Abstract
Heterotrophic bacteria are major contributors to biogeochemical cycles and influence water quality. Still, the lack of representative isolates and the few quantitative surveys leave the ecological role and significance of single bacterial populations to be revealed. Here we analyzed the diversity and dynamics of freshwater Flavobacteria populations in four eutrophic temperate lakes. From each lake, clone libraries were constructed using primers specific for either the class Flavobacteria or Bacteria. Sequencing of 194 Flavobacteria clones from 8 libraries revealed a diverse freshwater Flavobacteria community and distinct differences among lakes. Abundance and seasonal dynamics of Flavobacteria were assessed by quantitative PCR with class-specific primers. In parallel, the dynamics of individual populations within the Flavobacteria community were assessed with terminal restriction fragment length polymorphism analysis using identical primers. The contribution of Flavobacteria to the total bacterioplankton community ranged from 0.4 to almost 100% (average, 24%). Blooms where Flavobacteria represented more than 30% of the bacterioplankton were observed at different times in the four lakes. In general, high proportions of Flavobacteria appeared during episodes of high bacterial production. Phylogenetic analyses combined with Flavobacteria community fingerprints suggested dominance of two Flavobacteria lineages. Both drastic alterations in total Flavobacteria and in community composition of this class significantly correlated with bacterial production, emphasizing that resource availability is an important driver of heterotrophic bacterial succession in eutrophic lakes.
Over the last decades, information has accumulated regarding seasonal succession of phytoplankton in lakes. In general, diatoms dominate spring blooms in most eutrophic lakes, and massive developments of cyanobacteria (i.e., cyanobacterial blooms) are characteristic for late-summer conditions (17). On the contrary, our understanding of bacterioplankton dynamics in freshwaters is scarce, even if these mostly organoheterotrophic microorganisms are major contributors to processes that control water quality and the fate of pollutants. Recent studies have identified many uncultured and apparently widely distributed bacterial groups in freshwaters (see, for example, references 8, 13, and 46), but still little is known about the seasonality of bacterial taxa in such systems.
Three phyla, Proteobacteria, Actinobacteria, and Bacteriodetes, usually occur at high frequencies in clone libraries from lakes (for example, see references 8, 13, 22, and 46). Quantitative studies based on fluorescence in situ hybridization have revealed that members of the Bacteriodetes and Proteobacteria exhibit high temporal and spatial variation (10, 12, 19, 33, 34). It has been suggested that the ability of Bacteriodetes and Proteobacteria to rapidly exploit bioavailable organic matter and colonize aggregates could be responsible for their dynamic distribution patterns (1, 28, 44). Understanding of the role of bacterial populations relies on studies that simultaneously determine the distribution of single bacterial populations and properties of the environment. Only a few studies have provided first explanations for population patterns on spatial and temporal scales (see, for example, references 5, 18, and 23-25). For example, two studies based on PCR have shown that freshwater phytoplankton blooms foster bacterial communities where members of the class Flavobacteria are well represented (8, 20). Even if Flavobacteria are frequently observed in freshwater clone libraries (8, 13, 46), there are only a few freshwater systems where the distribution of groups within the Bacteroidetes has been assessed quantitatively (12, 24, 33, 34).
The present study reveals seasonal distribution patterns for the class Flavobacteria in four eutrophic lakes situated in the temperate zone of central Sweden. Culture-independent techniques (quantitative PCR [Q-PCR], terminal restriction fragment length polymorphism [T-RFLP], cloning, and sequence analysis) were applied in parallel to study the diversity and dynamics of Flavobacteria at the class and subclass levels. Dynamics of Flavobacteria communities were also compared to environmental-state variables to identify factors regulating this abundant but largely uncultured group of freshwater bacteria and elucidate their ecological roles in freshwater ecosystems.
MATERIALS AND METHODS
Water samples were collected from four lakes located in the same geographical region of central Sweden. Lake characteristics have been described in detail elsewhere (8). The four lakes differ in the types of Cyanobacteria that usually dominate their late-summer cyanobacterial blooms. Microcystis aeruginosa dominated in Lake Ekoln and Gloeotrichia echinulata dominated in Lake Erken, whereas multiple cyanobacterial species, such as the Anabaena and Aphanizomenon spp., dominated in the shallow lakes Limmaren and Vallentunasjön.
Sampling.
The four lakes were sampled once in 2002 and seven times between May and October in 2003. Single surface water samples (0 to 0.5 m) were collected from each lake using acid-rinsed and autoclaved polycarbonate bottles. Water samples were kept dark at in situ temperature for no more than 2 h before analysis.
Biotic and abiotic state variables.
Duplicate samples were preserved with borax-buffered formaldehyde (2% final concentration) for microscopic counts of bacterial abundance. Cells were stained with 4,6-diamidino-2-phenylindole (DAPI) prior to analysis by epifluorescence microscopy (27). At least 200 cells or a minimum of 10 fields of view were analyzed.
Bacterial production was analyzed by [3H]leucine incorporation (36) using protein-to-biomass conversion factors according to the method of Simon and Azam (35).
Chlorophyll a was measured spectrophotometrically for cells retained on Whatman GF/F filters (nominal pore size, 0.7 μm). Frozen filters were extracted in 96% ethanol for 6 h, followed by removal of particulate matter by 0.2-μm-membrane filtration. Chlorophyll a absorbance was determined at 665 nm with corrections for suspended solids at 750 nm (15). Dissolved organic carbon was analyzed by high-temperature catalytic oxidation on a Shimadzu TOC analyzer as previously described (7).
Nucleic acid extraction.
Bacterial cells for DNA extraction were captured by vacuum filtration (<30 kPa) of various volumes of lake water (150 to 500 ml) on 0.2-μm membrane filters (Supor, Gelman). Filters were immediately frozen and stored at −80°C until analysis. Bead beating and solid-phase extraction were used to isolate nucleic acids for PCR-based assays as detailed elsewhere (8). The quantity and average size of extracted nucleic acids were determined by 1% agarose gel electrophoresis (for more details, see reference 8). In short, gels were stained with ethidium bromide and extracted nucleic acids were quantified by comparison against a low-DNA-mass ladder (Invitrogen, Carlsbad, CA) using Gel-Pro Analyser version 3.1 (Media Cybernetics, Inc., Silver Spring MD). The concentration of genomic DNA in the extracts ranged from 2 to 20 ng μl−1.
Q-PCR.
Q-PCR reactions were prepared using Platinum SYBR Green qPCR SuperMix-UDS (Invitrogen, Carlsbad, CA) in 200-μl flat-lid PCR strips and an MX3000P thermocycler (Stratagene, La Jolla, CA). Results were analyzed with MxPro-Mx 3000P software (Version 3.00; Stratagene, La Jolla, CA). Reactions (20-μl volumes) contained 10 μl 2× Platinum SYBR Green PCR master mix, 250 nM of each primer (final concentration), 5 μl of template (2 to 20 ng DNA), 1 μl 20× bovine serum albumin, and UV-treated and 2-μl-filter-sterilized ultrapure Q-grade water. For each sample, two individual replicated Q-PCRs were performed using two different primer pairs. Primers used for amplification of class Flavobacteria 16S rRNA genes were 27f (5′-AGR GTT TGA TCM TGG CTC AG −3′) (42) and 588 r (5′-GGA CCC TTT AAA CCC AAG −3′) (modified from that described in reference 45). PCR was performed using a thermocycling program consisting of an initial step of 2 min at 95°C, followed by 45 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s with continuous acquisition of SYBR Green fluorescence signal. Total concentration of bacterial 16S rRNA genes was assessed using primers 27f (42) and 519r (5′-GWA TTA CCG CGG CKG CTG −3′) (21) and the same PCR assay as described above with the exception that an annealing temperature of 50°C was used. For all samples, melting profiles were done to check if the length of the resulting PCR product was homogenous. Positive controls and titers for the Q-PCR assay included genomic DNA from a Flavobacteria isolate obtained from Lake Vallentuna (isolate VA6, EF078979). This genomic DNA was diluted in 10-fold steps from 50 ng to 0.5 pg, and triplicates of each dilution were analyzed in parallel with the lake samples. Negative controls included samples lacking template DNA. The assay was not only tested with dilution series of the Flavobacteria strain, but also, a mixed environmental sample (Ekoln 13/08-2003) was serially diluted to three different concentrations. Simple regressions of the cycle threshold values for the DNA standards versus the log-transformed starting copy number were plotted for both the universal primer pair (total number of 16S rRNA genes) and the 16S rRNA genes targeted by the Flavobacteria-specific primer pair.
To provide information about reproducibility and sensitivity of the Q-PCR (37), the r2 value, the amplification efficiency, and the slope were reported for each standard curve (Table 1). Finally, values were corrected for their dilution factor and averaged to calculate the relative amount of Flavobacteria compared to the total bacterial 16S rRNA gene copies in a given sample. Results from the clone libraries corroborate the specific amplification of the class Flavobacteria with primer 588r (see Results, below). Absolute numbers of Flavobacteria were determined by multiplying the bacterial cell abundance (DAPI counts) and the relative contribution of the flavobacterial gene copy number to the total 16S rRNA gene copy number. Regression analysis revealed a significant relationship between the cycle threshold values of the Q-PCR using general bacterial primers and the number of bacteria used for DNA extraction (r2 = 0.35; P < 0.001).
TABLE 1.
Regression coefficient (r2), amplification efficiency (E), and slope of four standard curves for real-time PCR amplification of rRNA genesa
| Sample or isolate analyzed | r2 value | E value | Slope |
|---|---|---|---|
| Bacteria | |||
| VA 6 | 0.99 | 69.3 | 4.372 |
| EK13/8 | 0.99 | 70.4 | 4.321 |
| Flavobacteria | |||
| VA 6 | 0.98 | 78.8 | 4.142 |
| EK13/8 | 0.99 | 71.6 | 4.262 |
Either primer pair 27f and 519r (Bacteria) or pair 27f and 588r (Flavobacteria) was used. Standard curves for the environmental sample (EK13/8) and isolate VA 6 (EF078979) are shown.
T-RFLP.
T-RFLP analysis of Flavobacteria 16S rRNA genes was used to assess the composition of the Flavobacteria communities. Flavobacteria 16S rRNA genes were amplified from mixed genomic samples using PCR with the bacterial primer 27f (42) and the class Flavobacteria-specific primer 588r (modified from reference 45).
The forward primer was labeled with hexachlorofluorescein at the 5′ end (MWG Biotech) to enable fluorescence detection of terminal restriction fragments (ribotypes). Eight replicate PCRs of 20 μl (each) were amplified for each sample in a Stratagene Robocycler (Stratagene, La Jolla, CA) under the following PCR conditions: initial denaturation at 95°C for 8 min, followed by 20 cycles of amplification (1 min at 95°C, 1 min at 55°C, and 1 min at 72°C) and a final 4-min extension at 72°C.
Pseudoterminal fragments were eliminated by digestion of single-stranded DNA generated in the PCR with mung bean nuclease (6). The PCR products were then purified and concentrated using the QIAquick PCR purification kit (QIAGEN, Hilden, Germany). For each sample, aliquots containing approximately 50 ng of PCR product were digested with the endonuclease HhaI for at least 16 h according to instructions supplied by the manufacturer (Invitrogen). Fluorescently labeled fragments were separated by size in an ABI 3700 96-capillary sequencer running in GeneScan mode (Applied Biosystems, Foster City, CA). T-RFLP electropherograms were analyzed with the freeware GenScanView 4 (CRIBI group [http://grup.cribi.unipd.it]) using 1% of the total peak area as a lower cutoff for ribotypes to be included in the comparative analysis.
PCR and cloning.
Two clone libraries from each lake were constructed from PCR products based on the bacterial primer 27f (42) and either bacterial primer 1492r (5′-GGYTACCTTGTTACGACTT-3′) (21) or the Flavobacteria-specific primer 588r (modified from reference 45). The detailed procedures for generation of clone libraries using the universal bacterial primer pair are given elsewhere (8).
For PCR using Flavobacteria-specific primers, between 5 and 20 ng of genomic DNA from each sample was added to five replicate 20-μl reaction mixtures with PCR conditions as described for T-RFLP analysis. Replicate PCRs were pooled and precipitated with ethanol and sodium acetate (30), followed by agarose gel purification as described elsewhere (8). From each PCR product, 5 to 10 ng was cloned into the pCR 4-TOPO vector and transformed into competent Escherichia coli One Shot TOP10 using the TOPO TA cloning kit for sequencing as recommended by the manufacturer (Invitrogen, Carlsbad, CA).
All flavobacterial clone libraries (47 clones for each library) were screened for gene variants using a previously described high-resolution T-RFLP method (8). In contrast to the T-RFLP fingerprinting approach (described above), this method is based on parallel use of two endonucleases to cut a 16S rRNA gene fragment carrying different fluorescence tags at the two terminal ends. This procedure separates operational taxonomic units (OTUs) with 16S rRNA gene sequence similarities ranging from 73 to 99.9% for different taxonomic groups (8).
Sequencing and phylogenetic analysis.
At least one clone was sequenced for each OTU. Additional clones, chosen randomly, were also sequenced for the more abundant OTUs. From the universal clone libraries, a total of 558 clones were sequenced (for more details, see reference 8). From the four Flavobacteria clone libraries a total of 89 clones were sequenced on an ABI 3700 96-capillary sequencer (Applied Biosystems) using the primer M13f and the BigDye Terminator kit, version 3.1 (Applied Biosystems). Four sequences were ambiguous and hence discarded. All other sequences were high-quality reads of ∼550 bases. The RDPII Chimera Check program was used to identify sequences with a high probability of being chimeric. All cloned sequences were compared to GenBank entries using BLAST (Basic Local Alignment Tool; July 2006) in order to select reference sequences and obtain a preliminary phylogenetic affiliation of the clones. All sequences were imported into ARB and automatically aligned using the integrated aligner tool and the fast aligner option, followed by manual alignment of the sequences to closely related sequences with the secondary structure of rRNA taken into account. Phylogenetic analyses were performed with ARB (38). Trees were constructed using maximum likelihood, and tree topology was confirmed by maximum parsimony with 100 bootstrap replications and neighbor-joining with Jukes-Cantor distance correction (see reference 8 and references therein).
Data analyses.
For T-RFLP, the relative abundance of ribotypes was estimated from the peak area in percentage of total peak area. Relative ribotype peak areas were binned according to fragment length (∼1 bp). To investigate the degree of similarity between different Flavobacteria communities, T-RFLP patterns were converted to a binary (0/1) matrix. A dissimilarity matrix (Dxy) was calculated from the binary data with the equation Dxy = 1 − 2Nxy/(Nx + Ny), where Nx and Ny represent the number of peaks present either in sample x or y, respectively, and Nxy is the number of peaks present in both samples (7). Cluster analysis using the unpaired-pair-group-method-using-average-linkages algorithm was carried out in Statistica (Statsoft, Tulsa, OK). The Mantel test was used to correlate the dissimilarity matrix of bacterial community composition with dissimilarity matrices of chlorophyll a and bacterial production. Instead of using the mean, replicated measurements were pooled prior to ordinary least-square regression and correlation analyses to minimize the effects of measurement errors. We estimated the strength of the linear relationship (r2) and tested the association between dependent and independent variables (P value).
Nucleotide sequence accession numbers.
All sequences were deposited in GenBank under accession numbers EF060976 to EF061064.
RESULTS
Limnology of the sampled lakes.
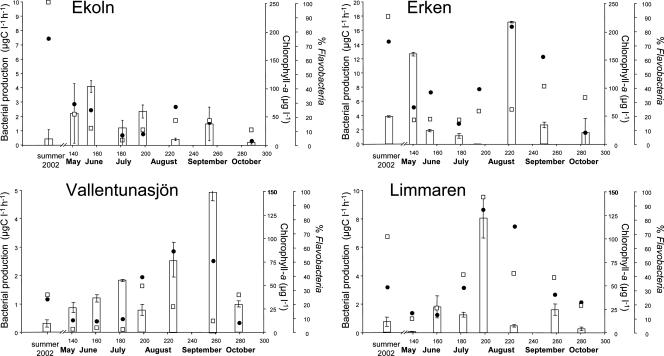
The four lakes are similar in their chemical and physical features (8). Dissolved organic carbon concentrations varied little over season in each lake (coefficient of variance for Lake Ekoln, 10.4%; for Lake Erken, 1.8%; for Lake Vallentunasjön, 6.6%; for Lake Limmaren, 1.7%), and there were only minor differences between the four studied systems, with averages for the entire study ranging from 9.8 to 14.6 mg C liter−1 (see Table S1 in the supplemental material). The chlorophyll a concentration was used as an approximation of phytoplankton biomass. The highest variation (50-fold) of the chlorophyll a concentration was observed for Lake Ekoln, and this lake also had the highest absolute chlorophyll a value (250 μg liter−1) (Fig. 1). Microscopic inspection revealed that this bloom was almost exclusively (>80%) composed of Microcystis aeruginosa colonies. In Lake Erken and Lake Limmaren, chlorophyll a at peak bloom was five- to sevenfold higher than the average for all samples. For the other lakes, microscopic analysis also confirmed that phytoplankton biomass was mainly comprised of Cyanobacteria during the summer period, with a dominance of Gloeotrichia echinulata in Lake Erken and Aphanizomenon spp. in Lake Limmaren. Summer phytoplankton communities in Lake Vallentunasjön consisted of Anabaena flos-aquae, Aphanizomenon flos-aquae, and Microcystis spp., with summer chlorophyll a levels three- to sevenfold higher than the chlorophyll a values measured in spring and autumn. Peaks of cyanobacterial blooms were accompanied by high abundances of heterotrophic bacteria, ranging from 1010 to 1011 cells liter−1 in the different lakes (see Table S1 in the supplemental material).
FIG. 1.
Flavobacterial 16S rRNA genes as a percentage of total bacterial 16S rRNA genes (% Flavobacteria; bars), changes in bacterial production (filled circles), and chlorophyll a (open squares) in four lakes. All values are averages from triplicate analyses, and error bars indicate standard deviations.
Diversity of class Flavobacteria in productive lakes.
In the present study, the diversity of the class Flavobacteria was first analyzed by screening universal bacterial clone libraries for each of the four lakes. Samples for these libraries were obtained during cyanobacterial bloom events in the year 2002. These four bacterial clone libraries were dominated by Cyanobacteria, although Bacteriodetes and Proteobacteria were also abundant (8). Of 1,461 clones, 105 were affiliated with the class Flavobacteria. The percentages of clones affiliated with Flavobacteria in bacterial clone libraries were 1.6% for Vallentunasjön, 2.7% for Lake Ekoln, 5.4% for Lake Erken and 23% for Lake Limmaren.
Clone libraries based on 16S rRNA genes amplified with primers specific to Flavobacteria were also prepared and analyzed for each lake. These libraries were prepared from samples collected in the summer of 2003 (Ekoln, 17/7; Erken, 13/8; Limmaren, 18/7; Vallentunasjön, 30/6). A total of 181 clones from the flavobacterial clone libraries were screened by high-resolution T-RFLP screening as described for the bacterial clone libraries (8). Twenty-four unique OTUs were detected, with 44 of the clones belonging to a single abundant OTU (B86; see Fig. S1 in the supplemental material). For B86, both T-RFLP fingerprinting and high-resolution T-RFLP screening revealed the same operationally defined group. In contrast, B88, operationally defined in the community T-RFLP fingerprint, represents a diverse group of Flavobacteria species as revealed by sequencing and high-resolution T-RFLP screening and includes several freshwater isolates (e.g., Flavibacterium columnare, Flavibacterium succinicans, and Flavibacterium aquatile). Using high-resolution T-RFLP screening, a total of five OTUs could be identified within B88. Similarly, clones that grouped into OTU B90 could be divided into two distinct groups (see Fig. S1 in the supplemental material).
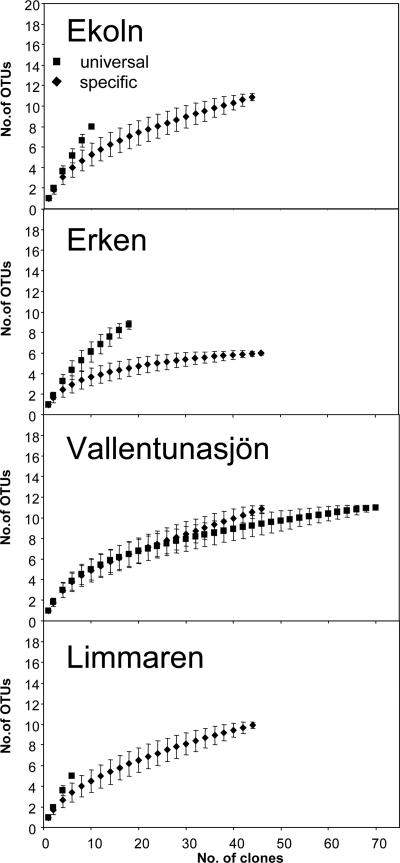
Phylogenetic analyses and rarefaction analyses (Fig. 2) of the four clone libraries revealed a wide diversity of Flavobacteria in the four productive lakes. A few freshwater Flavobacteria, which have 16S rRNA sequences similar to those of the OTUs detected in the present study, have previously been isolated and characterized (2, 39). For example, F. columnare is a well-described fish pathogen, and Flavobacterium limicola, which has been isolated from freshwater sediments, can degrade various organic polymers.
FIG. 2.
Rarefaction analyses of the eight clone libraries based on high-resolution terminal restriction fragment length polymorphism screening. Two clone libraries using either universal primers (27f and 1492r) or flavobacterium-specific primers (27f and 588r) are plotted in each graph.
Dynamics of class Flavobacteria.
The proportion of the bacterial community affiliated with Flavobacteria varied substantially both among lakes and over time (Fig. 1). The highest relative abundance was observed during the senescence and decline of a cyanobacterial bloom in Lake Erken (with Flavobacteria contributing to nearly 100% of the bacterioplankton community). Additional periods where the bacterioplankton community was dominated by Flavobacteria were observed during summer in both Lake Limmaren (approximately 80%) and Lake Vallentunasjön (approximately 90%) (Fig. 1).
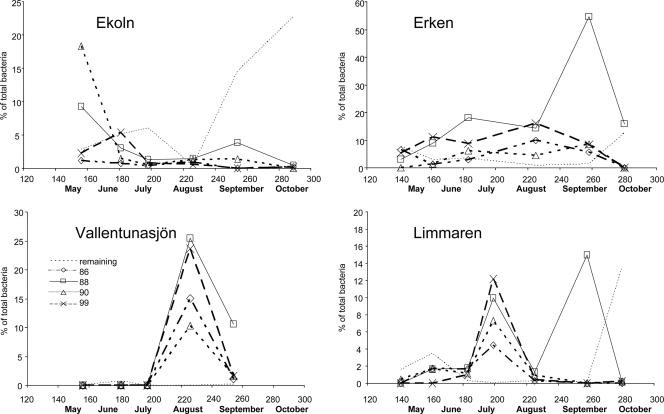
To increase the phylogenetic resolution of our PCR assay, T-RFLP fingerprinting was applied to follow the dynamics of operationally defined populations (ribotypes). T-RFLP results were compared by estimating the relative peak areas of individual ribotypes on different sampling occasions. Generally, two ribotypes, B88 and B99, were dominant in the T-RFLP electropherograms (Fig. 3). These populations were also mainly responsible for the observed Flavobacteria blooms, here defined as episodes where the contribution of Flavobacteria to total bacterioplankton exceeded 30%. During these blooms, multiple populations of Flavobacteria were often detected. In Lake Erken, ribotype B88 contributed more than 50% of the total 16S rRNA gene pool during the senescing phase of a cyanobacterial bloom. In Lake Vallentunasjön, the combined contribution of B88 and B99 to the bacterial 16S rRNA gene pool exceeded 50% during a bloom of filamentous cyanobacteria at the beginning of August. Also, in Lake Limmaren samples from late July, the Flavobacteria community was dominated by these two ribotypes during a filamentous cyanobacterial bloom. Ribotype B88 was observed, usually in high relative abundances, in all samples except in spring samples of Lake Vallentunasjön. Also, ribotypes B90, B99, and B86 could be detected in more than 50% of the samples and at least twice in each lake. These operationally defined populations were also the only ones detected in the clone libraries constructed from the four lakes, and they represent diverse lineages, as indicated in the phylogenetic tree (see Fig. S1 in the supplemental material). In addition, 29 rare or occasionally occurring ribotypes were also detected in the 4 lakes. These populations were infrequently detected but contributed substantially to the relative peak areas in single autumn samples (Fig. 3).
FIG. 3.
Temporal dynamics of dominant operationally defined Flavobacteria populations (terminal restriction fragments B86, B88, B90, and B99) and sum of all other peaks (remaining) from T-RFLP profiles of HhaI-digested PCR-amplified DNA from four lakes. y axis represents relative peak intensity multiplied by relative contribution of Flavobacteria to the total bacterial community. Phylogeny of each terminal restriction fragment is shown in Fig. S1 in the supplemental material.
DISCUSSION
In this study, we observed highly dynamic microbial communities, reflected by blooms of various Flavobacteria ribotypes in four eutrophic lakes. These blooms could be linked to bacterial production maxima and in some cases to cyanobacterial blooms (Fig. 1).
To determine the abundance of Flavobacteria, flavobacterium-specific primers and universal bacterial primers were applied in a Q-PCR. The flavobacterial primer 588r was modified to feature one mismatch at the 3′ end (guanine instead of thymidine) for most known freshwater Flavobacteria, with the exception of one group (indicated by B0 in Fig. S1 in the supplemental material). 16S rRNA genes from members of group B0 have two mismatches to primer 588r and were not amplified in the Flavobacteria class-specific PCR, whereas other Flavobacteria targets containing only the 3′ mismatch were amplified. At the same time, this modification prevents nonspecific amplification of the more than 9,000 bacterial nontargets in Ribosomal Database Project II, release 9, that feature only a single mismatch to the 5′ end of the original probe 588r (see Table S2 in the supplemental material). This was demonstrated by amplification of several single-mismatch Flavobacteria targets and lack of amplification of DNA from strains with two mismatches (data not shown). A regression between Q-PCR standard curves obtained using either the modified or unmodified primer 588r was highly significant, indicating that the two primers performed similarly in the quantification, except for the lower efficiency of the modified primer (see Table S2 and Fig. S2 in the supplemental material). The standard curves obtained from both isolate VA6 and an environmental sample suggested good linearity and no effect of dilution on quantification (Table 1). It is well known that 16S rRNA gene copy numbers can vary between different bacterial taxa and thus bias absolute quantification of bacterial abundance (see reference 37 and references therein). Also, amplification of plastid 16S rRNA genes can lead to an overestimation of total bacterial 16S rRNA genes. Results from the four bacterial clone libraries indicate that this bias is minor, since only between 1 and 5.5% of the clones in these libraries were affiliated with plastids (8). Comparison of the rarefaction curves obtained from the different libraries shows that the bacterial clone libraries exhibited steeper OTU rarefaction curves than the flavobacterial clone libraries for three of the lakes (Fig. 1). This corroborates that the Flavobacteria class-specific primers target a lower richness of OTUs within the Flavobacteria than the universal Bacteria primers (e.g., ribotype B0; see Fig. S1 in the supplemental material). However, since the clone libraries were constructed from different samples, it is not possible to make any direct estimates of the fraction of the Flavobacteria community that escape detection. Still, as an example, none of the Flavobacteria clones detected in the bacterial clone libraries of Vallentunasjön would have been amplified with the flavobacterium-specific primer (two mismatches or more to the modified primer 588r).
With these limitations in mind, estimated 16S rRNA genes of Flavobacteria contributed on average 16% to the total 16S rRNA gene pool for Lake Ekoln, 35% for Lake Erken, 18% for Lake Limmaren, and 27% for Lake Vallentunasjön. Hence, our study reveals proportions of Flavobacteria in freshwater bacterioplankton communities similar to those observed in previous studies (10, 12, 19, 24, 33, 34, 47). In several samples from the four lakes, Flavobacteria contributed more than 30% to the total 16S rRNA gene pool. High proportions and abundances of single heterotrophic bacterial populations have been described previously as a result of high grazing pressure (25). Several other studies suggest that phytoplankton blooms and high grazing pressure can favor members of the Bacteriodetes division (for example, see references 24, 33, and 34).
Multivariate statistical analyses, like principal component analysis and partial least-squares projection to latent structures (data not shown), did not result in any satisfactory model which could explain the dynamics of the class Flavobacteria. An exception was that the contribution of Flavobacteria seemed to follow the trends of bacterial production determined from leucine incorporation (Fig. 3). Correlation and ordinary least-square regression analyses using bacterial production as the independent variable and estimates of absolute abundance of Flavobacteria as the dependent variable (replicate measurements were pooled prior to analyses) showed a significant linear relationship (r2 = 0.42; P < 0.001; n = 32). There was also a significant relationship between bacterial production and total bacterial abundance (r2 = 0.40; P < 0.001; n = 32). This suggests that members of the class Flavobacteria are indeed favored by conditions that prevail during episodes of high heterotrophic activity and enhanced growth, but by no means does it imply that Flavobacteria are generally more competitive than other bacterioplankton under such conditions. The significant linear relationship between the chlorophyll a concentration and bacterial production (r2 = 0.50; P ≪ 0.001; n = 32) suggests that phytoplankton-derived organic matter is one of the main drivers of bacterial growth in the four studied lakes. A weak relationship between chlorophyll a and estimates of absolute abundance of Flavobacteria was observed (r2 = 0.18; P = 0.017; n = 32). Previous studies also indicate that Flavobacteria can be very abundant during phytoplankton blooms in marine systems (1, 10) and in freshwater environments (47). Further evidence that Flavobacteria are abundant during phytoplankton blooms, particularly during cyanobacterial blooms in freshwater environments, comes from high frequencies of 16S rRNA genes affiliated with this class in PCR-based gene surveys of bloom-associated communities and consortia linked to cyanobacterial colonies (8, 9, 20, 28). However, it is difficult to make any general conclusions about the dynamics of the class Flavobacteria during phytoplankton blooms from either the present or previous studies (1, 26, 28, 41, 47), since results are inconclusive.
Bacteria not only are regulated by autochthonous resources (3), but they are also tightly linked to allochthonous resource inputs (7, 40) and controlled by both eukaryote and virus predation in the aquatic environment (4, 5, 11, 26, 31-34, 43). Laboratory experiments suggest that certain groups of bacteria are more susceptible to flagellates grazing (16) and viral lysis (32) than others. Simek and colleagues (33, 34) reported that under enhanced grazing pressure, Cythophaga-Flavobacteria were overrepresented in grazing-resistant filamentous bacterial morphotypes (25).
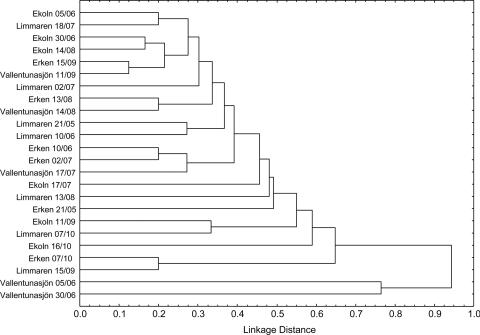
Another explanation for the weak relationship between Flavobacteria and environmental variables may be provided by the physiological and ecological diversity within the class. Even if T-RFLP analysis was applied to reveal dynamics at the subclass level (Fig. 3), the observed ribotypes do not represent operational taxonomic units that are monophyletic (see Fig. S1 in the supplemental material) and it is not likely that they are ecologically and physiologically uniform. It should be emphasized that even organisms with identical 16S rRNA genes can exhibit different physiologies and inhabit different niches (14, 29). Nevertheless, cluster analysis of T-RFLP patterns using binary data for dissimilarity analysis (Fig. 4) suggested a highly dynamic Flavobacteria community with low similarities between lakes and over seasons. Samples from different lakes and at different time points clustered together, giving no support for distinct Flavobacteria community characteristics for a certain lake or a certain seasonal succession stage. The average similarity of all samples collected from the four lakes was 44%, and the average similarity of samples collected from the same lake was 51% for Lake Ekoln, 55% for Lake Erken, 42% for Lake Limmaren, and 24% for Lake Vallentunasjön. Hence, it could not be concluded whether the greatest component of variation was related to differences either among lakes or over time (see also Fig. 4). The Flavobacteria community of Lake Vallentunasjön was the most variable among the four lakes. This could be a result of morphology, since Lake Vallentunasjön is the shallowest of the four lakes (8), and its bacterial community may be more influenced by wind-driven sediment resuspension and a high content of suspended particles. Additional statistical analysis using a Mantel test revealed no correlation between chlorophyll a and bacterial community composition. A significant correlation between dissimilarity matrices of bacterial production and bacterial community composition (r = 0.131; P = 0.02) was observed, suggesting that not only Flavobacteria abundance but also the composition of the class Flavobacteria is related to bulk bacterial growth conditions. Since bacterial production is equal to the amount of organic matter that can be transformed into bacterial biomass, the covariation further indicates that resource availability is likely to regulate Flavobacteria community composition. This is consistent with other studies that have shown correlations between resources and community composition (5, 26).
FIG. 4.
Cluster analysis of flavobacterial communities. Unweighted pair-grouped tree diagram of 24 samples that were analyzed by T-RFLP fingerprinting and subsequent statistical analyses as described elsewhere (8).
To conclude, eutrophic lakes harbor highly dynamic bacterioplankton communities which feature temporary blooms of Flavobacteria populations. The correlation between Flavobacteria dynamics and bacterial production suggests that the availability of resources is likely to shape heterotrophic bacterial seasonal succession.
Supplementary Material
Acknowledgments
We thank Edith Schallmeiner for assistance with setting up the Q-PCR assay.
This work was funded by the Swedish Research Council for Environment, Agricultural Sciences and Special Planning and the Swedish Research Council (grants to S.B.).
Footnotes
Published ahead of print on 13 April 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abell, G. C. J., and J. P. Bowman. 2005. Colonization and community dynamics of class Flavobacteria on diatom detritus in experimental mesocosms based on Southern Ocean seawater. FEMS Microbiol. Ecol. 53:379-391. [DOI] [PubMed] [Google Scholar]
- 2.Bernardet, J. F., P. Segers, M. Vancanneyt, F. Berthe, K. Kersters, and P. Vandamme. 1996. Cutting a Gordian knot: emended classification and description of the genus Flavobacterium, emended description of the family Flavobacteriaceae, and proposal of Flavobacterium hydatis nom. nov. (Basonym, Cytophaga aquatilis Strohl and Tait 1978). Int. J. Bacteriol. 46:128-148. [Google Scholar]
- 3.Cole, J. J., G. E. Likens, and D. L. Strayer. 1982. Photosynthetically produced dissolved organic carbon: an important carbon source for planktonic bacteria. Limnol. Oceanogr. 27:1080-1090. [Google Scholar]
- 4.Cottrell, M. T., and D. L. Kirchman. 2000. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl. Environ. Microbiol. 66:1692-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crump, B. C., G. W. Kling, M. Bahr, and J. E. Hobbie. 2003. Bacterioplankton community shifts in an arctic lake correlate with seasonal changes in organic matter source. Appl. Environ. Microbiol. 69:2253-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Egert, M., and M. W. Friedrich. 2003. Formation of pseudo-terminal restriction fragments, a PCR-related bias affecting terminal restriction fragment length polymorphism analysis of microbial community structure. Appl. Environ. Microbiol. 69:2555-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eiler, A., S. Langenheder, S. Bertilsson, and L. J. Tranvik. 2003. Heterotrophic bacterial growth efficiency and community structure at different natural organic carbon concentrations. Appl. Environ. Microbiol. 69:3701-3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eiler, A., and S. Bertilsson. 2004. Composition of freshwater bacterial communities associated with cyanobacterial blooms in four Swedish lakes. Environ. Microbiol. 6:1228-1243. [DOI] [PubMed] [Google Scholar]
- 9.Eiler, A., J. A. Olsson, and S. Bertilsson. 2006. Diurnal variation in the auto- and heterotrophic activity of cyanobacterial phycospheres (Gloeotrichia echinulata) and the identity of attached bacteria. Freshwater Biol. 51:298-311. [Google Scholar]
- 10.Fandino, L. B., L. Riemann, G. F. Steward, and F. Azam. 2005. Population dynamics of Cytophage-Flavobacteria during marine phytoplankton blooms analyzed by real-time quantitative PCR. Aquat. Microb. Ecol. 40:251-257. [Google Scholar]
- 11.Fuhrman, J. A. 1999. Marine viruses and their biogeochemical and ecological effects. Nature 399:541-548. [DOI] [PubMed] [Google Scholar]
- 12.Glöckner, F. O., B. M. Fuchs, and R. Amann. 1999. Bacterioplankton composition in lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl. Environ. Microbiol. 65:3721-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glöckner, F. O., E. Zaichikov, N. Belkova, L. Denissova, J. Pernthaler, A. Pernthaler, and R. Amann. 2000. Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of actinobacteria. Appl. Environ. Microbiol. 66:5053-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hahn, M. W., and M. Pöckl. 2005. Ecotypes of planktonic Actinobacteria with identical 16S rRNA genes adapted to thermal niches in temperate, subtropical, and tropical freshwater habitats. Appl. Environ. Microbiol. 71:766-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jespersen, A.-M., and K. Christoffersen. 1987. Measurements of chlorophyll a from phytoplankton using ethanol as extraction solvent. Arch. Hydrobiol. 109:445-454. [Google Scholar]
- 16.Jezbera, J., K. Hornak, and K. Simek. 2006. Prey selectivity of bacterivorous protists in different size fractions of reservoir water amended with nutrients. Environ. Microbiol. 8:1330-1339. [DOI] [PubMed] [Google Scholar]
- 17.Kalff, J., and R. Knoechel. 1978. Phytoplankton and their dynamics in oligotrophic and eutrophic lakes. Annu. Rev. Ecol. Syst. 9:475-479. [Google Scholar]
- 18.Kent, A. D., S. E. Jones, G. H. Lauster, J. M. Graham, R. J. Newton, and K. D. McMahon. 2006. Experimental manipulation of microbial food web interactions in a humic lake: shifting biological drivers of bacterial community structure. Environ. Microbiol. 8:1448-1459. [DOI] [PubMed] [Google Scholar]
- 19.Kirchman, D. L. 2002. The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Microbiol. Ecol. 39:91-100. [DOI] [PubMed] [Google Scholar]
- 20.Kolmonen, E., K. Sivonen, J. Rapala, and K. Haukka. 2004. Diversity of cyanobacteria and heterotrophic bacteria in cyanobacterial blooms in Lake Joutikas, Finland. Aquat. Microb. Ecol. 36:201-211. [Google Scholar]
- 21.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley and Sons, Chichester, United Kingdom.
- 22.Methe, B. A., W. D. Hiorns, and J. P. Zehr. 1998. Contrasts between marine and freshwater bacterial community composition: analyses of communities in Lake George and six other Adirondack lakes. Limnol. Oceanogr. 43:368-374. [Google Scholar]
- 23.Newton, R. J., A. D. Kent, E. W. Triplett, and K. D. McMahon. 2006. Microbial community dynamics in a humic lake: differential persistence of common freshwater phylotypes. Environ. Microbiol. 8:956-970. [DOI] [PubMed] [Google Scholar]
- 24.Pernthaler, J., F. O. Glöckner, S. Unterholzner, A. Alfreider, R. Psenner, and R. Amann. 1998. Seasonal community and population dynamics of pelagic bacteria and archaea in a high mountain lake. Appl. Environ. Microbiol. 64:4299-4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pernthaler, J., E. Zöllner, F. Warnecke, and K. Jürgens. 2004. Bloom of filamentous bacteria in a mesotrophic lake: identity and potential controlling mechanism. Appl. Environ. Microbiol. 70:6172-6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pinhassi, J., F. Azam, J. Hemphala, R. A. Long, J. Martinez, U. L. Zweifel, and A. Hagström. 1999. Coupling between bacterioplankton species composition, population dynamics, and organic matter degradation. Aquat. Microb. Ecol. 17:13-26. [Google Scholar]
- 27.Porter, K. G., and Y. S. Feig. 1980. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25:843-848. [Google Scholar]
- 28.Riemann, L., and A. Winding. 2001. Community dynamics of free-living and particle-associated bacterial assemblages during a freshwater phytoplankton bloom. Microb. Ecol. 42:274-285. [DOI] [PubMed] [Google Scholar]
- 29.Rocap, G., F. W. Larimer, J. Lamerdin, S. Malfatti, P. Chain, N. A. Ahlgren, A. Arellano, M. Coleman, L. Hauser, W. R. Hess, Z. I. Johnson, M. Land, D. Lindell, A. F. Post, W. Regala, M. Shah, S. L. Shaw, C. Steglich, M. B. Sullivan, C. S. Ting, A. Tolonen, E. A. Webb, E. R. Zinser, and S. W. Chisholm. 2003. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424:1042-1047. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., and D. W. Russel. 2000. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 31.Sanders, R. W., S. J. Bennett, and A. E. DeBiase. 1989. Seasonal dynamics of bacterivory by flagellates, ciliates, rotifers and cladocerans in a freshwater planktonic community. Limnol. Oceanogr. 34:673-687. [Google Scholar]
- 32.Schwalbach, M. S., I. Hewson, and J. A. Fuhrman. 2004. Viral effects on bacterial community composition in marine plankton mesocosms. Aquat. Microb. Ecol. 34:117-127. [Google Scholar]
- 33.Simek, K., P. Kojecká, J. Nedoma, P. Hartman, J. Vrba, and J. R. Dolan. 1999. Shifts in bacterial community associated with different microzooplankton size fractions in a eutrophic lake. Limnol. Oceanogr. 44:1634-1644. [Google Scholar]
- 34.Simek, K., J. Pernthaler, M. G. Weinbauer, K. Hornak, J. R. Dolan, J. Nedoma, M. Masin, and R. Amann. 2001. Changes in bacterial community composition and dynamics and viral mortality rates associated with enhanced flagellate grazing in a mesoeutrophic reservoir. Appl. Environ. Microbiol. 67:2723-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simon, M., and F. Azam. 1989. Protein content and protein synthesis rates of planktonic marine bacteria. Mar. Ecol. Prog. Ser. 51:201-213. [Google Scholar]
- 36.Smith, D. C., and F. Azam. 1992. A simple, economical method for measuring bacterial protein synthesis rates in seawater using 3H-leucine. Mar. Microb. Food Webs 6:107-114. [Google Scholar]
- 37.Smith, C. J., D. B. Nedwell, L. F. Dong, and A. M. Osborn. 2006. Evaluation of quantitative polymerase chain reaction-based approaches for determining gene copy and gene transcript numbers in environmental samples. Environ. Microbiol. 8:804-815. [DOI] [PubMed] [Google Scholar]
- 38.Strunk, O., and W. Ludwig. 1996. ARB: a software environment for sequence data, 2.1.1. Department of Microbiology, Technical University of Munich, Munich, Germany.
- 39.Tamaki, H., S. Hanada, Y. Kamagata, K. Nakamura, N. Nomura, K. Nakano, and M. Matsumura. 2003. Flavobacterium limicola sp. nov., a psychrophilic, organic-polymer-degrading bacterium isolated from freshwater sediments. Int. J. Syst. Evol. Microbiol. 53:519-526. [DOI] [PubMed] [Google Scholar]
- 40.Tranvik, L. J. 1988. Availability of dissolved organic carbon for planktonic bacteria in oligotrophic lakes of differing humic content. Microb. Ecol. 16:311-322. [DOI] [PubMed] [Google Scholar]
- 41.van Hannen, E. J., G. Zwart, M. P. van Agterveld, H. J. Gons, J. Ebert, and H. J. Laanbroek. 1999. Changes in bacterial and eukaryotic community structure after mass lysis of filamentous cyanobacteria associated with viruses. Appl. Environ. Microbiol. 65:795-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vergin, K. L., E. Urbach, J. L. Stein, E. F. DeLong, B. D. Lanoil, and S. J. Giovannoni. 1998. Screening a fosmid library of marine environmental genomic DNA fragments reveals four clones related to members of the order Planctomycetales. Appl. Environ. Microbiol. 64:3075-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weinbauer, M. G., and M. G. Höfle. 1998. Significance of viral lysis and flagellate grazing as factors controlling bacterioplankton production in a eutrophic lake. Appl. Environ. Microbiol. 64:431-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weiss, P., B. Schweitzer, R. Amann, and M. Simon. 1996. Identification in situ and dynamics of bacteria on limnetic organic aggregates (Lake Snow). Appl. Environ. Microbiol. 62:1998-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weller, R., F. O. Glöckner, and R. Amann. 2000. 16S rRNA target oligonucleotide probes for the in situ detection of members of the phylum Cytophaga-Flavobacterium-Bacteriodes. Syst. Appl. Microbiol. 23:107-114. [DOI] [PubMed] [Google Scholar]
- 46.Zwart, G., B. C. Crump, M. Kamst-van Agterveld, F. Hagen, and S.-K. Han. 2002. Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat. Microb. Ecol. 28:141-155. [Google Scholar]
- 47.Zwisler, W., N. Selje, and M. Simon. 2003. Seasonal patterns of the bacterioplankton community composition in a large mesotrophic lake. Aquat. Microb. Ecol. 31:211-225. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.