rRNA gene clones have been deposited in GenBank under accession no. DQ009933 to DQ009964 (CHUPS) and DQ335526 to DQ335537 and DQ813659 (PRODEPLOY). The amoA clone sequences have been deposited in GenBank under accession no. DQ519036 to DQ519048.
RESULTS AND DISCUSSION
Biogeochemical characteristics of the water column.
During this study, we identified some of the typical geochemical features that delineate the presence of an OMZ (Fig. 1). Vertical distributions of NO2− and NO3− for the CHUPS cruise data have been published by Castro-González and Farías (7). The water column was characterized by a steep oxycline (300 to ≤10 μM O2) situated within the top 50 and 70 m at the coastal stations CHBGQ and PRO2 and oceanic station CH120, respectively. Below the oxycline, oxygen-deficient waters (∼2 to 10 μM O2) were detected at all stations from the base of the oxycline until ∼400 m in depth. The oxygen-deficient waters presented a high accumulation of NO2− (up to 8.7 μM) in a prominent secondary nitrite maximum. This secondary nitrite maximum also coincided with NO3− concentrations (9.8 to 13.9 μM) lower than those expected from the Redfield stoichiometry (Fig. 1). Nitrogen deficits between 10 and 21 μM were estimated within the OMZ using the N* approach (14; data not shown), suggesting important nitrogen removal from this layer. The OMZ was also characterized by low NH4+ (≤0.14 μM) and N2O (≤32 nM) concentrations (data not shown). In contrast, the upper oxycline not only presented lower NO2− and higher NO3− concentrations (Fig. 1) but also showed higher NH4+ and N2O concentrations (up to 0.81 μM and up to 69.2 nM, respectively [data not shown]). N2O and NH4+ vertical distributions for the CHUPS cruise had been published by Castro-González and Farías (7) and have been submitted for publication by Molina and Farias, respectively.
These features reflect the strong nitrogen cycling associated with the OMZ off northern Chile, which appears to be influenced by denitrification and nitrification (7, 9a). In fact, during the CHUPS cruise, high rates of NO3− reduction (up to 9.1 μM day−1), denitrification (∼3.2 μM day−1), and the potential contribution of denitrifiers and nitrifiers to the local N2O production (92 and 8%, respectively) were measured (7). During the same cruise, high potential aerobic NH4+ oxidation and carbon fixation activity believed to be caused by the presence of AOB were found, with respective rates of up to 1.0 μM NH4+ day−1 and up to 0.22 μg C day−1 (Molina and Farias, submitted). These rates suggest that an AOB community is present and may contribute significantly to nitrogen and carbon cycles within the oxyclines and OMZ off northern Chile.
βAOB potential presence in the water column.
Positive PCR amplifications of the 16S rRNA and amoA genes related to βAOB were obtained in all instances when the nested PCR approach was used (see Materials and Methods). However, direct PCR on genomic DNA using NIT A-B, amoC305f-amoA2r, and amoA1f-2r primers generated positive results only in some of the samples. NIT A-B and amoA1f-2r products showed positive and more intense bands mainly in the samples retrieved from the upper part of the water column (2- to 100-m depth [data not shown]). PCR amplifications of 16S rRNA genes (using NITA-B and βΑΜΟ f-r primers) and amoA genes in the study of βAOB have been used as indicators of the presence of this group in environmental samples (20, 53, 54), and therefore the direct PCR amplification from genomic DNA obtained at some depths and with a higher intensity of the PCR product could indicate variability in the AOB target abundance in the water column. Indeed, Ward et al. (52) found vertical variation in the abundance of AOB in the water column off Peru by immunofluorescence staining against Nitrosomonas sp. (marine) and a Nitrosococcus oceanus strain, where the highest abundances were concentrated in the upper boundary of the OMZ, ∼55-m depth (1.8 × 105 cells liter−1). One must be cautious to infer the presence of βAOB using NIT A-B direct PCR results, as they could include non-βAOB sequences (see cloning results below). However, NIT A-B combined with amoA1f-2r direct PCR may provide further evidence that βAOB are more abundant in the upper part of the water column off northern Chile, including the surface oxic, hypoxic, and suboxic waters that comprise the upper part of the OMZ. Nevertheless, to determine the actual abundance and distribution of this group in the water column, the application of other techniques, such as fluorescence in situ hybridization or quantitative PCR, would be required.
16S rRNA gene cloning.
The number of βAOB in the 16S rRNA genes clone libraries derived from 341 f primer sequences analyses are listed in Table 2. In general, nested clone libraries constructed with βΑΜΟ f-r and NIT A-B PCR products showed a lower βAOB recovery compared to NIT A-B PCR products that originated directly from genomic DNA (i.e., ≤26% and ≥33%, respectively). All the βAOB-like sequences retrieved from βΑΜΟ f-r and NIT A-B clone libraries presented a sequence similarity after BLAST of ≥95% with Nitrosospira-like sequences. Most of the non-βAOB-like sequences were related to uncultured β- and γ-proteobacteria (data not shown). The low number of clones and sometimes absence of βAOB clone recovery within nested 16S rRNA genes clone libraries have been previously reported for planktonic samples and were potentially related to the abundance of this group in the water column (39). The results of our 16S rRNA clone libraries (Table 2) indicate the need to obtain PCR products directly from genomic DNA and also to generate larger clone libraries to study βAOB in environments where only a nested PCR approach can be used.
Except for some sequences retrieved from the OMZ (DQ009934, DQ009935, DQ009939, DQ009953, and DQ813659), the βAOB-like sequences obtained from the upper oxycline and OMZ were ≥99% similar (≤5 substitutions). The sequences DQ009934, DQ009935, DQ009939, and DQ009953 presented an unexpectedly high number of nucleotide substitutions at the beginning of the sequence compared with the rest of the βAOB-like sequences. These differences could be attributed to methodological biases during PCR and were excluded from further phylogenetic analyses. No chimeras were detected in the βAOB-like sequences analyzed. The sequence DQ813659 from the OMZ (PRO2-80 m) showed ∼95% similarity (∼50 substitutions) compared with other Nitrosospira-like sequences from our clone libraries.
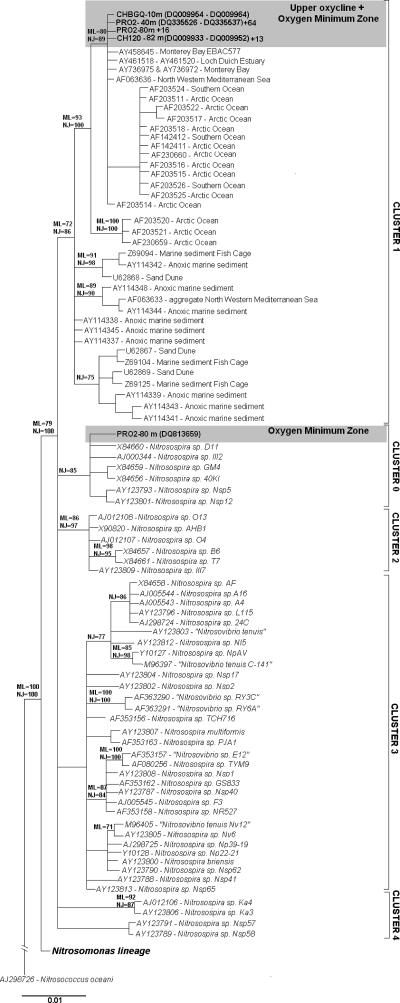
The phylogenetic relationships of the full βAOB-like 16S rRNA gene sequences from this study area are presented in a consensus tree in Fig. 2. Nitrosomonas and Nitrosospira lineages were clearly separated into the expected topology by application of the phylogenetic approaches used in the study by Purkhold et al. (40). Independent of the primer used (βΑΜΟf-r or NIT A-B), most of the βAOB-like sequences retrieved from the OMZ and all the sequences from the upper oxycline (hypoxic/oxic waters) fell within cluster 1 of Nitrosospira. This affiliation was congruent using neighbor-joining and maximum likelihood phylogenetic methods with bootstrap values of ≥72%. Cluster 1 exclusively contained sequences derived from uncultured representatives derived from 16S rRNA gene-based environmental and metagenomic surveys (e.g., clone AY458645 from a bacterial artificial chromosome library from Monterey Bay). All of the sequences of this cluster have been retrieved from different marine and marine-associated environments with variable oxygen conditions: i.e., sand dunes (21), anoxic sediments and aggregates (12, 13, 31, 39, 47), planktonic samples from polar oceans (3, 16), the northwestern Mediterranean sea (39), the Loch Duich estuary (13), and Monterey Bay (37).
FIG. 2.
Consensus phylogenetic tree based on 16S rRNA gene sequences related to ammonia-oxidizing bacteria retrieved from the study area and other cultured and yet-uncultured representatives of this group. The treeing methods were carried out with ≥1 kb of sequence, except for sequences that were generated with primer 341 f (∼700 bp), to confirm and compare βΑΜΟ f-r primer performance with NITA-B from CHUPS (82- and 10-m depths) and PRODEPLOY (80- and 300-m depths). The clones’ origins and accession numbers from this study are depicted in boldface. A plus sign indicates the number of identical sequences represented by each sequence in the corresponding clone library. The numbers in the branches indicate the bootstrap values (100 resamplings) associated with each treeing method used in the analyses (ML and NJ). The scale bar indicates the percentage of substitutions per site.
The sequence DQ813659 retrieved from the OMZ clone library (PRO2-80 m) was affiliated with cultured Nitrosospira sp. from cluster 0 (bootstrap values of ≥64%). Cultured Nitrosospira sequences from cluster 0 are mainly derived from sand, soil, and freshwater environments (20). To our knowledge, this is the first report of the presence of a sequence related to this cluster cloned from seawater. Our finding was also supported by sequences from amoA gene clone libraries from the OMZ (see below).
The dominance of Nitrosospira-like sequences in planktonic samples has been found previously (39). This result is not expected to be PCR bias against Nitrosomonas spp. because βAMO f-r and NIT A-B primer pairs have been designed to amplify 16S rRNA genes of all known βAOB (28, 50, 51). Indeed, environmental studies show that both primer sets could amplify βAOB representatives of Nitrosomonas spp. and Nitrosospira spp.: e.g., βAMO f-r (39) and NIT A-B (37). However, in an area further north of our sampling site (∼7 to 15°S), Ward et al. (52) detected Nitrosomonas-like βAOB by means of immunofluorescence assays using antiserum prepared with Nitrosomonas sp. (marine) strains. Unfortunately, the cross-reactivity of Nitrosospira spp. with Nitrosomonas-derived antibodies is not known (52). Therefore, it is possible that the Nitrosomonas-like counts obtained by Ward et al. could have also included Nitrosospira-like βAOB. On the other hand, Ward et al. (52) carried out immunofluorescence assays using whole-water samples, potentially including “particle-associated” βAOB, where Nitrosomonas-like bacteria have been found to be the predominant group (39). However, in the present study DNA was extracted from prefiltered seawater (see Materials and Methods).
amoA gene cloning.
Contrary to the results of 16S rRNA genes, the primers used for the amoA gene showed a high specificity. NCBI database screening of clone sequences originating from samples obtained at 40, 80, and 300 m using amoA34f-2r primers (35 sequences) and at 2, 10, 40, 80, and 300 m using amoA1f-2r (41 sequences) showed that all the amoA gene clones presented βAOB amoA-like sequences as the closest match. No differences related to the primer pair used for preparation of the clone libraries were found: i.e., amoA34f-2r versus amoA1f-2r from 40, 80, and 300 m.
The sequences retrieved from the OMZ (80 and 300 m) and from the surface and upper oxycline (2, 10, and 40 m) presented a low similarity: ∼73% and ∼87% for both nucleotide and amino acid analyses, respectively.
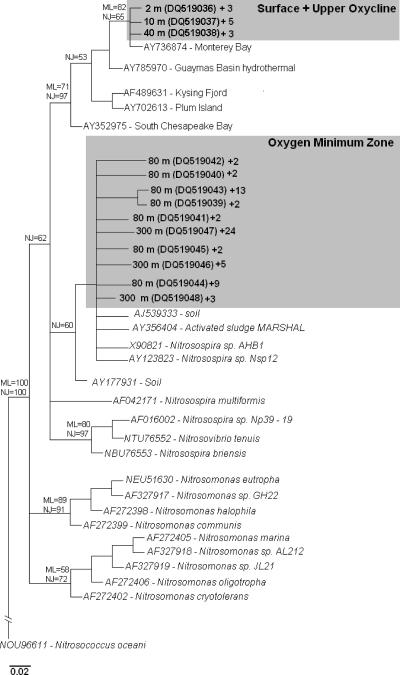
The relationships among the βAOB-like amoA amino-acid sequences from this study are presented in a consensus tree in Fig. 3. The monophyly of the Nitrosospira lineage could only be recovered with low bootstrap values (i.e., 62% and 33% by neighbor joining and maximum likelihood, respectively), whereas the Nitrosomonas lineage monophyly was only obtained with the maximum likelihood approach with a bootstrap value of 40%. These results were expected for the amoA gene marker phylogeny, since it generally shows a lower bootstrap support of Nitrosospira and Nitrosomonas lineages compared to 16S rRNA genes (40). The amoA gene sequences retrieved from the OMZ were affiliated within the Nitrosospira lineage, with bootstrap values of 60% and 29% (NJ and ML, respectively). The OMZ amoA gene sequence subcluster also contains sequences from other yet-uncultured βAOB from soil (1; M. Grimm, unpublished data [GenBank]) and from an aerated-anoxic Orbal process waste treatment plant (38), as well as sequences from cultured Nitrosospira strains isolated from soil, Nitrosospira sp. strain AHB1 (43), and Nitrosospira sp. strain Nsp12 (40). Within the cultured representatives, the OMZ amoA gene sequences presented a higher similarity (∼91% and ∼95%, nucleotide and amino acid, respectively) with Nitrosospira sp. strain Nsp12. According to the 16S rRNA gene phylogeny nomination of Purkhold et al. (51), Nsp12 affiliates with Nitrosospira cluster 0.
FIG. 3.
Consensus phylogenetic tree based on ≥149-amino-acid-residue sequences of the ammonia monooxygenase active subunit gene (amoA) related to ammonia-oxidizing bacterial sequences retrieved from the study area and other cultured and yet-uncultured representatives of this group. The clone origins and accession numbers from this study are depicted in boldface. A plus sign indicates the number of identical sequences represented by each sequence in the corresponding clone library. The numbers in the branches indicate the bootstrap values (100 resamplings) associated with each treeing method used in the analyses (ML and NJ). The scale bar indicates the percentage of sequence divergence.
The surface and upper-oxycline amoA gene sequences grouped in a different subcluster within the Nitrosospira lineage, supported by bootstrap values of ≥71% (NJ and ML). This subcluster consists exclusively of yet-uncultured sequences retrieved from different marine environments: i.e., Monterey Bay (37), South Chesapeake Bay (10), Plum Island (4), Kysing Fjord (34), Guaymas Basin (P. Lam, unpublished data [GenBank]).
Environmental covariates, such as salinity, pH, temperature, and fertilizers, have been found to correlate with richness and diversity of amoA genes in other ecosystems (2, 4). Therefore, the differences observed by the amoA gene marker within the surface and upper oxycline and OMZ off northern Chile could indicate that oxygen and/or other variables linked with this environmental factor in the water column (e.g., NH4+ and NO2−) might control vertical variability in the βAOB community structure.
16S rRNA and amoA gene phylogeny inference.
Similarities in the topology of the amoA and 16S rRNA gene phylogenies have been found in other marine environments (5, 37). In our study, the amoA gene sequences from the surface and upper oxycline were grouped in a subcluster together with sequences from marine uncultured organisms, analogous to sequences from 16S RNA genes into Nitrosospira cluster 1. A similar 16S rRNA and amoA gene phylogeny agreement was found by O′Mullan and Ward (37) in Monterey Bay. The combined results support a potential connection between the widespread Nitrosospira-like cluster 1 16S rRNA gene sequences and the functional gene amoA subcluster.
All of the amoA gene sequences that originated from the OMZ were closely affiliated with culture Nitrosospira from clusters 0 and 2. Also, a single 16S rRNA gene sequence affiliated with cluster 0 was also found in the OMZ, a coincident result with OMZ amoA gene sequences. Furthermore, no amoA gene sequences related to uncultured marine Nitrosospira were found in the OMZ, despite Nitrosospira-like cluster 1 being dominant in 16S rRNA gene libraries retrieved from this layer. One explanation for this discrepancy would be a methodological bias related to the PCR. It is possible that the nested approach used to obtain amoA gene products from the OMZ may have favored one target over another. However, in the case of the upper-oxycline DNA samples, the direct and nested amoA gene PCR products generated the same βAOB group, which differed from the ones found in the OMZ. Hence, a bias in the PCR does not seem to be a likely explanation.
Another possibility could be that the amoA marker in the clone libraries reflects the βAOB abundance in the water column and that the βAOB group within the OMZ presented a higher number of amoA copies. Cultured βAOB have between two and three amoA copies in their genome (20). Therefore, in the OMZ a higher number of amoA gene targets might be present, potentially related to the single 16S RNA gene type affiliated to Nitrosospira cluster 0 rather than Nitrosospira-like cluster 1. Moreover, the amoA found in the OMZ might be related to the 16S RNA genes of Nitrospira-like cluster 1. In this case, a potential divergence at the functional level in the OMZ might be an indication of an evolutionary pressure, resulting in the presence of distinct ecotypes of Nitrosospira-like βAOB. Further studies must be conducted in order to establish the connection between 16S RNA and functional genes of βAOB in this and other marine environments, for example, by using metagenomic approaches.
Both of the latter explanations about the discrepancy between 16S RNA and amoA genes are linked to the presence of a “functional community shift,” based on the clear separation between the βAOB communities from the upper oxycline and the OMZ layers when using the amoA gene, a marker of functional diversity according to Castro et al. (6a). The upper-oxycline βAOB community should be able to cope with the strong physicochemical gradients and high NH4+ availability characteristic of this layer. For example, a strong NH4+ cycling by bacterioplankton and flagellate communities has been found in the upper oxycline off northern Chile (29). In contrast, the OMZ βAOB community should be adapted to the more stable but oxygen-NH4+-deficient conditions. The presence of a Nitrosospira-like cluster 0 sequence in the OMZ suggests also an additional biogeochemical role for the βAOB. For example, members of Nitrosospira cluster 0 showed high rates of nitrifier denitrification (45). However, this process has not yet been measured in OMZs.
In summary, analyses of the functional amoA gene showed that the OMZ was clearly differentiable from the oxycline and oxic surface layer, but this was not the case when the 16S rRNA gene approach was used. On the other hand, the 16S rRNA and amoA genes showed the presence of βAOB sequences closely affiliated with Nitrosospira cluster 0 in the OMZ. These OMZ sequences of amoA were not novel, as was, for example, the case for the nirS sequences of denitrifiers (6a), but until now they have not been reported in marine environments. Finally, the 16S rRNA gene analyses confirm the dominant presence of the still-uncultured Nitrosospira-like cluster 1 in clone libraries of βAOB in different marine environments, which now include OMZs. The ubiquity and apparent metabolic plasticity of Nitrosospira-like cluster 1 point to the need to determine their ecology and biogeochemical role.
Acknowledgments
We acknowledge G. Alarcón, C. Burghardt, M. Castro-González, K. M. Fuentes, F. González, O.-S. Kim, J. F. Santibañez, and K. Wiedenhoeft for their help in the laboratory and in the field and the crews of the Abate Molina and Purihaalar. We thank H. Stevens and H. Bouman for comments that improved the manuscript and W. Rojas for providing the map of the study area.
This research was financed by the Comisión Nacional de Investigaciones Científicas y Tecnológicas (CONICYT) through FONDECYT grant no. 1030741 and FONDAP grant no. 150100007 and was carried out partly during DAAD (2004) and by an MPI (2005) short-stay visitor's grant in the MPI für Limnologie. V.M. was supported by Fundación Andes and MECESUP UCO002 Ph.D. grants and by a Marine Genomics postdoctoral fellowship (PBCT RUE 004).
Footnotes
Published ahead of print on 6 April 2007.
REFERENCES
- 1.Avrahami, S., and R. Conrad. 2005. Cold-temperate climate: a factor for selection of ammonia oxidizers in upland soil? Can. J. Microbiol. 51:709-714. [DOI] [PubMed] [Google Scholar]
- 2.Avrahami, S., W. Liesack, and R. Conrad. 2003. Effects of temperature and fertilizer on activity and community structure of soil ammonia oxidizers. Environ. Microbiol. 5:691-705. [DOI] [PubMed] [Google Scholar]
- 3.Bano, N., and J. T. Hollibaugh. 2000. Diversity and distribution of DNA sequences with affinity to ammonia-oxidizing bacteria of the β subdivision of the class Proteobacteria in the Arctic Ocean. Appl. Environ. Microbiol. 66:1960-1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernhard, A. E., T. Donn, A. E. Giblin, and D. A. Stahl. 2005. Loss of diversity of ammonia-oxidizing bacteria correlates with increasing salinity in an estuary system. Environ. Microbiol. 7:1289-1297. [DOI] [PubMed] [Google Scholar]
- 5.Caffrey, J. M., N. Harrington, I. Solem, and B. B. Ward. 2003. Biogeochemical processes in a small California estuary. 2. Nitrification activity, community structure and role in nitrogen budgets. Mar. Ecol. Prog. Ser. 248:27-40. [Google Scholar]
- 6.Casciotti, K. L., D. M. Sigman, and B. B. Ward. 2003. Linking diversity and stable isotope fractionation in ammonia-oxidizing bacteria. Geomicrobiol. J. 20:335-353. [Google Scholar]
- 6a.Castro-Gonzalez, M., G. Braker, L. Farias, and O. Ulloa. 2005. Communities of nirS-type denitrifiers in the water column of the oxygen minimum zone in the eastern South Pacific. Environ. Microbiol. 7:1298-1306. [DOI] [PubMed] [Google Scholar]
- 7.Castro-Gonzalez, M., and L. Farias. 2004. N2O cycling at the core of the oxygen minimum zone off northern Chile. Mar. Ecol. Prog. Ser. 280:1-11. [Google Scholar]
- 8.Codispoti, L., and J. Christensen. 1985. Nitrification, denitrification and nitrous oxide cycling in the eastern tropical South Pacific Ocean. Mar. Chem. 16:277-300. [Google Scholar]
- 9.Daffonchio, D., S. Borin, T. Brusa, L. Brusetti, P. van der Wielen, H. Bolhuis, M. M. Yakimov, G. D'Auria, L. Giuliano, D. Marty, C. Tamburini, T. J. McGenity, J. E. Hallsworth, A. M. Sass, K. N. Timmis, A. Tselepides, G. J. de Lange, A. Hubner, J. Thomson, S. P. Varnavas, F. Gasparoni, H. W. Gerber, E. Malinverno, C. Corselli, J. Garcin, B. McKew, P. N. Golyshin, N. Lampadariou, P. Polymenakou, D. Calore, S. Cenedese, F. Zanon, and S. Hoog. 2006. Stratified prokaryote network in the oxic-anoxic transition of a deep-sea halocline. Nature 440:203-207. [DOI] [PubMed] [Google Scholar]
- 9a.Farias, L., A. Paulmier, and M. Gallegos. 2007. Nitrous oxide and N-nutrient cycling in the oxygen minimum zone off northern Chile. Deep-Sea Res. Part I 54:164-180. [Google Scholar]
- 10.Francis, C., G. O′Mullan, and B. B. Ward. 2003. Diversity of ammonia monooxygenase (amoA) genes across environmental gradients in Chesapeake Bay sediments. Geobiology 1:129-140. [Google Scholar]
- 11.Francis, C. A., K. J. Roberts, J. M. Beman, A. E. Santoro, and B. B. Oakley. 2005. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. USA 102:14683-14688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freitag, T. E., and J. I. Prosser. 2003. Community structure of ammonia-oxidizing bacteria within anoxic marine sediments. Appl. Environ. Microbiol. 69:1359-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freitag, T. E., and J. I. Prosser. 2004. Differences between betaproteobacterial ammonia-oxidizing communities in marine sediments and those in overlying water. Appl. Environ. Microbiol. 70:3789-3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gruber, N., and J. L. Sarmiento. 1997. Global patterns of marine nitrogen fixation and denitrification. Global Biogeochem. Cycles 11:235-266. [Google Scholar]
- 15.Helly, J. J., and L. A. Levin. 2004. Global distribution of naturally occurring marine hypoxia on continental margins. Deep-Sea Res. Part I 51:1159-1168. [Google Scholar]
- 16.Hollibaugh, J. T., N. Bano, and H. W. Ducklow. 2002. Widespread distribution in polar oceans of a 16S rRNA gene sequence with affinity to Nitrosospira-like ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 68:1478-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holmes, R. M., A. Aminot, R. Kerouel, B. A. Hooker, and B. J. Peterson. 1999. A simple and precise method for measuring ammonium in marine and freshwater ecosystems. Can. J. Fish. Aquat. Sci. 56:1801-1808. [Google Scholar]
- 17a.Jayakumar, D. A., C. A. Francis, S. W. A. Naqvi, and B. B. Ward. 2004. Diversity of nitrite reductase genes (nirS) in the denitrifying water column of the coastal Arabian Sea. Aquat. Microb. Ecol. 34:69-78. [Google Scholar]
- 18.Kamykowski, D., and S.-J. Zentara. 1990. Hypoxia in the world ocean as recorded in the historical data set. Deep-Sea Res. Part A 37:1861-1874. [Google Scholar]
- 19.Koops, H.-P., U. Purkhold, A. Pommerening-Röser, G. Timmermann, and M. Wagner. 2003. The lithotrophic ammonia oxidizing bacteria. Springer-Verlag, New York, NY.
- 20.Kowalchuk, G. A., and J. R. Stephen. 2001. Ammonia-oxidizing bacteria: a model for molecular microbial ecology. Annu. Rev. Microbiol. 55:485-529. [DOI] [PubMed] [Google Scholar]
- 21.Kowalchuk, G. A., J. R. Stephen, W. De Boer, J. I. Prosser, T. M. Embley, and J. W. Woldendorp. 1997. Analysis of ammonia-oxidizing bacteria of the β subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR-amplified 16S ribosomal DNA fragments. Appl. Environ. Microbiol. 63:1489-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 23.Kuypers, M. M. M., G. Lavik, D. Woebken, M. Schmid, B. M. Fuchs, R. Amann, B. B. Jorgensen, and M. S. M. Jetten. 2005. Massive nitrogen loss from the Benguela upwelling system through anaerobic ammonium oxidation. Proc. Natl. Acad. Sci. USA 102:6478-6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lane, D. J. 1991. 16S/23S rRNA sequencing. John Wiley & Sons, New York, NY.
- 25.Liesack, W., H. Weyland, and E. Stackebrandt. 1991. Potential risks of gene amplification by PCR as determined by 16S rDNA analysis of a mixed-culture of strict barophilic bacteria. Microb. Ecol. 21:191-198. [DOI] [PubMed] [Google Scholar]
- 26.Lipschultz, F., S. C. Wofsy, B. B. Ward, L. A. Codispoti, G. Friedrich, and J. W. Elkins. 1990. Bacterial transformations of inorganic nitrogen in the oxygen-deficient waters of the eastern tropical South-Pacific Ocean. Deep-Sea Res. Part A 37:1513-1541. [Google Scholar]
- 27.Ludwig, W., O. Strunk, S. Klugbauer, N. Klugbauer, M. Weizenegger, J. Neumaier, M. Bachleitner, and K. H. Schleifer. 1998. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis 19:554-568. [DOI] [PubMed] [Google Scholar]
- 28.McCaig, A. E., T. M. Embley, and J. I. Prosser. 1994. Molecular analysis of enrichment cultures of marine ammonia oxidizers. FEMS Microbiol. Lett. 120:363-367. [DOI] [PubMed] [Google Scholar]
- 29.Molina, V., L. Farias, Y. Eissler, L. A. Cuevas, C. E. Morales, and R. Escribano. 2005. Ammonium cycling under a strong oxygen gradient associated with the oxygen minimum zone off northern Chile (∼23 °S). Mar. Ecol. Prog. Ser. 288:35-43. [Google Scholar]
- 30.Morrison, J. M., L. A. Codispoti, S. L. Smith, K. Wishner, C. Flagg, W. D. Gardner, S. Gaurin, S. W. A. Naqvi, V. Manghnani, L. Prosperie, and J. S. Gundersen. 1999. The oxygen minimum zone in the Arabian Sea during 1995. Deep-Sea Res. Part II 46:1903-1931. [Google Scholar]
- 31.Mortimer, R. J. G., S. J. Harris, M. D. Krom, T. E. Freitag, J. I. Prosser, J. Barnes, P. Anschutz, P. J. Hayes, and I. M. Davies. 2004. Anoxic nitrification in marine sediments. Mar. Ecol. Prog. Ser. 276:37-51. [Google Scholar]
- 32.Muyzer, G., E. C. De Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naqvi, S. W. A., T. Yoshinari, D. A. Jayakumar, M. A. Altabet, P. V. Narvekar, A. H. Devol, J. A. Brandes, and L. A. Codispoti. 1998. Budgetary and biogeochemical implications of N2O isotope signatures in the Arabian Sea. Nature 394:462-464. [Google Scholar]
- 34.Nicolaisen, M. H., and N. B. Ramsing. 2002. Denaturing gradient gel electrophoresis (DGGE) approaches to study the diversity of ammonia-oxidizing bacteria. J. Microbiol. Methods 50:189-203. [DOI] [PubMed] [Google Scholar]
- 35.Nold, S. C., J. Zhou, A. H. Devol, and J. M. Tiedje. 2000. Pacific Northwest marine sediments contain ammonia-oxidizing bacteria in the β subdivision of the Proteobacteria. Appl. Environ. Microbiol. 66:4532-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Norton, J. M., J. J. Alzerreca, Y. Suwa, and M. G. Klotz. 2002. Diversity of ammonia monooxygenase operon in autotrophic ammonia-oxidizing bacteria. Arch. Microbiol. 177:139-149. [DOI] [PubMed] [Google Scholar]
- 37.O'Mullan, G. D., and B. B. Ward. 2005. Relationship of temporal and spatial variabilities of ammonia-oxidizing bacteria to nitrification rates in Monterey Bay, California. Appl. Environ. Microbiol. 71:697-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park, H. D., J. M. Regan, and D. R. Noguera. 2002. Molecular analysis of ammonia-oxidizing bacterial populations in aerated-anoxic Orbal processes. Water Sci. Technol. 46:273-280. [PubMed] [Google Scholar]
- 39.Phillips, C. J., Z. Smith, T. M. Embley, and J. I. Prosser. 1999. Phylogenetic differences between particle-associated and planktonic ammonia-oxidizing bacteria of the β subdivision of the class Proteobacteria in the northwestern Mediterranean Sea. Appl. Environ. Microbiol. 65:779-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Purkhold, U., M. Wagner, G. Timmermann, A. Pommerening-Roser, and H. P. Koops. 2003. 16S rRNA and amoA-based phylogeny of 12 novel betaproteobacterial ammonia-oxidizing isolates: extension of the dataset and proposal of a new lineage within the nitrosomonads. Int. J. Syst. Evol. Microbiol. 53:1485-1494. [DOI] [PubMed] [Google Scholar]
- 41.Richards, F. 1965. Anoxic basins and fjords, vol. VI. Academic Press, New York, NY.
- 42.Rogers, A. D. 2000. The role of the oceanic oxygen minima in generating biodiversity in the deep sea. Deep-Sea Res. Part II 47:119-148. [Google Scholar]
- 43.Rotthauwe, J. H., W. De Boer, and W. Liesack. 1995. Comparative-analysis of gene-sequences encoding ammonia monooxygenase of Nitrosospira sp. Ahb1 and Nitrosolobus multiformis C-71. FEMS Microbiol. Lett. 133:131-135. [DOI] [PubMed] [Google Scholar]
- 44.Rotthauwe, J. H., K. P. Witzel, and W. Liesack. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63:4704-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shaw, L. J., G. W. Nicol, Z. Smith, J. Fear, J. I. Prosser, and E. M. Baggs. 2006. Nitrosospira spp. can produce nitrous oxide via a nitrifier denitrification pathway. Environ. Microbiol. 8:214-222. [DOI] [PubMed] [Google Scholar]
- 46.Reference deleted.
- 47.Stephen, J. R., A. E. McCaig, Z. Smith, J. I. Prosser, and T. M. Embley. 1996. Molecular diversity of soil and marine 16S rRNA gene sequences related to β-subgroup ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 62:4147-4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stevens, H., M. Stubner, M. Simon, and T. Brinkhoff. 2005. Phylogeny of Proteobacteria and Bacteroidetes from oxic habitats of a tidal flat ecosystem. FEMS Microbiol. Ecol. 54:351-365. [DOI] [PubMed] [Google Scholar]
- 49.Thamdrup, B., T. Dalsgaard, M. M. Jensen, O. Ulloa, L. Farías, and R. Escribano. 2006. Anaerobic ammonium oxidation in the oxygen-deficient waters off northern Chile. Limnol. Oceanogr. 51:2145-2156. [Google Scholar]
- 50.Utaker, J. B., and I. F. Nes. 1998. A qualitative evaluation of the published oligonucleotides specific for the 16S rRNA gene sequences of the ammonia-oxidizing bacteria. Syst. Appl. Microbiol. 21:72-88. [DOI] [PubMed] [Google Scholar]
- 51.Voytek, M. A., and B. B. Ward. 1995. Detection of ammonium-oxidizing bacteria of the beta-subclass of the class Proteobacteria in aquatic samples with the PCR. Appl. Environ. Microbiol. 61:1444-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ward, B. B., H. E. Glover, and F. Lipschultz. 1989. Chemoautotrophic activity and nitrification in the oxygen minimum zone off Peru. Deep-Sea Res. Part A 36:1031-1051. [Google Scholar]
- 53.Ward, B. B., D. P. Martino, M. C. Diaz, and S. B. Joye. 2000. Analysis of ammonia-oxidizing bacteria from hypersaline Mono Lake, California, on the basis of 16S rRNA sequences. Appl. Environ. Microbiol. 66:2873-2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ward, B. B., M. A. Voytek, and R. P. Witzel. 1997. Phylogenetic diversity of natural populations of ammonia oxidizers investigated by specific PCR amplification. Microb. Ecol. 33:87-96. [DOI] [PubMed] [Google Scholar]
- 55.West, N., and D. J. Scanlan. 1999. Niche-partitioning of Prochlorococcus populations in a stratified water column in the eastern North Atlantic Ocean. Appl. Environ. Microbiol. 65:2585-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wuchter, C., B. Abbas, M. J. L. Coolen, L. Herfort, J. van Bleijswijk, P. Timmers, M. Strous, E. Teira, G. J. Herndl, J. J. Middelburg, S. Schouten, and J. S. Sinninghe Damsté. 2006. Archaeal nitrification in the ocean. Proc. Natl. Acad. Sci. USA 103:12317-12322. [DOI] [PMC free article] [PubMed] [Google Scholar]