Abstract
To investigate quorum sensing in rhizosphere soil, a whole-cell biosensor, Agrobacterium tumefaciens(pAHL-Ice), was constructed. The biosensor responded to all N-acyl homoserine lactones (AHLs) tested, except C4 homoserine lactone, with a minimum detection limit of 10−12 M, as well as to both exogenously added AHLs and AHL-producing bacterial strains in soil. This highly sensitive biosensor reveals for the first time the increased AHL availability in intact rhizosphere microbial communities compared to that in bulk soil.
For many bacterial species, population behavior is controlled by quorum sensing (QS), a cell density-dependent secretion, detection, and response to small molecules (25, 29). Bacteria must reach a quorum to perform ecologically relevant and often vital functions, including biofilm formation, virulence, symbiosis, and extracellular enzyme production (5, 29, 32). Small spatial scales and the heterogeneity of microbial habitats make it difficult to study QS in nature by classical destructive methods, such as extraction of signals or signal producers; whole-cell biosensors address this problem by examining biogeography and ecology at microbial scales (2, 6, 14, 17).
QS in soil may indicate locations where signal molecules cannot disperse away from the producers (26), which is critical in stressful, nutrient-limiting environments. The patchy distribution of bacterial colonization and nutrient availability in soil and on plants (22, 23, 27) suggests that the expression of QS will be similarly heterogeneous. We thus developed a biosensor to examine QS in the rhizosphere, where root exudates quickly foster high microbial activity and altered community structure (19, 36), conditions that are likely to include the activation of QS in some populations.
The Agrobacterium tumefaciens(pAHL-Ice) biosensor was constructed as a transcriptional fusion of the A. tumefaciens tra box promoter region (traG-proximal region) with the inaZ reporter gene and includes a copy of the A. tumefaciens N-acyl homoserine lactone (AHL) receptor TraR gene on the same plasmid. The tra box promoter region was PCR amplified from A. tumefaciens C58 genomic DNA based on the published sequence (9), using primers designed with Primer3 (28). The tra box fragment was cloned into the broad-host-range vector pPROBE-KI′ upstream of the promoterless inaZ gene, which encodes an outer membrane-localized ice nucleation protein that catalyzes ice formation (16, 21). Using freeze-thaw transformation (12), pAHL-Ice was mobilized into A. tumefaciens strain C58C1 (strain C58 cured of its Ti plasmid) (34), and the resulting strain was designated A. tumefaciens(pAHL-Ice). Quantification of AHL-dependent ice nucleation was measured by a droplet-freezing assay (18).
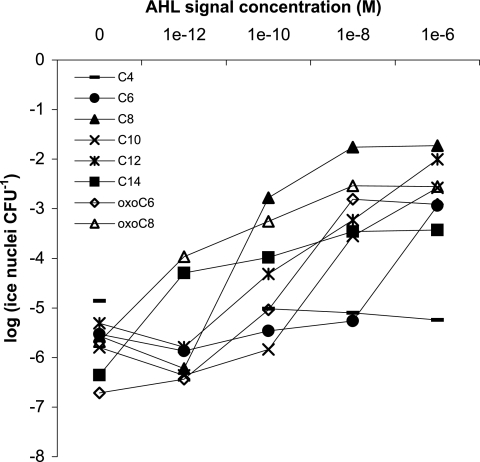
Given that considerable cross talk between species using different AHLs may occur in a mixed microbial community, a biosensor that is relatively nonspecific in its detection of AHLs is a useful tool. A. tumefaciens(pAHL-Ice) responds to a wide range of AHLs (Fig. 1), exhibiting increased ice nucleation activity over 6 orders of magnitude with increasing concentrations of oxo-octanoyl homoserine lactone (oxo-C8-HSL), the cognate QS signal (Fig. 1). The biosensor also exhibited increased ice nucleation activity with increasing concentrations of hexanoyl (C6), ketocaproyl (oxo-C6), octanoyl (C8), decanoyl (C10), dodecanoyl (C12), and tetradecanoyl (C14) HSLs (Fig. 1). No concentration of butyryl (C4) HSL tested was sufficient to activate the biosensor above the background. Surprisingly, C14-HSL, like oxo-C8-HSL, activated the biosensor at very low concentrations (10−12 M) but conferred a level of ice nucleation that was only about twofold higher than the background. The ice nucleation activity of A. tumefaciens(pAHL-Ice) remained stable for up to 20 h when cells were incubated in the presence of oxo-C6-HSL, and when they were transiently (5 min) exposed to oxo-C6-HSL, biosensor ice nucleation activity peaked by 2 h and decreased with time (data not shown). These data suggest that the AHL biosensor is suitable for detecting QS in rhizosphere soils, where the signals are likely to vary strongly for species, concentration, and time.
FIG. 1.
Dose-response curves for a range of AHLs and the biosensor A. tumefaciens(pAHL-Ice) in minimal medium. In each case, the biosensor was grown to about 109 cells ml−1 and then split into 5-ml subcultures that already had a range of concentrations of one of the signals shown. Standard error bars were left off the graph for clarity, but standard errors were as follows: C4, 0.172 to 0.371; C6, 0.179 to 1.026; oxo-C6, 0.08 to 0.946; C8, 0.19 to 0.683; oxo-C8, 0.435 to 0.97; C10, 0.362 to 0.798; C12, 0.254 to 0.582; and C14, 0.168 to 0.638. Biosensor activity is reported as log(ice nuclei CFU−1), and four biological replicates were performed for each concentration.
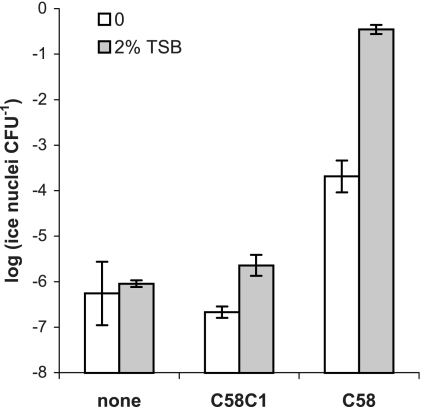
Since soil microbes are generally considered carbon limited (1), it was important to establish that our AHL biosensor could function in bulk soil, which is considered a low-carbon environment. AHL levels in soil were altered by introducing two isogenic strains of A. tumefaciens differing in AHL production before inoculation with the biosensor. Wild-type A. tumefaciens strain C58 produces oxo-C8-HSL, while C58C1 is cured of the Ti plasmid and is incapable of making AHLs. Soils were collected during the spring growing season from under the growing zones of the annual graminoid Avena fatua at the University of California Hopland Research and Extension Center (Hopland, CA). The A. tumefaciens(pAHL-Ice) biosensor responded to AHLs in soil and was not dependent upon exogenous nutrient addition (Fig. 2). Previous use of A. tumefaciens as a host for a nitrate biosensor demonstrated that inaZ and gfp reporter gene products are detectable in the bulk soil without the addition of nutrients (6), which was also the case here.
FIG. 2.
The ability of the biosensor to respond to QS signals in soil was assessed by adding A. tumefaciens strain C58C1, which is cured of its Ti plasmid and cannot make QS signals, or the QS signal-producing A. tumefaciens strain C58. Bulk soil assays were conducted with soil slurries of about 0.5 g soil ml−1 in water alone (control) or in 2% tryptic soy broth (TSB) (nutrient addition), which were placed into tubes in which AHL stocks in ethyl acetate were previously dispensed and allowed to dry. For inoculation into soil, overnight cultures of biosensor cells in M9 medium were washed and resuspended in KPO4 buffer at a concentration of 1010 cells ml−1. Biosensor cells were added to the soil solution and incubated for 4 h. Soil slurries were then macerated to disperse cells before serial dilution and ice nucleation assays. Ice nucleation activities are reported as the logarithms of the numbers of ice nuclei measured per CFU of biosensor grown on selective plates. Error bars represent 1 standard error of the mean for four biological replicates.
Nutrient additions that mimicked root exudates caused a slight increase of biosensor activity, either by increasing AHL production of the native soil bacterial population or by enhancing the biosensor's ability to respond to the endogenous AHLs in the soil. Root exudates stimulate growth of rhizosphere soil microbial communities (14, 33), estimated at about 1010 cells g−1 soil (11). Bacteria capable of QS are more common in the rhizosphere than in bulk soil (8), so they were likely stimulated by the 2% tryptic soy broth addition. While nutrient addition probably stimulated C58 AHL production as well as the responsiveness of the biosensor, exogenous nutrients were not necessary to detect AHLs.
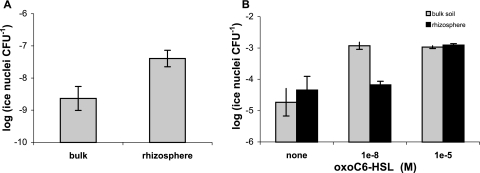
To understand the capacity of the biosensor to respond to AHLs in heterogeneous soil environments, we examined the response of the biosensor to native rhizosphere soil compared to that to bulk soil as well as rhizosphere and bulk soils to which oxo-C6-HSL was added. Rhizosphere studies were conducted in microcosms planted with Avena fatua (wild oat; Valley Seed Service, Fresno, CA). Two greenhouse experiments were conducted using soils collected at different times of year, planted with seeds from the same seed stock, as previously described (14). Both experiments resulted in higher ice nucleation activity in the rhizosphere than in bulk soil (Fig. 3A and B).
FIG. 3.
(A) QS signal availability was measured in greenhouse microcosms, with roots and soils being spray inoculated with A. tumefaciens(pAHL-Ice) and ice nucleation activity being measured 12 h later. Background ice nucleation activity in soils was insignificant. Soils that adhered to the roots after gentle shaking were considered rhizosphere soil, while bulk soil was excised from intact microcosms at least 4 mm away from any roots. The difference in activities was significant (Student's t test; P = 0.0012). (B) To understand if the biosensor was capable of detecting increased signals, the QS signal oxo-C6-HSL was added at 10−8 M and 10−5 M to growth chamber microcosms before the application of the biosensor. Error bars represent 1 standard error of the mean for three biological replicates corresponding to separate microcosms.
The biosensor responded when low (10−8 M) levels of oxo-C6-HSL were added exogenously to bulk soil but not to rhizosphere soil (Fig. 3B), which may indicate enhanced rates of AHL degradation in the rhizosphere compared to those in bulk soil. QS signal quenching is common in many soils (35). Measured rates of 10−8 moles C6-HSL h−1 g soil−1 would be sufficient to reduce the low (10−8 M) but not the high (10−5 M) AHL levels to below the detection limit of our biosensor (Fig. 3B). AHL-degrading strains are found in the Proteobacteria and Bacillus groups (7, 13, 37), and A. tumefaciens harbors the lactonase gene aiiB on its Ti plasmid as well as attM on the At plasmid (4, 20). In our A. tumefaciens biosensor, the presence of attM on the At plasmid might have resulted in moderate AHL degradation, although a previous examination of C58 compared to the same strain without the At plasmid revealed only a 10% loss of signal via attM (4). These data suggest that while rhizosphere soil contains 10 times more AHLs than bulk soil, the standing AHL pool in the rhizosphere represents an equilibrium in a dynamic system of production, dilution, and degradation of signals.
Limitations of previously tested AHL biosensors precluded conclusions regarding the occurrence of QS in the rhizosphere compared to that in bulk soils. For example, a Pseudomonas putida lasB::gfp reporter strain was activated in nonsterile soil, indicating that at least 10−8 M oxo-C12-HSL was present (31), although this study did not compare AHL levels in rhizosphere and bulk soils. Another study used a gfp-based Vibrio fischeri biosensor, stimulated by adding plant litter, to detect AHLs in compost soil (3), demonstrating that QS could be enhanced by adding a nutrient source. Interestingly, the AHL biosensor used in that study seldom detected AHLs in nonamended soils, revealing either its lack of sensitivity for AHLs, the relatively low levels of these signal molecules in soils lacking nutrient additions, or both.
The A. tumefaciens(pAHL-Ice) biosensor developed here is among the most sensitive AHL biosensors constructed, permitting examination of rhizosphere concentrations of AHLs for the first time. The analogous A. tumefaciens traG::lacZ biosensor has a minimum detection limit for a given AHL of at least 10-fold higher than that for A. tumefaciens(pAHL-Ice) (Fig. 1) (30). A traI::lacZ biosensor with traR overexpressed via the T7 promoter responds to oxo-C8-HSL at concentrations down to 10−12 M (38). A. tumefaciens(pAHL-Ice) has the same lower detection limit due to the higher sensitivity of inaZ than that of lacZ (15, 18). The biosensor permits us to estimate rhizosphere AHL levels at 10−8 M oxo-C8-HSL equivalents, in contrast to <10−12 M for bulk soil (Fig. 3B); this is potentially 100-fold higher given the low response of the biosensor to noncognate AHLs (Fig. 1). These concentrations are as high as or higher than estimated signal concentrations in soil (10, 24) and agree well with reported levels of AHLs required to activate QS systems in vitro (29).
The heterogeneity of the soil environment makes biosensors powerful and essential tools for understanding QS in natural systems. QS is fast becoming understood as a central process influencing a myriad of plant-microbe interactions, and QS likely mediates microbial behaviors unique to the rhizosphere. The ice nucleation-based biosensor described here should be a useful tool for testing for AHL signaling in any natural environment, especially in heterogeneous soil environments.
Acknowledgments
We thank D. Dahlbeck for providing the A. tumefaciens strains and D. Dahlbeck and S. K. Farrand for helpful comments regarding the construction of the whole-cell biosensor.
Support from the EPA Science To Achieve Results (EPA STAR) program and an NSF doctoral dissertation improvement grant are also acknowledged.
Footnotes
Published ahead of print on 30 March 2007.
REFERENCES
- 1.Brimecombe, M. J., F. A. DeLeij, and J. M. Lynch. 2001. The effect of root exudates on rhizosphere microbial populations, p. 95-140. In R. Pinton, Z. Varanini, and P. Nannipieri (ed.), The rhizosphere: biochemistry and organic substances at the soil-plant interface. Marcel-Dekker, Inc., New York, NY.
- 2.Bringhurst, R. M., Z. G. Cardon, and D. J. Gage. 2001. Galactosides in the rhizosphere: utilization by Sinorhizobium meliloti and development of a biosensor. Proc. Natl. Acad. Sci. USA 98:4540-4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burmolle, M., L. H. Hansen, and S. J. Sorensen. 2005. Use of a whole-cell biosensor and flow cytometry to detect AHL production by an indigenous soil community during decomposition of litter. Microb. Ecol. 50:221-229. [DOI] [PubMed] [Google Scholar]
- 4.Carlier, A., S. Uroz, B. Smadja, R. Fray, X. Latour, Y. Dessaux, and D. Faure. 2003. The Ti plasmid of Agrobacterium tumefaciens harbors an attM-paralogous gene, aiiB, also encoding N-acyl homoserine lactonase activity. Appl. Environ. Microbiol. 69:4989-4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chernin, L. S., M. K. Winson, J. M. Thompson, S. Haran, B. W. Bycroft, I. Chet, P. Williams, and G. S. A. B. Stewart. 1998. Chitinolytic activity in Chromobacterium violaceum: substrate analysis and regulation by quorum sensing. J. Bacteriol. 180:4435-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeAngelis, K. M., P. S. Ji, M. K. Firestone, and S. E. Lindow. 2005. Two novel bacterial biosensors for detection of nitrate availability in the rhizosphere. Appl. Environ. Microbiol. 71:8537-8547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong, Y. H., A. R. Gusti, Q. Zhang, J. L. Xu, and L. H. Zhang. 2002. Identification of quorum-quenching N-acyl homoserine lactonases from Bacillus species. Appl. Environ. Microbiol. 68:1754-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elasri, M., S. Delorme, P. Lemanceau, G. Stewart, B. Laue, E. Glickmann, P. M. Oger, and Y. Dessaux. 2001. Acyl-homoserine lactone production is more common among plant-associated Pseudomonas spp. than among soilborne Pseudomonas spp. Appl. Environ. Microbiol. 67:1198-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farrand, S. K., I. Hwang, and D. M. Cook. 1996. The tra region of the nopaline-type Ti plasmid is a chimera with elements related to the transfer systems of RSF1010, RP4, and F. J. Bacteriol. 78:4233-4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gantner, S., M. Schmid, C. Durr, R. Schuhegger, A. Steidle, P. Hutzler, C. Langebartels, L. Eberl, A. Hartmann, and F. B. Dazzo. 2006. In situ quantitation of the spatial scale of calling distances and population density-independent N-acylhomoserine lactone-mediated communication by rhizobacteria colonized on plant roots. FEMS Microbiol. Ecol. 56:188-194. [DOI] [PubMed] [Google Scholar]
- 11.Herman, D. J., K. K. Johnson, C. H. Jaeger, E. Schwartz, and M. K. Firestone. 2006. Root influence on nitrogen mineralization and nitrification in rhizosphere soil of slender wild oats. Soil Sci. Soc. Am. J. 70:1504-1511. [Google Scholar]
- 12.Holsters, M., D. Dewaele, A. Depicker, E. Messens, M. Vanmontagu, and J. Schell. 1978. Transfection and transformation of Agrobacterium tumefaciens. Mol. Gen. Genet. 163:181-187. [DOI] [PubMed] [Google Scholar]
- 13.Huang, J. J., J. I. Han, L. H. Zhang, and J. R. Leadbetter. 2003. Utilization of acyl-homoserine lactone quorum signals for growth by a soil pseudomonad and Pseudomonas aeruginosa PAO1. Appl. Environ. Microbiol. 69:5941-5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaeger, C. H., S. E. Lindow, S. Miller, E. Clark, and M. K. Firestone. 1999. Mapping of sugar and amino acid availability in soil around roots with bacterial sensors of sucrose and tryptophan. Appl. Environ. Microbiol. 65:2685-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindow, S. E. 1995. The use of reporter genes in the study of microbial ecology. Mol. Ecol. 4:555-566. [Google Scholar]
- 16.Lindow, S. E., E. Lahue, A. G. Govindarajan, N. J. Panopoulos, and D. Gies. 1989. Localization of ice nucleation activity and the iceC gene product in Pseudomonas syringae and Escherichia coli. Mol. Plant-Microbe Interact. 2:262-272. [DOI] [PubMed] [Google Scholar]
- 17.Loper, J. E., and S. E. Lindow. 1994. A biological sensor for iron available to bacteria in their habitats on plant surfaces. Appl. Environ. Microbiol. 60:1934-1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loper, J. E., and S. E. Lindow. 1996. Reporter gene systems useful in evaluating in situ gene expression by soil- and plant-associated bacteria, p. 482-491. In C. J. Hurst (ed.), Manual of environmental microbiology. American Society for Microbiology, Washington, DC.
- 19.Lynch, J. M., and J. M. Whipps. 1990. Substrate flow in the rhizosphere. Plant Soil 129:1-10. [Google Scholar]
- 20.Madigan, M. T., J. M. Martinko, and J. Parker. 2003. Brock biology of microorganisms, 10th ed. Prentice Hall, Upper Saddle River, NJ.
- 21.Miller, W. G., J. H. J. Leveau, and S. E. Lindow. 2000. Improved gfp and inaZ broad-host-range promoter-probe vectors. Mol. Plant-Microbe Interact. 13:1243-1250. [DOI] [PubMed] [Google Scholar]
- 22.Monier, J. M., and S. E. Lindow. 2005. Spatial organization of dual-species bacterial aggregates on leaf surfaces. Appl. Environ. Microbiol. 71:5484-5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mummey, D., W. Holben, J. Six, and P. Stahl. 2006. Spatial stratification of soil bacterial populations in aggregates of diverse soils. Microb. Ecol. 51:404-411. [DOI] [PubMed] [Google Scholar]
- 24.Pierson, E. A., D. W. Wood, J. A. Cannon, F. M. Blachere, and L. S. Pierson. 1998. Interpopulation signaling via N-acyl-homoserine lactones among bacteria in the wheat rhizosphere. Mol. Plant-Microbe Interact. 11:1078-1084. [Google Scholar]
- 25.Pierson, L. S., D. W. Wood, and E. A. Pierson. 1998. Homoserine lactone-mediated gene regulation in plant-associated bacteria. Annu. Rev. Phytopathol. 36:207-225. [DOI] [PubMed] [Google Scholar]
- 26.Redfield, R. J. 2002. Is quorum sensing a side effect of diffusion sensing? Trends Microbiol. 10:365-370. [DOI] [PubMed] [Google Scholar]
- 27.Ritz, K., W. McNicol, N. Nunan, S. Grayston, P. Millard, D. Atkinson, A. Gollotte, D. Habeshaw, B. Boag, C. D. Clegg, B. S. Griffiths, R. E. Wheatley, L. A. Glover, A. E. McCaig, and J. I. Prosser. 2004. Spatial structure in soil chemical and microbiological properties in an upland grassland. FEMS Microbiol. Ecol. 49:191-205. [DOI] [PubMed] [Google Scholar]
- 28.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365-386. [DOI] [PubMed] [Google Scholar]
- 29.Schauder, S., and B. L. Bassler. 2001. The languages of bacteria. Genes Dev. 15:1468-1480. [DOI] [PubMed] [Google Scholar]
- 30.Shaw, P. D., G. Ping, S. L. Daly, C. Cha, J. E. Cronan, K. L. Rinehart, and S. K. Farrand. 1997. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc. Natl. Acad. Sci. USA 94:6036-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steidle, A., K. Sigl, R. Schuhegger, A. Ihring, M. Schmid, S. Gantner, M. Stoffels, K. Riedel, M. Givskov, A. Hartmann, C. Langebartels, and L. Eberl. 2001. Visualization of N-acylhomoserine lactone-mediated cell-cell communication between bacteria colonizing the tomato rhizosphere. Appl. Environ. Microbiol. 67:5761-5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teplitski, M., J. B. Robinson, and W. D. Bauer. 2000. Plants secrete substances that mimic bacterial N-acyl homoserine lactone signal activities and affect population density-dependent behaviors in associated bacteria. Mol. Plant-Microbe Interact. 13:637-648. [DOI] [PubMed] [Google Scholar]
- 33.Tesfaye, M., N. S. Dufault, M. R. Dornbusch, D. L. Allan, C. P. Vance, and D. A. Samac. 2003. Influence of enhanced malate dehydrogenase expression by alfalfa on diversity of rhizobacteria and soil nutrient availability. Soil Biol. Biochem. 35:1103-1113. [Google Scholar]
- 34.Van Larebeke, N., G. Engler, M. Holsters, S. Van den Elsacker, I. Zaenen, R. A. Schilperoort, and J. Schell. 1974. Large plasmid in Agrobacterium tumefaciens is essential for crown gall-inducing ability. Nature 252:169-170. [DOI] [PubMed] [Google Scholar]
- 35.Wang, Y. J., and J. R. Leadbetter. 2005. Rapid acyl-homoserine lactone quorum signal biodegradation in diverse soils. Appl. Environ. Microbiol. 71:1291-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang, C.-H., and D. E. Crowley. 2000. Rhizosphere microbial community structure in relation to root location and plant iron nutritional status. Appl. Environ. Microbiol. 66:345-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang, W. W., J. I. Han, and J. R. Leadbetter. 2006. Utilization of homoserine lactone as a sole source of carbon and energy by soil Arthrobacter and Burkholderia species. Arch. Microbiol. 185:47-54. [DOI] [PubMed] [Google Scholar]
- 38.Zhu, J., Y. R. Chai, Z. T. Zhong, S. P. Li, and S. C. Winans. 2003. Agrobacterium bioassay strain for ultrasensitive detection of N-acylhomoserine lactone-type quorum-sensing molecules: detection of autoinducers in Mesorhizobium huakuii. Appl. Environ. Microbiol. 69:6949-6953. [DOI] [PMC free article] [PubMed] [Google Scholar]