Abstract
The diversity population of microorganisms with the capability to use selenate as a terminal electron acceptor, reducing it to selenite and elemental selenium by the process known as dissimilatory selenate reduction, is largely unknown. The overall objective of this study was to gain an in-depth understanding of anaerobic biotransformation of selenium in the environment, particularly anaerobic respiration, and to characterize the microorganisms catalyzing this process. Here, we demonstrate the isolation and characterization of four novel anaerobic dissimilatory selenate-respiring bacteria enriched from a variety of sources, including sediments from three different water bodies in Chennai, India, and a tidal estuary in New Jersey. Strains S5 and S7 from India, strain KM from the Meadowlands, NJ, and strain pn1, categorized as a laboratory contaminant, were all phylogenetically distinct, belonging to various phyla in the bacterial domain. The 16S rRNA gene sequence shows that strain S5 constitutes a new genus belonging to Chrysiogenetes, while strain S7 belongs to the Deferribacteres, with greater than 98% 16S rRNA gene similarity to Geovibrio ferrireducens. Strain KM is related to Malonomonas rubra, Pelobacter acidigallici, and Desulfuromusa spp., with 96 to 97% 16S rRNA gene similarity. Strain pn1 is 99% similar to Pseudomonas stutzeri. Strains S5, S7, and KM are obligately anaerobic selenate-respiring microorganisms, while strain pn1 is facultatively anaerobic. Besides respiring selenate, all these strains also respire nitrate.
Anaerobic microbial respiration of terminal electron acceptors, such as nitrate, iron, sulfate, and carbonate, plays a major role in the oxidation of organic carbon in sediments and also contributes to the biogeochemical cycling of these elements in nature. Other oxyanions, including selenate, arsenate, chlorate, and chromate, may also be used as terminal electron acceptors during microbial respiration depending on their availabilities in different environments (4, 14, 26, 45). The solubilities of these elements in aquatic systems are dependent on their redox states, which also govern their toxicities. Therefore, these redox-sensitive elements can either be released into the water column or be sequestered into the sediments depending on whether they are oxidized or reduced.
The presence of toxic selenium oxyanions in agricultural wastewaters draining from seleniferous soils is widespread and a serious environmental problem in the western United States (10). The accumulation of toxic selenium oxyanions has led to major ecological damage in the San Joaquin Valley, CA (53), with reproductive defects, such as depressed egg hatchability and embryonic deformity of the eyes, beak, and limbs, occurring in waterfowl (37). Mining effluents and residue from coal-fired power plants are another major source of selenium contamination of aquatic systems (21, 34).
Selenium in the environment exists as selenate [Se(VI)] and selenite [Se(IV)], which are soluble; elemental selenium [Se(0)], a solid; and selenide [Se(−II)], which is gaseous. Various physical, chemical, and biological reactions facilitate the conversion of selenium from one form to another, mediating the cycling of selenium in nature. Although physical processes such as dissolution, volatilization, and adsorption contribute to abiotic transformations, the biogeochemical cycling of selenium in the environment is predominantly governed by microorganisms which play an important role in oxidation, reduction, methylation, and volatilization (7, 8, 9, 23, 39, 42).
Selenate can be used as a terminal electron acceptor for respiration, a process termed dissimilatory selenate reduction (38). Selenate reduction is thus a major sink for selenium oxyions present (as contaminants) in aquatic ecosystems, and bioremediation of selenate-contaminated drainage water has been demonstrated in pilot-scale studies using the selenate-respiring bacterium Thauera selenatis (3, 30). Although a large number of microorganisms are known to aerobically reduce selenate or selenite to elemental selenium (see, for example, references 25, 50, and 51), to date, only a few anaerobic dissimilatory selenate-respiring microorganisms have been isolated and studied in pure culture (45) and very little is known about these microorganisms.
The primary goal of this study was to examine the taxonomic diversity of selenate-respiring bacteria and thereby to gain a fundamental understanding of their role in the biogeochemical cycling of selenium. We enriched and isolated selenate-respiring microorganisms from geographically and characteristically different sediments. Four novel bacterial strains that can carry out dissimilatory selenate reduction, two of which can respire selenate to elemental selenium with only a transient selenite accumulation, were isolated. Our results indicate that selenate-respiring microorganisms are physiologically and phylogenetically diverse.
MATERIALS AND METHODS
Sediment enrichment.
Sediment samples, the details of which are given in Table 1, were collected from different water bodies in Chennai, India, and New Jersey. The samples were stored at 4°C until used. Enrichment cultures were established with a 10% (wt/vol) sediment inoculum as a slurry in anaerobic minimal salt medium (11) by using a strict anaerobic technique (19) with Na2SeO4 (10 mM) as the sole electron acceptor and 4-hydroxybenzoate (250 μM) or pyruvate (5 mM) or both as electron donors and carbon sources. Cultures were incubated in the dark, statically at 28°C. Once stable selenate-reducing primary enrichment cultures were established, sequential transfers (1:10 dilution) were made into fresh medium with the goal of enriching and isolating pure cultures.
TABLE 1.
Selenate-reducing activity in sediment enrichment cultures
| Sediment source | Selenate reduction present in primary enrichment culturea | Isolate obtainedb |
|---|---|---|
| India | ||
| Kovam | ||
| Egmore | + | |
| Chetput | + | |
| Aminjikarai | + | |
| Buckingham canal | ||
| Canal bank | + | |
| Chepauk | + | Strain S5 |
| Adyar river | ||
| Kotturpuram | + | |
| Jafferkhanpet | + | Strain S7 |
| Anagaputhur | + | |
| New Jersey | ||
| Sawmill Creek | ||
| Spartina sp. rhizosphere | + | |
| Phargmites sp. rhizosphere | + | |
| Mudflat | + | |
| Kearny marsh | + | Strain KM |
| Chesquake | ||
| Spartina sp. rhizosphere | + | |
| Phargmites sp. rhizosphere | + | |
| Arthur Kill, New York-New Jersey harbor | + (21) | Sedimenticola selenatireducens strain AK4OH1 (38) |
| Laboratory contaminant | NA | Strain pn1 |
Selenate as electron acceptor and 4-hydroxybenzoate or pyruvate or both as electron donors. +, selenate reduction detected; NA, not applicable.
All strains were isolated and maintained on pyruvate except S. selenatireducens, which was maintained on 4-hydroxybenzoate.
Isolation.
Active selenate-reducing enrichment cultures were transferred to fresh medium and used for isolation in soft-agar shake tubes (0.4% Noble agar; Difco) with 10 mM selenate as the electron acceptor and 5 mM pyruvate as the electron donor. Single colonies were picked and rediluted in soft-agar shake tubes multiple times to ensure the purity of the isolated colonies. Further verification of purity was ensured by microscopy, colony morphology, and 16S rRNA gene sequencing.
Growth experiments.
To demonstrate the growth of strains coupled to selenate reduction, a washed cell suspension was inoculated into medium with 10 mM Na2SeO4 as an electron acceptor and 10 mM acetate as an electron donor. Controls were established under the same conditions without an electron acceptor or without an electron donor. Cell-free controls were also established to detect any abiotic loss. The experiment was done in triplicate.
Utilization of alternate electron acceptors and electron donors.
To test the metabolic potentials of isolated strains, a range of electron donors and electron acceptors were tested for growth. Cultures were also tested for fermentation of pyruvate (10 mM) without an electron acceptor, as were appropriate abiotic controls. Cultures previously grown on pyruvate and selenate were used for this experiment. Both the loss of carbon substrate and transformation of selenate to selenite or selenium were used as indicators for utilization of carbon substrate. The utilization of an electron acceptor was considered positive when there was over 70% loss of the electron acceptor [except for anthraquinone disulfonate (AQDS), for which the criterion was a color change to orange, and Fe(III), for which the criterion was a color change to black] and no loss in abiotic controls.
Analytical techniques.
The selenium oxyanions, sulfate, nitrate, nitrite, chlorate, and arsenate were analyzed using ion chromatography (DX120; Dionex, Sunnyvale, CA) and benzoates with high-performance liquid chromatography as described previously (19). Organic acids acetate, citrate, lactate and pyruvate were measured using the same high-performance liquid chromatography and UV detection at 210 nm with a HPX-87H organic acid column (Bio-Rad, Hercules, CA), heated to 60°C. The mobile phase was 4 mM H2SO4 at a flow rate of 0.6 ml/min. The formations of anthrahydroquinone disulfonate (AHQDS) due to AQDS reduction (29) and sulfide due to elemental sulfur reduction (5) were estimated as described previously. The reduction of Fe(III) to Fe(II) resulted in the production of a black precipitate of reduced iron sulfide. Biomass was measured as the increase in protein concentration according to Bradford's assay using a microtiter assay technique with bovine serum albumin as the standard (Bio-Rad, Hercules, CA). Nessler's reagent was used to qualitatively determine if nitrate reduction led to ammonia production.
Phylogenetic analysis.
The 16S rRNA gene of each strain was amplified using the eubacterial primers 27F and 1522R (20) and the purified PCR product (QIAGEN PCR purification kit) directly used for sequencing with the primers 27F, 704F, 1242F, 685R, 907R, 1220R, and 1552R (18, 20). The sequence data were compiled in Contig Express (Vector NTI Suite; Informax). The 16S rRNA gene sequences of related microorganisms as determined by BLAST (1) analysis were downloaded from GenBank, and sequences were aligned using ClustalX (49). A phylogenetic analysis was performed using the parsimony and distance functions with Phylo_Win (13). A bootstrap analysis was performed for all completed trees.
Electron microscopy.
Electron microscopy was performed at the electron microscopy facility at the Nelson Biology Laboratory, Rutgers University, as described previously (35). Scanning electron microscopy was performed by directly mounting the sample on filter discs.
Nucleotide sequence accession numbers.
The nearly complete 16S rRNA gene sequences of strains KM, S5, S7, and pn1 have been deposited in GenBank under accession numbers DQ991964 to DQ991967.
RESULTS AND DISCUSSION
Primary selenate- and selenite-reducing enrichment cultures.
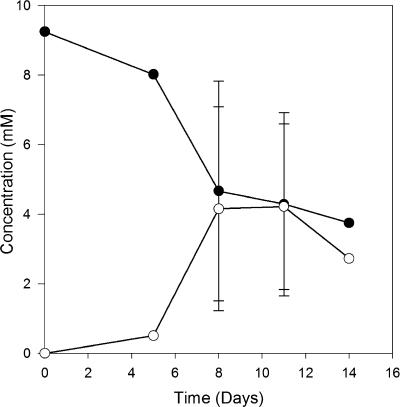
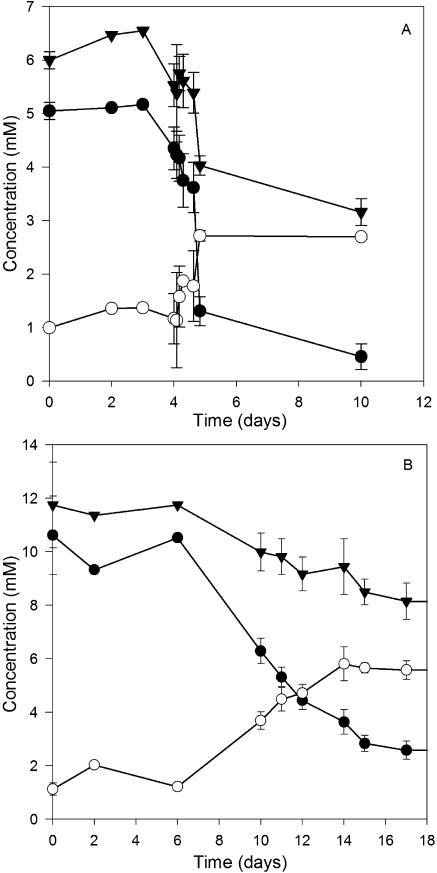
Selenate and selenite reductions were promoted within four weeks in each of the 17 enrichment cultures established from different aquatic sediments with selenate as the electron acceptor and pyruvate or 4-hydroxybenzoate or both as electron donors (Table 1). Only transient selenite accumulation was observed, indicating reductions of both selenate and selenite, along with the formation of a bright red reduced elemental-selenium precipitate. A typical selenate and selenite reduction trend exemplified by a Kearny Marsh culture is shown in Fig. 1. An active population of selenate-reducing bacteria developed within 7 days. Autoclaved sediment controls did not show selenate loss, indicating that the activity was microbially mediated. Stable selenate-reducing enrichment cultures were maintained for six of the original sediment sources through repeated subculturing, after which followed isolation of dissimilatory selenate-respiring microorganisms.
FIG. 1.
Selenate reduction in Kearny Marsh sediment enrichment culture. •, selenate; ○, selenite.
Isolation of selenate-respiring bacteria.
After sequential transfers into soft-agar shake tubes and dilution to extinction, pure cultures were isolated as listed in Table 1. Colonies were bright red in agar shake tubes due to elemental selenium, and we could note distinguishable colony morphologies from different enrichment cultures. Strain pn1 had a unique isolation history. While working with a previously characterized selenate-respiring bacterium, Sedimenticola selenatireducens strain AK4OH1 (35), we noticed that many of the culture tubes were contaminated with another bacterium which could also respire selenate. Based on the capability of strain AK4OH1 to utilize 4-hydroxybenzoate and the inability of strain pn1 to do so, we could purify the two strains. We performed an in-depth physiological, phylogenetic, and metabolic analysis on all of these strains to better understand the physiology of these selenate-reducing microorganisms and determine their taxonomic status.
Phylogenetic analysis.
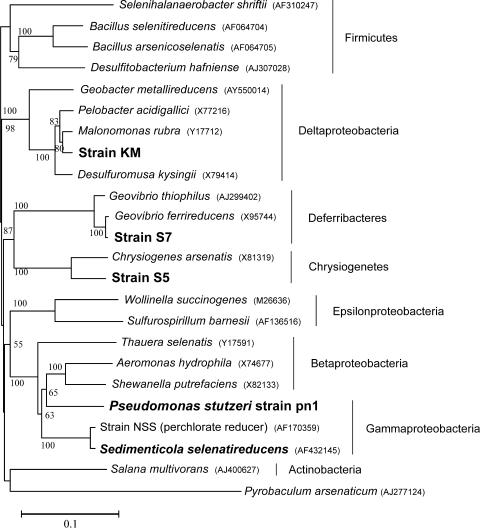
The strains isolated in the present study were identified by their 16S rRNA gene sequences. The phylogenetic tree of the 16S rRNA genes (Fig. 2) shows the relationship of the new dissimilatory selenate-reducing strains with other known selenate reducers, as well as their taxonomically related species. Strain pn1 was identified as a member of the species Pseudomonas stutzeri, with greater than 99% 16S rRNA gene sequence similarity. This identification was further confirmed by cellular fatty acid analysis (data not shown). Strain S5 formed a new deeply branching clade and grouped with Chrysiogenes arsenatis. The 16S rRNA gene sequences of strain S5 and C. arsenatis are 94% similar, and strain S5 thus represents a new genus and species within Chrysiogenetes (32). Strain S7 is about 98% similar to Geovibrio ferrireducens (3) and belongs to the phylum Deferribacteres (17). Further studies, such as studies with DNA-DNA hybridization and comparisons of fatty acid compositions of cell membranes, might help establish the taxonomy of strain S7. Nevertheless, strain S7 does merit attention because of its representation in the phylum Deferribacteres, previously unknown for its dissimilatory selenate-respiring capability. Strain KM belongs to the class Deltaproteobacteria and clusters within the family Geobacteraceae. Its 16S rRNA gene sequence is 97% similar to that of Malonomonas rubra and 96% similar to those of a number of Pelobacter spp. (43) and Desulfuromusa spp. (22). Strain KM thus appears to represent a new species with this group of metabolically similar bacteria within the Deltaproteobacteria. The phylogenetic tree (Fig. 2) shows that bacteria capable of dissimilatory selenate respiration are diverse and distributed over multiple phyla in the bacterial domain. Previously known isolates include two well-characterized selenate reducers, Thauera selenatis (29, 31) and Sulfurospirillum barnesii (44); a thermophillic archeon, Pyrobaculum arsenaticum (16); a halophile, Selenihalanaerobacter shriftii; an alkaliphile, Bacillus arsenicoselenatis (37, 46); a Bacillus species (12); Salana multivorans isolated from anaerobic bioreactor (52); and the recently described bacterium Sedimenticola selenatireducens (19, 35). Some dehalorespiring bacteria, such as a species of Desulfitobacterium (36), have also been shown to respire metals and metalloids, such as selenate. Interestingly, none of our new isolates belong to the already existing groups of bacteria known for their dissimilatory selenate-respiring capabilities.
FIG. 2.
Neighbor-joining phylogenetic tree, based on 16S rRNA gene sequences, showing selenate-respiring isolates strain KM, strain S5, strain S7, and P. stutzeri strain pn1 with their closely related strains and along with other selenate-respiring microorganisms. The tree was constructed with unambiguously aligned sequences of 1,118 bp in length. Bootstrap values expressed as percentages are given at each node.
Selenate respiration.
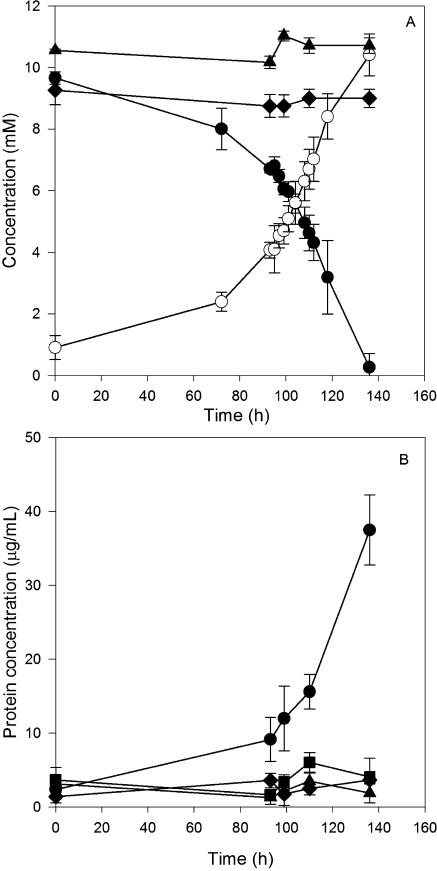
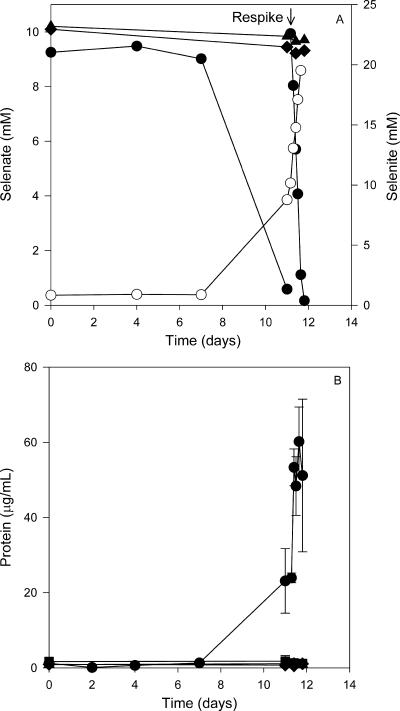
To clearly demonstrate the process of growth-dependent selenate respiration, cultures were incubated with or without an electron acceptor (selenate) and an electron donor (acetate or lactate). Selenate reduction followed a stoichiometric accumulation of selenite in strains S7 and pn1 as shown in Fig. 3 and 4. Strain S7 and pn1 grown on acetate reduced 10 mM selenate, with an accumulation of approximately 10 mM selenite. Cultures without an electron donor or abiotic controls showed no loss of selenate (Fig. 3A and 4A). A second spike of selenate to the pn1 culture resulted in the accumulation of 20 mM selenite. A significant increase in the protein concentration (measured as an indicator for increases in biomass) over time was observed only in cultures fed with both acetate as an electron donor and selenate as an electron acceptor (Fig. 3B and 4B). No protein increase was observed in cultures fed with only selenate or acetate or in abiotic controls. Strain pn1 also reduced selenate to elemental selenium when grown aerobically on tryptic soy broth (data not shown) independent of growth. The ability of Pseudomonas stutzeri to respire selenate contrasts with findings by Lortie et al. (24), who showed that their P. stutzeri strain only reduced selenate independent of growth. Our data indicate that the P. stutzeri pn1 strain can switch between nonrespiratory selenate reduction and anaerobic selenate respiration. This versatile physiology may enable it to adapt to various growth conditions.
FIG. 3.
Selenate respiration (A) and protein increase (B) in strain S7 with acetate as the carbon source. (A) Cultures were fed with both selenate and acetate. •, selenate; ○, selenite; ⧫, selenate in cultures without acetate. (B) Protein concentrations in cultures fed with both selenate and acetate (•), fed with acetate only (▪), fed with selenate only (⧫), and in an abiotic control (▴).
FIG. 4.
Selenate respiration (A) and protein increase (B) in Pseudomonas stutzeri strain pn1 with acetate as the carbon source. (A) Cultures were fed with both selenate and acetate. •, selenate; ○, selenite; ⧫, selenate in cultures without acetate. (B) Protein concentrations in cultures fed with both selenate and acetate (•), fed with acetate only (▪), fed with selenate only (⧫), and in an abiotic control (▴).
Strain KM exhibited a physiology that was very different from those of strains S7 and pn1. Strain KM grew with lactate as an electron donor both with and without selenate as an electron acceptor, indicating that strain KM was capable of fermentative metabolism. Lactate was fermented to propionate and acetate concomitant with robust growth, with cell density increasing severalfold, as observed from turbidity measurements (not shown). In cultures fed with both lactate and selenate, a very rapid reduction of selenate and selenite occurred (Fig. 5A), along with the formation of a bight red precipitate of elemental selenium. Hence, to clearly demonstrate whether strain KM was indeed respiring selenate, acetate was used as an electron donor, as it cannot energetically be fermented (48). As shown in Fig. 5B, although the reductions of both selenate and selenite were slower with acetate than with lactate, a considerable decrease in the total soluble selenium (i.e., the sum of the amounts of selenate and selenite), along with a red precipitate, was observed over time, indicating that both selenate and selenite were reduced to elemental selenium. In comparison to lactate, acetate served as a poor substrate for growth; cells adhered to the walls of the culture vial along with the precipitated selenium, forming a biofilm. This greatly hampered protein estimation (see below).
FIG. 5.
Selenate reduction by strain KM with lactate (A) or acetate (B) as the electron donor. •, selenate; ○, selenite; ▴, sum of selenate and selenite.
We further established an electron balance for growth with acetate and selenate for strains pn1, S7, and KM (Table 2). The utilization of 3.7 mM acetate by strain pn1 was coupled to the reduction of 8.7 mM selenate to selenite and the production of 23 mg/liter protein. Assuming that 50% of the cell dry weight is protein, the growth yield (increase in protein concentration) (28) was estimated to be 26% for strain pn1. Based on the stoichiometric equations shown in Table 2, we obtained an electron balance of 81% after accounting for substrate conversion to biomass. Similarly, the electron balance for strain S7 was 103% after accounting for substrate conversion to biomass. Hence, this clearly establishes that selenate respiration is coupled to acetate utilization in both strain pn1 and strain S7.
TABLE 2.
Stoichiometry of selenate respiration by strains KM, S7, and pn1a
| Strain | Acetate utilized (mM) | Protein increase (mg/liter)b | Carbon conversion to cells (%)c | Electrons produced (mM)d | Selenate reduced (mM) | Selenite produced (mM) | Selenium produced (mM) | Electrons consumed (mM)e | Electron balance (%)f |
|---|---|---|---|---|---|---|---|---|---|
| pn1 | 3.7 ± 1 | 23.1 ± 8 | 26 ± 4 | 21.9 ± 5.6 | 8.7 ± 1.4 | 7.9 ± 1.8 | 0 | 17.3 ± 2.9 | 81 ± 11 |
| S7 | 3.8 ± 0.2 | 35 ± 4.4 | 39 ± 5 | 18.3 ± 2.2 | 9.3 ± 0.5 | 9.6 ± 0.9 | 0 | 18.6 ± 1 | 102 ± 8 |
| KM | 5.4 ± 1.1 | ND | 32g | 29.1 ± 5.7 | 7.3 ± 0.5 | 4.6 ± 0.5 | 2.7 ± 0.2h | 25.2 ± 1 | 88 ± 15 |
All values are means ± standard deviations.
The increase in cell carbon was estimated to be equal to the increase in protein concentration (28). ND, not determined.
Percentage of acetate used to produce new cells (calculated from the measured protein concentration).
Corrected for carbon conversion to biomass, the remaining substrate is assumed to be oxidized to CO2.
Amounts of electrons used up for reduction of selenate to selenite or selenium based on measured concentrations of electron acceptors.
Calculated on the basis of the following stoichiometric equations as percentages of electrons consumed/electrons produced: C2H4O2 + 2H2O → 2CO2 + 8H+ + 8e−; SeO42− + 2H+ + 2e− → SeO32− + 4H2O; and SeO42− + 8H+ + 6e− → Se0 + 4H2O.
Protein could not be estimated due to the formation of a biofilm with precipitated selenium firmly adhering to the bottom of the culture vial. Hence, we assumed 32% carbon conversion to the cell based on an average of values obtained with strain S7 and strain pn1. We obtained about 60% electron balance when we did not account for biomass produced.
Calculated as difference of total selenate and soluble selenium as measured. Further confirmation for elemental selenium formation was demonstrated by XANES analysis.
The stoichiometry was, however, difficult to estimate for strain KM due to the inability to accurately measure protein after growth on acetate. (The culture grows as a biofilm on the bottom of the flask, and representative subsamples could not be obtained.) By calculating the electron donor and acceptor stochiometries with a portion of the soluble selenium oxyanions converted to selenium and without considering substrate conversion to biomass, we obtained a modest 60% electron balance. If we assume that carbon conversion to biomass is approximately 30%, as was the case with strains pn1 and S7, we can estimate an electron balance of 88%, as shown in Table 2. This establishes that strain KM does indeed respire selenate and selenite to elemental selenium with acetate as the electron donor. X-ray absorption near-edge spectroscopic (XANES) analysis showed the presence of selenium in its elemental form (unpublished data).
Although the ability to reduce selenate to selenite appears not to be uncommon, we know very little about organisms that respire selenite. Strain KM and strain S5 are the first organisms isolated so far that can carry out respiration of selenate completely to selenium. Bacillus selenitireducens can respire only selenite (46), although a coculture of Bacillus selenitireducens and B. arsenicoselenatis mediated elemental selenium precipitation from selenate (46). In contrast, selenite reduction in T. selenatis occurred only during active denitrification (6), suggesting the involvement of a nitrite reductase for selenite reduction. Washed cell suspensions of S. barnesii had the capability to reduce selenite, although this could not be coupled to growth (39). An intriguing factor was that strain KM could not reduce selenite as a sole electron acceptor, while strain S5 could reduce small quantities (1 to 2 mM) of selenite in the absence of selenate. It appears that selenate reduction was a prerequisite for selenite reduction to occur in strains KM and S5. It is possible that a high dose (5 mM) of spiked selenite was toxic, as opposed to the gradual transient accumulation of selenite during selenate reduction. B. selenitireducens still remains the only bacterium capable of respiring large quantities (10 mM) of selenite to elemental selenium (46). It is also possible that strain KM reduces selenate or selenite only to remove excess reducing power, similar to nitrate reduction in Clostridium perfringens (15). However, the near-stoichiometric electron balance suggests that strain KM indeed couples growth with the reductions of selenate and selenite (Table 2). It is unclear if the same enzyme that is catalyzing selenate reduction is also catalyzing selenite reduction, but definitely the cells do not need to undergo active nitrate reduction, as in T. selenatis or S. barnesii (6, 40) for selenite reduction to proceed.
Morphology and formation of elemental selenium granules.
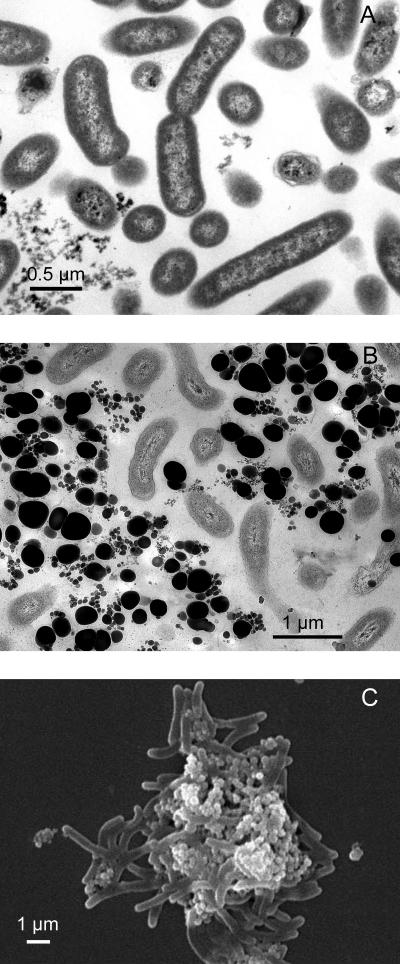
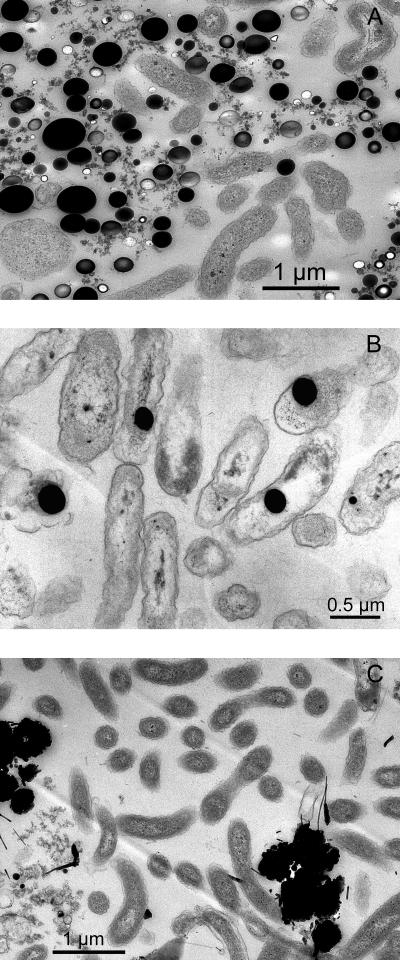
Cells of selenate-respiring cultures produced abundant elemental-selenium granules closely associated with the cells as shown by transmission and scanning electron micrographs. Strain KM is a rod-shaped bacterium (Fig. 6A and B). Strains S7 and S5 are vibroids (Fig. 7A and C). The electron micrographs of strain KM (Fig. 6B and C) show elemental selenium granules of 0.1 to 0.3 μm in diameter closely associated with the cells. The granules are uniformly circular and are neatly packed around the cells. This is further evidence for the ability of strain KM to reduce selenate to elemental selenium. As shown from thin sections (Fig. 6B), these selenium granules were generally not observed within the cells and were completely absent in cultures grown without selenate (Fig. 6A). These extracellular granules appear to be similar to the spheres produced by the selenate- and selenite-respiring bacteria S. barnesii, B. selenitireducens, and S. shriftii, whose structural and spectral features were demonstrated recently (41). In the case of strains pn1 and S7, although selenate was not reduced completely to selenium, there are still a few selenium particles associated with the cells (Fig. 7A and B). In strain pn1, the selenium spheres appear to be internal. These granules could also be seen with light microscopy (not shown).
FIG. 6.
Electron micrographs of selenate-respiring isolate strain KM. (A) Transmission electron microscopy when the strain was grown without selenate. (B) Transmission electron microscopy when the strain was grown in the presence of selenate. (C) Scanning electron microscopy of cells grown in the presence of selenate.
FIG. 7.
Transmission electron micrographs of selenate-respiring isolates strain S7 (A), strain pn1 (B), and strain S5 (C) are shown.
Metabolic diversity.
The ability of the selenate-respiring strains to utilize a range of electron donors and acceptors was tested to gain an understanding of their metabolic diversity (Table 3). Strain KM has the capability to reduce poorly crystalline Fe(III), nitrate, chlorate, sulfur, and AQDS, in addition to selenate, with pyruvate as an electron donor. Interestingly, arsenate, chromate, and nitrite completely inhibited even fermentative growth on pyruvate, whereas in the presence of sulfate, growth of strain KM occurred fermentatively without the loss of any sulfate. Although selenate was reduced via selenite to elemental selenium, strain KM could not reduce selenite (5 mM) as the sole electron acceptor. Strain S7 could respire only selenate and nitrate. Strain S5 was able to respire selenate, arsenate, Fe(III) citrate, and nitrate. Notably, strains S5 differed from the other bacteria in its ability to use 1 to 2 mM selenite as an electron acceptor in the absence of selenate, further evidence of which was obtained by XANES analysis (not shown). Strain pn1 could respire only selenate, nitrate, nitrite, and oxygen.
TABLE 3.
Comparison of metabolic properties of selenate-respiring microorganisms isolated in this study compared with those of Sedimenticola selenatireducens strain AK4OH1a
| Properties | Strain pn1 | Strain S7 | Strain KM | Strain S5 | Sedimenticola selenatireducens strain AK4OH1b |
|---|---|---|---|---|---|
| Ability to use indicated electron donorc | |||||
| 4-Hydroxybenzoate | − | − | − | − | + |
| 3-Hydroxybenzoate | − | − | − | ND | + |
| Benzoate | − | − | − | ND | + |
| Acetate | + | + | + | − | + |
| Pyruvate | + | + | + | + | + |
| Lactate | + | + | + | − | + |
| Citrate | + | − | + | ND | − |
| Fermentation ability | − | − | + | − | − |
| Ability to respire indicated electron acceptord | |||||
| Oxygen | + | − | − | − | − |
| Fumarate | ND | − | + | ND | ND |
| Fe(III) citrate | − | − | + | + | − |
| Poorly crystalline Fe(III) | ND | − | + | ND | ND |
| Nitrate | + | + | + | + | + |
| Nitrite | + | − | −e | −f | + |
| Selenate | + | + | + | + | + |
| Selenite | − | − | +g | + | − |
| Arsenate | − | − | − | + | − |
| Chromate | ND | − | − | ND | ND |
| Chlorate | − | − | + | ND | − |
| Sulfate | − | − | − | ND | − |
| Sulfur | ND | ND | + | ND | ND |
| AQDS | − | − | + | ND | − |
+, the strain has the indicated property; −, the strain does not have the indicated property; ND, not determined.
Data obtained from our previous study (35).
Electron donors were tested with selenate (10 mM) as an electron acceptor except for in the case of strain S5, for which nitrate (10 mM) was used.
With pyruvate or lactate as the electron donor.
Nitrite does not accumulate when grown on nitrate.
Nitrite does not accumulate when grown on nitrate.
KM reduces selenite only when preceded by selenate reduction.
Conclusions.
In summary, an important fact that has emerged from this and previous studies is that selenate-respiring bacteria are phylogenetically and physiologically diverse and appear to be ubiquitous in aquatic sediments. However, even close to two decades after the first description of a novel sulfate-independent dissimilatory selenate respiration pathway by Oremland et al. (38), we know very little about this process and the microorganisms harboring this metabolic capability. It is interesting that we have yet to reisolate strains of the same species or even the same genus of already existing dissimilatory selenate-respiring prokaryotes, as all the studies to date have found novel organisms (Fig. 2). Furthermore, under similar conditions with respect to media, electron donors, and electron acceptors, we have shown that enrichment cultures from different sediments result in entirely different populations of selenate-respiring bacteria. These are both metabolically and taxonomically diverse in the taxa Gammaproteobacteria, Deltaproteobacteria, Deferribacteres, and Chrysiogenetes. Other than the unique metabolic capability to respire selenate for which they were enriched, the isolates from our study are completely unrelated to each other, and we are yet to unearth the true diversity of this group of microorganisms. What contributes or drives the existence of these organisms in nature is a challenging and intriguing question. Further biochemical characterization of the selenate reductase enzymes from these isolates would shed some light on the evolution of this respiratory process.
Acknowledgments
We are highly indebted to Purvaja Ramachandran (Anna University) for sharing the sediment samples from India. We thank Nathan Yee for help with XANES analysis at Brookhaven National Laboratory and Valentin Starovoytov for electron micrography. We acknowledge Brookhaven National Laboratory for resources and beam line training by personnel.
The work was funded in part by the New Jersey Water Resources Research Institute.
Footnotes
Published ahead of print on 13 April 2007.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caccavo, F., Jr., J. D. Coates, R. A. Rossello-Mora, W. Ludwig, K. D. Schleifer, D. R. Lovley, and M. J. McInerney. 1996. Geovibrio ferrireducens, a phylogenetically distinct dissimilatory Fe(III)-reducing bacterium. Arch. Microbiol. 165:370-376. [DOI] [PubMed] [Google Scholar]
- 3.Cantafio, A. W., K. D. Hagen, G. E. Lewis, T. L. Bledsoe, K. M. Nunan, and J. M. Macy. 1996. Pilot-scale selenium bioremediation of San Joaquin drainage water with Thauera selenatis. Appl. Environ. Microbiol. 62:3298-3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coates, J. D., U. Michaelidou, R. A. Bruce, S. M. O'Connor, J. N. Crespi, and L. A. Achenbach. 1999. Ubiquity and diversity of dissimilatory (per)chlorate-reducing bacteria. Appl. Environ. Microbiol. 65:5234-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cord-Ruwisch, R. 1985. A quick method for the determination of dissolved and precipitated sulfides in cultures of sulfate reducing bacteria. J. Microbiol. Methods 4:33-36. [Google Scholar]
- 6.DeMoll-Decker, H., and J. M. Macy. 1993. The periplasmic nitrite reductase of Thauera selenatis may catalyse the reduction of selenite to elemental selenium. Arch. Microbiol. 160:241-247. [Google Scholar]
- 7.Doran, J. W. 1982. Microorganisms and the biological cycling of selenium. Adv. Microb. Ecol. 6:1-32. [Google Scholar]
- 8.Dowdle, P. R., and R. S. Oremland. 1998. Microbial oxidation of elemental selenium in soil slurries and bacterial cultures. Environ. Sci. Technol. 32:3749-3755. [Google Scholar]
- 9.Dungan, R. S., S. R. Yates, and W. T. Frankenberger. 1999. Transformation of selenate and selenite by Stenotrophomonas maltophila isolated from seleniferous agricultural drainage pond sediment. Environ. Microbiol. 5:287-295. [DOI] [PubMed] [Google Scholar]
- 10.Engberg, R. A., D. W. Westcot, M. Delamore, and D. D. Holz. 1998. Federal and state perspectives on regulation and remediation of irrigation-induced selenium problems, p. 1-27. In W. T. Frankenberger, Jr. and S. Benson (ed.), Selenium in the environment. Marcel Dekker, Inc., New York, NY.
- 11.Fennell, D. E., S. K. Rhee, Y. B. Ahn, M. M. Häggblom, and L. J. Kerkhof. 2004. Detection and characterization of a dehalogenating microorganism by terminal restriction fragment length polymorphism fingerprinting of 16S rRNA in a sulfidogenic, 2-bromophenol-utilizing enrichment. Appl. Environ. Microbiol. 70:1169-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujita, M., M. Ike, S. Nishimoto, K. Takahashi, and M. Kashiwa. 1997. Isolation and characterization of a novel selenate-reducing bacterium, Bacillus sp. SF-1. J. Ferment. Bioeng. 83:517-522. [Google Scholar]
- 13.Galtier, N., M. Gouy, and C. Gautier. 1996. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biol. Sci. 12:543-548. [DOI] [PubMed] [Google Scholar]
- 14.Ghiorse, W., C., and J. T. Wilson. 1988. Microbial ecology of the terrestrial subsurface. Adv. Appl. Microbiol. 33:107-172. [DOI] [PubMed] [Google Scholar]
- 15.Hasan, S. M., and J. B. Hall. 1975. The physiological function of nitrate reduction in Clostridium perfringens. J. Gen. Microbiol. 87:120-127. [DOI] [PubMed] [Google Scholar]
- 16.Huber, R., M. Sacher, A. Vollmann, H. Huber, and D. Rose. 2000. Respiration of arsenate and selenate by hyperthermophilic archaea. Syst. Appl. Microbiol. 23:305-314. [DOI] [PubMed] [Google Scholar]
- 17.Janssen, P. H., W. Liesack, and B. Schink. 2002. Geovibrio thiophilus sp. nov., a novel sulfur-reducing bacterium belonging to the phylum Deferribacteres. Int. J. Syst. Evol. Microbiol. 52:1341-1347. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, J. L. 1994. Similarity analyses of rRNAs, p. 683-700. In P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.), Methods for general and molecular bacteriology. ASM Press, Washington, DC.
- 19.Knight, V. K., I. Nijenhuis, L. J. Kerkhof, and M. M. Häggblom. 2002. Degradation of aromatic compounds coupled to selenate reduction. Geomicrobiol. J. 19:77-86. [Google Scholar]
- 20.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acids techniques in bacterial sytematics. Wiley, New York, NY.
- 21.Lemly, A. D. 1997. Environmental implications of excessive selenium: a review. Biomed. Environ. Sci. 10:415-435. [PubMed] [Google Scholar]
- 22.Liesack, W., and K. Finster. 1994. Phylogenetic analysis of five strains of gram-negative obligately anerobic, sulfur-reducing bacteria and description of Desulfuromusa gen. nov., including Desulfuromusa kysingii sp. nov., Desulfuromusa bakii sp. nov., and Desulfuromusa succinoxidans sp. nov. Int. J. Syst. Bacteriol. 44:753-758. [Google Scholar]
- 23.Lipman, J. G., and S. A. Waksman. 1923. The oxidation of elemental selenium by a new group of autotrophic microorganisms. Science 57:1463. [DOI] [PubMed] [Google Scholar]
- 24.Lortie, L., W. D. Gould, S. Rajan, R. G. L. McCready, and K. J. Cheng. 1992. Reduction of selenate and selenite to elemental selenium by a Pseudomonas stutzeri isolate. Appl. Environ. Microbiol. 58:4042-4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Losi, M. E., and W. T. Frankenberger, Jr. 1997. Reduction of selenium oxyanions by Enterobacter cloacae SLD 1a-I: isolation and growth of the bacterium and its expulsion of selenium particles. Appl. Environ. Microbiol. 63:3079-3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lovley, D. R. 1993. Dissimilatory metal reduction. Annu. Rev. Microbiol. 47:263-290. [DOI] [PubMed] [Google Scholar]
- 27.Lovley, D. R., J. D. Coates, E. L. Blunt-Harris, E. J. P. Phillips, and J. C. Woodward. 1996. Humic substances as electron acceptors for microbial respiration. Nature 382:445-448. [Google Scholar]
- 28.Luria, S. E. 1960. The bacterial protoplasm: composition and organization, p. 1-34. In I. C. Gunsalus and R. Y. Stainer (ed.), The bacteria, vol. 1. Academic Press, New York, NY. [Google Scholar]
- 29.Macy, J. M., T. A. Michel, and D. G. Kirsch. 1989. Selenate reduction by a Pseudomonas species: a new mode of anaerobic respiration. FEMS Microbiol. Lett. 52:195-198. [DOI] [PubMed] [Google Scholar]
- 30.Macy, J. M., S. Lawson, and H. Demoll-Decker. 1993. Bioremediation of selenium oxyions in San Joaquin drainage water using Thauera selenatis in a biological reactor system. Appl. Microbiol. Biotechnol. 40:588-594. [Google Scholar]
- 31.Macy, J. M., S. Rech, G. Auling, M. Dorsch, E. Stackebrandt, and L. I. Sly. 1993. Thauera selenatis gen. nov., sp., nov., a member of the beta subclass of the Proteobacteria with a novel type of anaerobic respiration. Int. J. Syst. Bacteriol. 43:135-142. [DOI] [PubMed] [Google Scholar]
- 32.Macy, J. M., K. Nunan, K. D. Hagen, D. R. Dixon, P. J. Harbour, M. Cahill, and L. I. Sly. 1996. Chrysiogenes arsenatis gen. nov., sp. nov., a new arsenate-respiring bacterium isolated from gold mine wastewater. Int. J. Syst. Bacteriol. 46:1153-1157. [DOI] [PubMed] [Google Scholar]
- 33.Masscheleyn, P. H., and W. H. Patrick, Jr. 1993. Biogeochemical processes affecting selenium cycling in wetlands. Env. Toxicol. Chem. 12:2235-2243. [Google Scholar]
- 34.Muscatello, J. R., P. M. Bennett, K. T. Himbeault, A. M. Belknap, and D. M. Janz. 2006. Larval deformities associated with selenium accumulation in northern pike (Esox lucius) exposed to metal mining effluent. Environ. Sci. Technol. 40:6506-6512. [DOI] [PubMed] [Google Scholar]
- 35.Narasingarao, P., and M. M. Häggblom. 2006. Sedimenticola selenatireducens, gen. nov., sp. nov., a novel anaerobic selenate-respiring bacterium isolated from estuarine sediment. Syst. Appl. Microbiol. 29:382-388. [DOI] [PubMed] [Google Scholar]
- 36.Niggemyer, A., S. Spring, E. Stackebrandt, and R. F. Rosenzweig. 2001. Isolation and characterization of a novel As(V)-reducing bacterium: implications for arsenic mobilization and the genus Desulfitobacterium. Appl. Environ. Microbiol. 67:5568-5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohlendorf, H. M., D. J. Hoffman, M. K. Saiki, and T. W. Aldrich. 1986. Embryonic mortality and abnormalities of aquatic birds: apparent impacts of selenium from irrigation drain water. Sci. Total Environ. 52:49-63. [Google Scholar]
- 38.Oremland, R. S., J. T. Hollibaugh, A. S. Maest, T. S. Presser, L. G. Miller, and C. W. Culbertson. 1989. Selenate reduction to elemental selenium by anaerobic bacteria in sediments and culture: biogeochemical significance of a novel sulfate-independent respiration. Appl. Environ. Microbiol. 55:2333-2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oremland, R. S., J. Switzer Blum, C. W. Culbertson, P. T. Visscher, L. G. Miller, P. Dowdle, and F. E. Strohmaier. 1994. Isolation, growth, and metabolism of an obligately anaerobic, selenate-respiring bacterium, strain SES-3. Appl. Environ. Microbiol. 60:3011-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oremland, R. S., J. Switzer-Blum, A. B. Bindi, P. R. Dowdle, M. Herbel, and J. F. Stolz. 1999. Simultaneous reduction of nitrate and selenate by cell suspensions of selenium-respiring bacteria. Appl. Environ. Microbiol. 65:4385-4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oremland, R. S., M. J. Herbel, J. Switzer-Blum, S. Langley, T. J. Beveridge, P. M. Ajayan, T. Sutto, A. V. Ellis, and S. Curran. 2004. Structural and spectral features of selenium nanospheres produced by Se-respiring bacteria. Appl. Environ. Microbiol. 70:52-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sarathchandra, S. U., and J. H. Watkinson. 1981. Oxidation of elemental selenium to selenite by Bacillus megaterium. Science 21:600-601. [DOI] [PubMed] [Google Scholar]
- 43.Schink, B., and N. Pfennig. 1982. Fermentation of trihydroxybenzenes by Pelobacter acidigallici gen. nov. sp. nov., a new strictly anaerobic, non-spore forming bacterium. Arch. Microbiol. 133:195-201. [Google Scholar]
- 44.Stolz, J. F., D. J. Ellis, J. S. Blum, D. Ahmann, D. R. Lovley, and R. S. Oremland. 1999. Sulfurospirillum barnesii sp. nov. and Sulfurospirillum arsenophilum sp. nov., new members of the Sulfurospirillum clade of the epsilon Proteobacteria. Int. J. Syst. Bacteriol. 49:1177-1180. [DOI] [PubMed] [Google Scholar]
- 45.Stolz, J. F., and R. S. Oremland. 1999. Bacterial respiration of arsenic and selenium. FEMS Microbiol. Rev. 23:615-627. [DOI] [PubMed] [Google Scholar]
- 46.Switzer-Blum, J., A. B. Bindi, J. Buzzelli, J. F Stolz, and R. Oremland. 1998. Bacillus arsenicoselenatis, sp. nov., and Bacillus selenitireducens, sp. nov.: two haloalkaliphiles from Mono Lake, California that respire oxyanions of selenium and arsenic. Arch. Microbiol. 171:19-30. [DOI] [PubMed] [Google Scholar]
- 47.Switzer-Blum, J., J. F. Stolz, A. Oren, and R. S. Oremland. 2001. Selenihalanaerobacter shriftii gen. nov., sp. nov., a halophilic anaerobe from Dead Sea sediments that respires selenate. Arch. Microbiol. 175:208-219. [DOI] [PubMed] [Google Scholar]
- 48.Thauer, R. K., K. Jungermann, and K. Decker. 1977. Energy conservation in chemotrophic bacteria. Bacteriol. Rev. 41:100-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The clustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tomei, F. A., L. L. Barton, C. L. Lemanski, and T. G. Zocco. 1992. Reduction of selenate and selenite to elemental selenium by Wolinella succinogenes. Can. J. Microbiol. 38:1328-1333. [Google Scholar]
- 51.Tomei, F. A., L. L. Barton, C. L. Lemanski, T. G. Zocco, N. H. Fink, and L. O. Sillerud. 1995. Transformation of selenate and selenite to elemental selenium by Desulfovibrio desulfuricans. J. Ind. Microbiol. 14:329-336. [Google Scholar]
- 52.Wintzingerode, F., U. B. Göbel, R. A. Siddiqui, U. Rosick, P. Schumann, A. Fruhling, M. Rohde, R. Pukall, and E. Stackebrandt. 2001. Salana multivorans gen. nov., sp. nov., a novel actinobacterium isolated from an anaerobic bioreactor and capable of selenate reduction. Int. J. Syst. Evol. Microbiol. 51:1653-1661. [DOI] [PubMed] [Google Scholar]
- 53.Wu, L. 2004. Review of 15 years of research on ecotoxicology and remediation of land contaminated by agricultural drainage sediment rich in selenium. Ecotoxicol. Environ. Safety 57:257-269. [DOI] [PubMed] [Google Scholar]