Abstract
A gene cluster responsible for the biosynthesis of anticancer agent FK228 has been identified, cloned, and partially characterized in Chromobacterium violaceum no. 968. First, a genome-scanning approach was applied to identify three distinctive C. violaceum no. 968 genomic DNA clones that code for portions of nonribosomal peptide synthetase and polyketide synthase. Next, a gene replacement system developed originally for Pseudomonas aeruginosa was adapted to inactivate the genomic DNA-associated candidate natural product biosynthetic genes in vivo with high efficiency. Inactivation of a nonribosomal peptide synthetase-encoding gene completely abolished FK228 production in mutant strains. Subsequently, the entire FK228 biosynthetic gene cluster was cloned and sequenced. This gene cluster is predicted to encompass a 36.4-kb DNA region that includes 14 genes. The products of nine biosynthetic genes are proposed to constitute an unusual hybrid nonribosomal peptide synthetase-polyketide synthase-nonribosomal peptide synthetase assembly line including accessory activities for the biosynthesis of FK228. In particular, a putative flavin adenine dinucleotide-dependent pyridine nucleotide-disulfide oxidoreductase is proposed to catalyze disulfide bond formation between two sulfhydryl groups of cysteine residues as the final step in FK228 biosynthesis. Acquisition of the FK228 biosynthetic gene cluster and acclimation of an efficient genetic system should enable genetic engineering of the FK228 biosynthetic pathway in C. violaceum no. 968 for the generation of structural analogs as anticancer drug candidates.
FK228 (C24H36N4O6S2; molecular weight, 540.2) (Fig. 1), also known as FR901228 or depsipeptide and registered as NSC 630176 or romidepsin, is a natural product discovered in the fermentation broth of Chromobacterium violaceum no. 968 in a screening program for agents that reverse the malignant phenotype of a Ha-ras oncogene-transformed NIH 3T3 cell line (51, 52). It exhibited outstanding anticancer activities against an array of tumor cell lines, including many members of a standard panel of 60 cell lines from the U.S. National Cancer Institute (18, 53). FK228 has entered extensive clinical trials and has shown promising properties as a new type of anticancer drug (5, 30, 35, 36, 41). A multinational pivotal trial of FK228 for the treatment of cutaneous T-cell lymphoma has been launched by Gloucester Pharmaceuticals, Inc., and the company plans to file for U.S. Food and Drug Administration approval in late 2007.
FIG. 1.
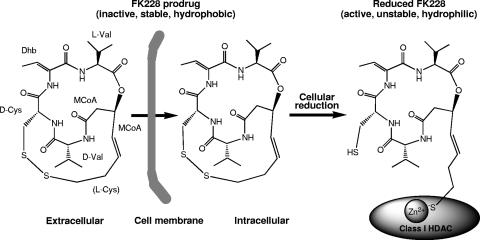
FK228 structure and mode of action (modified from reference 17 with permission of the publisher). Hydrophobic FK228 can diffuse across the cell membrane. Inside cells, FK228 is activated by cellular reduction, and a freed sulfhydryl group chelates Zn2+ inside the catalytic pocket of the preferred class I HDACs and therefore inhibits the enzyme activities.
Structurally, FK228 is a bicyclic depsipeptide that features a 16-membered macrolactone ring containing an ester linkage and a 17-membered ring containing the same ester linkage and a disulfide bond, the latter of which endows FK228 with an unprecedented molecular scaffold (Fig. 1). Its structure was determined by spectroscopic and X-ray crystallographic analyses (45) and was confirmed by total synthesis (27). A close examination of the FK228 structure identified building blocks of three amino acids (d-cysteine, d-valine, and l-valine), an amino acid derivative (2,3-dehydro-2-aminobutanoic acid; also called 2,3-dehydrothreonine), and a complex l-(S,E)-3-hydroxy-7-mercaptohept-4-enoic acid moiety that is likely built from one Cys and two C2 units derived from malonyl coenzyme A (MCoA). These observations suggest a hybrid nonribosomal peptide (NRP)-polyketide (PK)-NRP nature for FK228.
Mechanistically, FK228 was originally discovered as an anti-ras agent (51, 52); later, it was found to interfere with mitogen-induced signaling pathways (38, 42, 43), and more recently it has been identified as a potent histone deacetylase (HDAC) inhibitor (17, 33). Histone acetylation catalyzed by histone acetyltransferases is an important component of chromatin remodeling and gene expression regulation; histone hypoacetylation mediated by HDACs is often associated with the onset and progression of cancer (24, 57). HDAC inhibitors are a diverse group of molecules that can induce growth arrest, differentiation, apoptosis, and autophagocytic cell death of cancer cells (10, 24, 31, 57). Interestingly, FK228 has an intramolecular disulfide bond, which makes it structurally distinct from other known HDAC inhibitors, such as hydroxamic acids, apicidin, and trapoxin. This disulfide bond has been postulated to mediate a novel mechanism of cytotoxic action of FK228 (Fig. 1). Furumai and coworkers showed that FK228 serves as a stable prodrug and is activated by intracellular reduction of the disulfide bond after uptake into cells or organisms. The freed sulfhydryl group on the longer aliphatic tail of reduced FK228 fits inside the catalytic pocket of preferred class I HDACs, chelating Zn2+, and thus inhibits the enzyme activities (17). Xiao and coworkers also independently detected more active metabolites in rat plasma and human plasma following their incubation with FK228 in the presence of glutathione (56). The 50% inhibitory concentration of FK228 was found to be nanomolar for inducing apoptosis in cells from patients with chronic lymphocytic leukemia (6). Research on FK228 has been expanding rapidly in recent years.
HDAC inhibitors are prime agents for the development of novel anticancer drugs (1, 10, 18, 24, 31, 35, 57). One HDAC inhibitor, Zolinza (vorinostat or suberoylanilide hydroxamic acid), was approved by the U.S. Food and Drug Administration in October 2006, and at least nine other HDAC inhibitors, including FK228, are in various stages of clinical trials as monotherapies or in combinations with other agents (18). Due to its outstanding anticancer activities and novel structural characteristics, as well as certain levels of undesirable cardiac toxicity, FK228 may serve as an excellent molecular scaffold for the generation of structural analogs, from which compounds with improved anticancer properties may be identified. However, chemical synthesis of FK228 has been difficult (27), and derivatization of FK228 by chemical synthesis has not been reported.
We set out to take an alternative approach to making FK228 analogs by means of pathway engineering, combinatorial biosynthesis, or chemoenzymatic synthesis. As the first essential step towards this goal, we identified, cloned, and partially characterized a biosynthetic gene cluster (designated dep for depsipeptide) responsible for FK228 biosynthesis. Here we report the identification of candidate biosynthetic genes by a genome-scanning approach, the adaptation of a Pseudomonas aeruginosa gene replacement system to create targeted gene- inactivated mutant strains, and the subsequent cloning and characterization of an unusual hybrid nonribosomal peptide synthetase (NRPS)-polyketide synthase (PKS)-NRPS pathway for FK228 biosynthesis in C. violaceum no. 968. Acquisition of the dep gene cluster and acclimation of an efficient genetic system should provide a solid foundation for the generation of FK228 analogs by engineered biosynthesis strategies (for recent comprehensive reviews, see references 20 and 54).
MATERIALS AND METHODS
Bacterial strains, culture conditions, and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. The FK228-producing strain, C. violaceum no. 968, was cultured in nutrient broth (1% Difco nutrient broth and 1% glucose) at 30°C for genomic DNA preparation and in fermentation medium (nutrient broth supplemented with 5% Diaion HP-20 resin [Supelco, Pennsylvania]) at 30°C for FK228 production. The vectors pEX18Tc and pPS858, originally developed for P. aeruginosa genetics (23), were adopted and applied successfully in C. violaceum.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain(s) or plasmid(s) | Description | Source or reference(s) |
|---|---|---|
| Chromobacterium violaceum strains | ||
| No. 968 (= FERM BP-1968) | Wild type, FK228 producing, Apr Thiora | IPODb |
| Cv56a/b/c | Serial mutants with an internal part of pP3-B6 DNA replaced by the FRT cassette (Gmr GFP+) from pPS858, FK228 producing | This study |
| Cv57a/b/c | Serial mutants with an internal part of pP4-B4 DNA (on depD gene) replaced by the FRT cassette (Gmr GFP+) from pPS858, non-FK228 producing | This study |
| Cv58a/b/c | Serial mutants with an internal part of pP4-G7 DNA (on depD gene) replaced by the FRT cassette (Gmr GFP+) from pPS858, non-FK228 producing | This study |
| Escherichia coli strains | ||
| DH5α | General cloning host | 40 |
| XL1-Blue MR | Host strain for cosmid library construction | Stratagene |
| S17-1 | Host strain for interspecies conjugation | 46 |
| ET12567(pUZ8002) | Alternative host strain (methylation deficient) for conjugation | 25, 28 |
| MT670(pRK600) | Alternative host strain for conjugation | 14 |
| Plasmids | ||
| pGEM-3Zf | Apr, general cloning vector | Promega |
| pGEM-T Easy | Apr, general cloning vector | Promega |
| pP3-A6 | 2.8-kb random genomic DNA of C. violaceum cloned into pGEM-T Easy, sequenced | This study |
| pP4-B4 | 3.6-kb random genomic DNA of C. violaceum cloned into pGEM-T Easy, sequenced | This study |
| pP4-G7 | 2.6-kb random genomic DNA of C. violaceum cloned into pGEM-T Easy, sequenced | This study |
| pPS858 | Apr Gmr GFP+, source of the FRT cassette | 23 |
| pYC03-56a | Apr Gmr GFP+, replacement of an internal 1.8-kp EcoRV fragment on pP3-A6 by a 1.8-kb SmaI fragment (containing the FRT cassette) from pPS858 | This study |
| pYC03-57a | Apr Gmr GFP+, replacement of an internal 1.1-kb BglII/NruI fragment on pP4-B4 (blunt ended) by a 1.8-kb SmaI fragment (containing the FRT cassette) from pPS858 | This study |
| pYC03-58a | Apr Gmr GFP+, replacement of two adjacent internal NruI fragments (456 and 489 bp) on pP4-G7 (blunt ended) by a 1.8-kb SmaI fragment (containing the FRT cassette) from pPS858 | This study |
| pEX18Tc | TcroriT+sacB+, gene replacement vector, conjugative | 23 |
| pYC03-56b | Conjugative construct with a 2.8-kb NotI fragment (blunt ended, containing the FRT cassette and flanking DNAs) from pYC03-56a ligated into the SmaI site of pEX18Tc | This study |
| pYC03-57b | Conjugative construct with a 4.3-kb NotI fragment (blunt ended, containing the FRT cassette and flanking DNAs) from pYC03-57a ligated into the SmaI site of pEX18Tc | This study |
| pYC03-58b | Conjugative construct with a 3.7-kb PstI/SphI fragment (blunt ended, containing the FRT cassette and flanking DNAs) from pYC03-58a ligated into the SmaI site of pEX18Tc | This study |
| SuperCos 1 | Apr Kanr, cosmid vector | Stratagene |
| Cosmid 18 | Cosmid clone containing the FK228 biosynthetic gene cluster (dep) and flanking DNAs, shotgun sequenced | This study |
| Cosmid 2 | Cosmid clone containing a partial dep gene cluster | This study |
| pCos2S1 to pCos2S5 | BamHI fragments (4.0, 0.8, 6.2, 4.5, and 7.7 kb, respectively) of cosmid 2 inserted into the same site of pGEM-3Zf, sequenced by the primer walking method | This study |
Thior, thiostrepton resistance.
IPOD, International Patent Organism Depositary, Tsukuba, Japan.
DNA manipulations, genome library construction, and DNA sequencing.
General DNA manipulations, including plasmid preparation, restriction enzyme digestion, agarose gel electrophoresis, subcloning, and bacterial transformation, were done according to standard protocols (40) or the manufacturer's instructions (New England BioLabs; QIAGEN). Genomic DNA of a C. violaceum wild-type or mutant strain was prepared from an overnight culture with a Genomic-tip 500/G kit (QIAGEN) or with an UltraClean microbial DNA isolation kit (MO BIO Labs). For construction of a genome sampling library (58), high-molecular-weight C. violaceum genomic DNA was mechanically sheared with a nebulization device (Invitrogen). DNA molecules that were 2 to 4 kb long were recovered from an agarose gel and end repaired with T4 DNA polymerase and Klenow enzyme in the presence of deoxynucleoside triphosphates (1 mM each). The ends of resultant DNA molecules were adenylated using Taq DNA polymerase with dATP, ligated to the pGEM-T Easy vector, and transformed into Escherichia coli DH5α cells. Four 96-well plates of clones were subjected to template DNA preparation by PCR amplification and purification with a PerfectPrep PCR Cleanup 96 kit (Eppendorf), and end sequencing with BigDye chemistry and SP6 as the primer was performed with an ABI 3730 automated DNA sequencer (Applied Biosystems) at the University of Wisconsin-Madison Biotechnology Center. DNA oligonucleotides were synthesized by Operon Biotechnologies, Inc., and DNA sequencing by primer walking was performed by standard procedures (40). A cosmid library was constructed in the SuperCos 1 vector using previously described procedures (8). Southern blotting, labeling of DNA as a probe, hybridization, and detection were performed according to the manufacturer's protocols (Roche). Shotgun sequencing of cosmid 18 and contig assembly were performed by a service company (ACGT Inc.). Local sequence analysis was performed with the Lasergene program package (DNASTAR, Inc.) and by a homology search against the GenBank database using the BLAST algorithms (2). The domain organization of biosynthetic enzymes was analyzed as described by Ansari et al. (3), with manual intervention.
General strategy for the construction of targeted gene-inactivated mutants of C. violaceum no. 968.
To mutate a candidate gene by a gene replacement strategy, an internal part of the DNA of a genomic DNA clone (ampicillin resistant) was replaced by a 1.8-kb FRT cassette (gentamicin resistant) from pPS858 to make an intermediate construct (Apr Gmr). The FRT cassette, along with two flanking genomic DNAs for homologous DNA recombination, was excised and subcloned into pEX18Tc to make a final conjugation construct (Gmr and tetracycline resistant). The conjugation construct was introduced into E. coli S17-1 cells and subsequently transferred into C. violaceum cells by conjugation as follows. Two bacterial strains were grown in LB media supplemented with appropriate antibiotics (10 μg/ml gentamicin and 10 μg/ml tetracycline for E. coli S17-1 [a conjugation construct] and 200 μg/ml ampicillin for C. violaceum, which is naturally resistant to ampicillin) at 37 or 30°C with shaking until the late mid-log phase (6 to 8 h). Cells from 1 ml of each culture were collected by centrifugation at 4,000 × g for 15 min at 4°C, and the cell pellets were washed once with 1 ml LB medium. Cells were collected again by centrifugation and resuspended in 100 μl LB medium. Cell suspensions of two bacterial strains were pooled and spread evenly on a wet 0.45-μm nitrocellulose membrane (Whatman) on LB agar. After the plate had been incubated at 30°C for 12 to 16 h, the membrane seeded with bacteria was used to print several LB agar plates containing 200 μg/ml ampicillin, 50 μg/ml gentamicin, and 5% sucrose to select for exconjugants.
FK228 production and detection by LC-MS.
Wild-type and mutant strains of C. violaceum were grown in 25 ml of fermentation medium at 30°C for 3 days with constant agitation (200 rpm). Cells and resins were then collected together by centrifugation at 4,000 × g for 20 min at the ambient temperature and lyophilized to dryness. A crude FK228 preparation was obtained by eluting the dried cell debris and resins with 10 ml ethyl acetate. Twenty microliters of this preparation was injected into an Agilent 1100 series LC/MSD Trap mass spectrometer (MS) (Agilent) for detection of the positive ion signals of FK228. The liquid chromatography (LC) program included a linear gradient from buffer A (20% methanol with 0.1% formic acid) to buffer B (80% methanol with 0.1% formic acid) in 15 min and constant elution in buffer B for 5 min, followed by a linear return to buffer A in 5 min. Samples were fractionated by using a Zorbax Eclipse XDB-C18 column (2.1 by 110 mm; Agilent) with a flow rate of 0.25 ml/min.
Nucleotide sequence accession numbers.
The nucleotide sequences of the inserts in pP3-B6, pP4-B4, pP4-G7, and cosmid 18 have been deposited in the GenBank database under accession numbers EF015612, EF015613, EF015614, and EF210776, respectively.
RESULTS AND DISCUSSION
Identification of candidate natural product biosynthetic genes in C. violaceum no. 968.
The hybrid NRP-PK-NRP nature of FK228 (Fig. 1) suggests that FK228 is likely biosynthesized by a hybrid NRPS-PKS-NRPS assembly line, probably with an additional enzymatic activity for the formation of an intramolecular disulfide bond. The biosynthesis of NRPs, PKs, and hybrid NRP-PK or PK-NRP natural products via successive condensation of simple building blocks, such as amino acids, amino acid derivatives, and short carboxylic acids, catalyzed by NRPSs, PKSs, and hybrid NRPS-PKS or PKS-NRPS systems, respectively, has been well studied (for recent comprehensive reviews, see references 15, 16, and 22). For ester bond formation in depsipeptide natural products, the involvement of a discrete d-hydroxyisovalerate dehydrogenase in enniatin biosynthesis by Fusarium sambucinum (26) or a novel NRPS module containing an adenylation (A) domain to activate an α-keto acid and an embedded α-ketoreductase (KR) to reduce the tethered substrate to an α-hydroxyacyl intermediate (and presumably a downstream condensation [C] domain acting as a chiral ester synthase rather than an amide synthase) in cereulide and valinomycin biosynthesis in actinomycetes (29) has been experimentally established. However, whether intramolecular disulfide bond formation in natural products (such as FK228) is an enzymatic reaction or a spontaneous chemical oxidation is unknown. Therefore, our search for candidate FK228 biosynthetic genes focused initially on the genes encoding an obvious NRPS, PKS, or, in particular, hybrid NRPS-PKS or PKS-NRPS system.
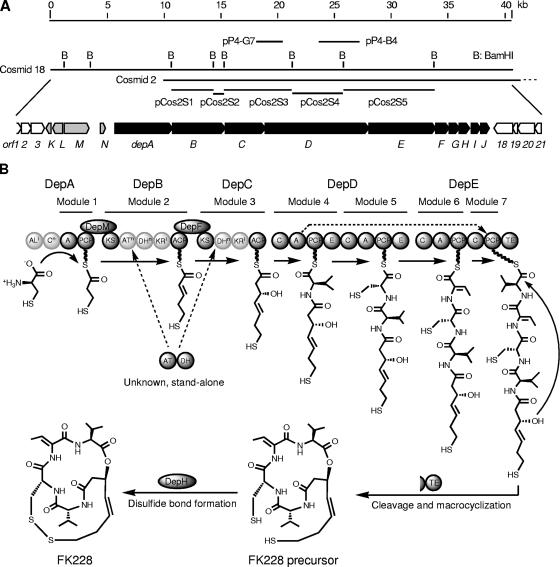
Among 360 valid sequence tags obtained from sequencing of the genome sampling library of C. violaceum (see Materials and Methods), three distinctive sequence tags, P3-A6-SP6, P4-B4-SP6, and P4-G7-SP6, were identified to be parts of genes encoding PKS, NRPS, and a hybrid PKS-NRPS system, respectively (Table 2). Genes that contain these three tags were considered candidate natural product biosynthetic genes, possibly involved in FK228 biosynthesis. Further primer walking sequencing revealed the complete sequences of the corresponding inserts in pP3-A6, pP4-B4, and pP4-G7. The insert in pP3-A6 contains a 2,826-bp DNA that includes a partial PKS gene (not named), and its translated amino acid sequence has homology to the β-ketoacyl synthase and acyltransferase (AT) domains of type I PKSs (44). Three signature motifs (QTRTAQ, GHSYG, and AAFH) were identified in the AT domain, and these motifs are similar to the motifs of ATs using MCoA as a substrate (39). The insert in pP4-B4 contains a 3,612-bp DNA that includes a partial gene (designated depD) (Table 3 and Fig. 2A), and its translated amino acid sequence has homology to the A, peptidyl carrier protein (PCP), and epimerase domains of type A NRPSs (32). The “NRPS substrate specificity code” of the A domain was identified as DLFEMSLIWK, and this A domain is predicted to activate l-Cys, according to Ansari et al. (3), Challis et al. (7), and Stachelhaus et al. (47). The insert in pP4-G7 contains a 2,599-bp DNA that includes two partial genes (designated depC and depD) (Table 3 and Fig. 2A), and their translated amino acid sequences have homology to the KR and acyl carrier protein (ACP) domains of PKSs, followed by the C and A domains of NRPSs, indicating that there is a hybrid PKS-NRPS system (11). The A domain is incomplete; therefore, the “NRPS substrate specificity codes” cannot be extracted for prediction of substrate specificity. Inserts in pP4-B4 and pP4-G7 cover different parts of the same depD gene.
TABLE 2.
Properties of three sequence tags and their associated candidate (partial) genes
| Sequence tag | Recombinant plasmid | Insert size (bp) | Associated gene(s)a | Protein homolog(s) (accession no.) | Domain organizationb | Protein classification | Signature motif(s) or substrate specificity | Predicted substrate specificity | Necessary for FK228 biosynthesis |
|---|---|---|---|---|---|---|---|---|---|
| P3-A6-SP6 | pP3-A6 | 2,826 | NNc | JamL (AAS98783) | KS-ATi | Type I PKS | QTRTAQ, GHSYG, and AAFH in AT domain | MCoA | No |
| P4-B4-SP6 | pP4-B4 | 3,612 | depD | BmyB (CAE11249) | Ci-A-PCP-E | Type A NRPS | DLFEMSLIWK in A domain | l-Cys | Yes |
| P4-G7-SP6 | pP4-G7 | 2,599 | depC, depD | AmphI (AAK73501), NosC (AAF17280) | KRi-ACP, C-Ai | PKS, NRPS | NAd | NA | Yes |
See Fig. 2.
A superscript i indicates incomplete. KS, β-ketoacyl synthase; E, epimerase.
NN, not named.
NA, not available.
TABLE 3.
Deduced functions of open reading frames and genes in the dep gene cluster and flanking regions
| Open reading frame or gene | Protein size (amino acids) | Protein homolog | Accession no. | % Identity/ % similarity | Origin | Proposed functiona |
|---|---|---|---|---|---|---|
| orf1b | 150c | CV_3386 | AAQ61050 | 87/93 | C. violaceum ATCC 12472 | 16S rRNA pseudouridine synthase |
| orf2 | 163 | CV_3385 | AAQ61049 | 66/76 | C. violaceum ATCC 12472 | MutT/nudix family phosphohydrolase |
| orf3 | 190 | CV_3384 | AAQ61048 | 88/94 | C. violaceum ATCC 12472 | Transcription elongation factor GreB |
| depK | 85 | CCO_1235 | EAL57087 | 36/52 | Campylobacter coli RM2228 | Conserved hypothetical protein, function unknown |
| depL | 155 | CV_3383 | AAQ61047 | 68/78 | C. violaceum ATCC 12472 | Helix-turn-helix transcriptional regulator, MarR family |
| depM | 389 | PFL_4362 | AAY93617 | 59/73 | Pseudomonas fluorescens Pf-5 | Aminotransferase, class I and II family protein |
| depN | 65 | PCPn | ||||
| depA | 1,697 | SafB | AAC44128 | 31/45 | Myxococcus xanthus strain Mx ×48 | NRPS: ALi-Cn-ACys-PCP |
| depB | 1,553 | CurG | AAT70102 | 45/61 | Lyngbya majuscula | PKS: KS-ATn-DHn-KRi-ACP |
| depC | 1,183 | CrpB | ABM21570 | 44/64 | Nostoc sp. strain ATCC 53789 | PKS: KS-DHn-KRi-ACP |
| depD | 3,057 | PvdI | AAX16361 | 36/51 | Pseudomonas aeruginosa | NRPS: C-AVal-PCP-E-C-ACys-PCP-E |
| depE | 1,892 | McyB | BAA83993 | 35/52 | Microcystis aeruginosa | NRPS: C-ADhb-PCP-C-PCP-TE |
| depF | 390 | PP_2437 | AAN68049 | 38/56 | Pseudomonas putida KT2440 | FadE2-like acyl coenzyme A dehydrogenase |
| depG | 321 | PSPTO_2724 | AAO56225 | 32/53 | Pseudomonas syringae pv. tomato DC3000 | Phosphotransferase |
| depH | 319 | PA4170 | AAG07557 | 56/70 | Pseudomonas aeruginosa PAO1 | Flavin adenine dinucleotide-dependent pyridine nucleotide-disulfide oxidoreductase |
| depI | 304 | RRSL_03772 | EAP73858 | Ralstonia solanacearum UW551 | Putative esterase/lipase | |
| depJ | 254 | LnmN | AAN85527 | 43/58 | Streptomyces atroolivaceus S-140 | Type II thioesterase |
| orf18 | 312 | CV_3378 | AAQ60142 | 87/93 | C. violaceum ATCC 12472 | Hydrogen peroxide-inducible gene activator OxyR |
| orf19 | 85 | CV_3377 | AAQ60141 | 92/98 | C. violaceum ATCC 12472 | Cell division topological specificity factor MinE |
| orf20 | 270 | CV_3376 | AAQ61040 | 92/98 | C. violaceum ATCC 12472 | Septum site-determining protein MinD |
| orf21b | 107c | CV_3375 | AAQ61039 | 93/97 | C. violaceum ATCC 12472 | Septum formation inhibitor MinC |
Subscripts indicate the substrate specificities of enzymes. Superscripts indicate inactive (i) or nonfunctional (n). Dhb, 2,3-dehydro-2-aminobutanoic acid.
Incomplete.
Truncated.
FIG. 2.
FK228 biosynthetic (dep) gene cluster and a proposed model for FK228 biosynthesis. (A) Physical map of clones and genes. pP4-G7 and pP4-B4 are positively identified genome sampling clones, and each contains part of the depD gene. The insert in pP4-G7 was labeled as a DNA probe to obtain cosmids 18 and 2. Cosmid 18 was shotgun sequenced. pCos2S1 to pCos2S5 are subclones of cosmid 2 and were sequenced by a primer walking method. Predicted genes in the dep gene cluster are designated depA to depN, and open reading frames outside the dep gene cluster are designated orf1 to orf3 and orf18 to orf21. Genes indicated by solid bars (depA to depJ) were predicted to be in the dep gene cluster with confidence; genes indicated by gray bars (depK to depN) were predicted to be in the dep gene cluster with less confidence. (B) Proposed model of FK288 biosynthesis by a hybrid NRPS-PKS-NRPS assembly line, including accessory activities of discrete proteins. PKS and NRPS domains are described in the text. A superscript “i” indicates that a domain is inactive; a superscript “n” indicates that a domain is nonfunctional. Inactive and nonfunctional domains are light gray. AL, acyl coenzyme A ligase; KS, β-ketoacyl synthase; E, epimerase.
Adaptation of a P. aeruginosa genetic system in C. violaceum no. 968 to create targeted gene-inactivated mutant strains.
To test whether the identified candidate genes are necessary for FK228 biosynthesis, we inactivated the individual genes (except depC, which has only a very short segment on the insert of pP4-G7) in C. violaceum no. 968. C. violaceum strains belong to the gram-negative β-proteobacteria. Although isolates of C. violaceum produce many products with biotechnological and pharmaceutical utility (13) and the genome of a representative strain, C. violaceum ATCC 12472, has been sequenced (4), a genetic system for targeted gene inactivation in C. violaceum has not been reported prior to this study. Here, a broad-host-range Flp-FRT recombination system originally developed for P. aeruginosa genetics (23) was adopted and successfully applied to C. violaceum no. 968.
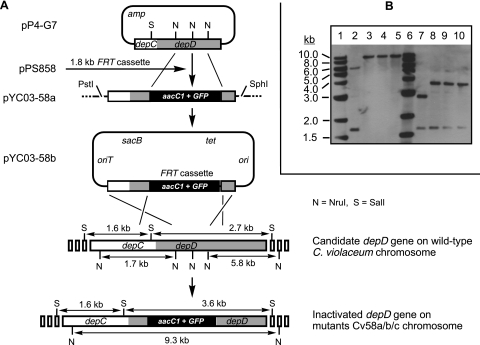
To inactivate the P4-G7-SP6-associated depD gene (depD was chosen as an example for full description here because it encodes part of a hybrid PKS-NRPS system that is of prime interest) (Fig. 3A), two internal NruI fragments (0.46 and 0.49 kb) of the pP4-G7 insert were removed and replaced by a 1.8-kb SmaI fragment of the FRT cassette from pPS858 to make an intermediate construct, pYC03-58a. A 3.7-kb PstI/SphI fragment containing the FRT cassette with flanking DNAs from pYC03-58a was recovered, end repaired, and inserted into the SmaI site of pEX18Tc to make a final construct, pYC03-58b. Plasmid pYC03-58b was introduced into E. coli S17-1 cells and subsequently transferred into C. violaceum cells by conjugation. In the designed selection medium (see Materials and Methods), ampicillin at a concentration of 200 μg/ml suppresses the growth of E. coli S17-1 cells, gentamicin at a concentration of 50 μg/ml selects for the presence of the FRT cassette, and sucrose at a concentration of 5% counterselects for the loss of a functional sacB+ gene on the vector. Collectively, this experiment strongly selected for double-crossover mutants of C. violaceum with part of the targeted depD replaced by the FRT cassette. Hundreds of exconjugants appeared on a typical selection plate after incubation at 30°C for 2 days. The efficiency of conjugation and gene recombination was estimated to be in the range from 10−6 to 10−5 per cell.
FIG. 3.
Creation of depD-inactivated mutant strains by targeted gene replacement. (A) Construction of gene replacement vector pYC03-58b and homologous recombination via double crossover between the vector and the bacterial chromosome to generate a mutant genotype. (B) Southern analysis of the genotypes of wild-type and depD-inactivated mutant strains of C. violaceum, using the labeled 2.6-kb insert DNA of pP4-G7 as a probe. Genomic DNA was digested with NruI (lanes 2 to 5) or SalI (lanes 7 to 10). Lanes 1 and 6, 1-kb DNA ladders (New England Biolabs), hybridized with their digoxigenin-labeled probes; lanes 2 and 7, DNA from the wild-type strain; lanes 3 to 5 and 8 to 10, DNA from independent mutant strains Cv58a, Cv58b, and Cv58c, respectively. A change in the pattern of positively hybridized DNA bands indicates targeted gene inactivation.
Southern analysis (Fig. 3B) clearly showed that when genomic DNA of C. violaceum strains was digested with NruI (lanes 2 to 5), the wild-type strain showed two bands (1.7 and 5.8 kb; 0.46- and 0.49-kb DNA fragments ran off the gel during electrophoresis) that hybridized to the probe made from the 2.6-kb insert of pP4-G7. Considering that there are three internal NruI sites in the 2.6-kb insert of pP4-G7 and that one central NruI site was removed and two other sites were destroyed during the construction of pYC03-58a, insertion of the 1.8-kb FRT cassette via double-crossover DNA recombination was expected to result in a 9.3-kb (1.7 kb + 5.8 kb + 1.8 kb) hybridized band in the mutant genotype. Three of eight random exconjugants were proven in this experiment to have the correct genotype, and they were designated independent depD-inactivated mutant strains Cv58a, Cv58b, and Cv58c (collectively designated the Cv58a/b/c mutants). Similarly, when genomic DNA was digested with SalI (lanes 7 to 10), the size of a 2.7-kb hybridized band in the wild-type strain increased to 3.6 kb (2.7 kb − 0.49 kb − 0.46 kb + 1.8 kb) in the mutant strains, as expected. The 1.6-kb band in the wild-type strain remained unchanged in mutant strains because the DNA fragment is located outside the gene replacement region.
The same strategy was used to inactivate the P3-A6-SP6-associated gene (not named) and the P4-B4-SP6-associated depD gene (3′ part), to create mutant strains Cv56a/b/c and Cv57a/b/c, respectively, and their genotypes were verified by Southern analyses as well (data not shown).
During the course of method development, two other conjugation systems were also tested. One method used the methylation-deficient strain E. coli ET12567(pUZ8002) (25, 28) and the other used E. coli MT607(pRK600) (14) as donor strains to mobilize a conjugation construct (such as pYC03-58b) into C. violaceum cells. Both systems generated exconjugants, but they were at least 10-fold less efficient than the E. coli S17-1 strain-mediated conjugation between E. coli and C. violaceum cells (data not shown). In addition, it was noticed that, since the FRT cassette contains a functional GFP gene that encodes the green fluorescent protein (GFP), E. coli and C. violaceum colonies or cultures with the FRT cassette present on a replicable plasmid or integrated into the chromosome were distinguishable from the wild-type bacteria by a greenish color (data not shown). Therefore, bacterial exconjugants carrying the FRT cassette could be identified by direct observation or by a simple GFP assay. Furthermore, the marker genes (aacC1 and GFP in the FRT cassette) integrated into the mutant chromosome could be excised precisely by a FLP recombinase encoded by the pFLP2 plasmid in the Flp-FRT system to create unmarked mutants (23). Unmarked mutants could be mutated at different loci sequentially to create multiple gene deletions or gene replacements. This feature could be very useful for future pathway engineering and combinatorial biosynthesis studies.
Confirmation of the necessity of the depD gene for FK228 biosynthesis in C. violaceum no. 968.
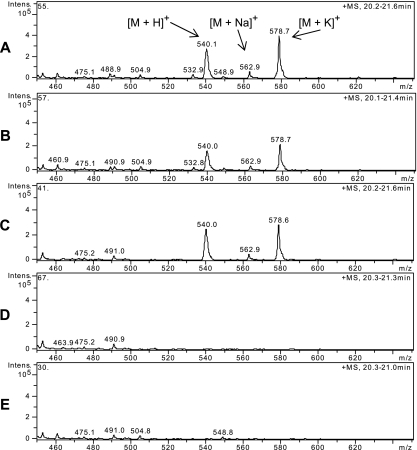
The FK228 productivity of the wild-type and mutant (Cv56a/b/c, Cv57a/b/c, and Cv58a/b/c) strains of C. violaceum was examined by fermentation and LC-MS analysis. FK228 does not produce a characteristic UV spectrum because it lacks a chromophore, but its positive ion signals are strong and appeared near 20.8 min under the chromatographic conditions tested (Fig. 4). The calculated positive ion signal of FK228 is [M + H]+ at m/z 541.2, and its ion adducts are [M + Na]+ at m/z 563.2 and [M + K]+ at m/z 580.2 for an authentic FK228 sample, but the actual observed signals were m/z 540.1, m/z 562.9, and m/z 578.7, respectively. The small mass differences between the calculated and observed values were likely due to inadequate instrument calibration. The samples from wild-type and Cv56a/b/c mutant strains yielded almost the same signals as the authentic FK228. However, no FK228 ion signal was detected in samples from Cv57a/b/c or Cv58a/b/c mutant strains. These results suggest that inactivation of depD, but not inactivation of the P3-A6-SP6-associated gene, completely abolished FK228 production, which confirmed the necessity of depD for FK228 biosynthesis in C. violaceum no. 968.
FIG. 4.
Detection of FK228 positive ion signals by LC-MS. Samples were obtained from an authentic FK228 standard (A), wild-type C. violaceum (B), Cv56a/b/c mutants with the pP3-A6-associated gene inactivated (C), Cv57a/b/c mutants with the pP4-B4-associated depD gene (3′ part) inactivated (D), and Cv58a/b/c mutants with the pP4-G7-associated depD gene inactivated (E). For each mutation three mutants (a, b, and c) yielded identical results; therefore, only one data profile for each mutation is presented.
Cloning, sequencing, and in silico analysis of the FK228 biosynthetic (dep) gene cluster.
A series of overlapping cosmid clones were obtained by colony hybridization with digoxigenin-labeled insert DNA of pP4-G7 as a probe. Cosmid end sequencing indicated that, among these clones, cosmid 18 appears to contain the entire dep gene cluster; therefore, the nucleotide sequence of cosmid 18 was determined by the shotgun method, which revealed a 40,434-bp contig (Fig. 2A). Due to concern about the irregularity of the deduced protein domain organizations (see below for details), cosmid 2, which covers most but not all of the dep gene cluster, was also sequenced by a subcloning and primer walking strategy (Fig. 2A). A cosmid clone carrying a partial dep gene cluster was chosen for sequencing verification purposes because a partial gene cluster cloned from the gram-negative bacterium C. violaceum into another gram-negative bacterium, E. coli, should not result in acquired toxicity, minimizing possible gene deletion or recombination. The sequences of the overlapped region in cosmid 18 and cosmid 2 agreed perfectly, confirming the shotgun sequence quality and reliability.
The assembled contig contains 21 apparent genes or open reading frames (two partial sequences at the ends) (Table 3 and Fig. 2A). Bioinformatic analyses further predicted that the dep gene cluster consists of 14 genes, designated depA through depN, flanked by several housekeeping genes (orf1 through orf3 and orf18 through orf21), although the exact boundaries of the dep gene cluster have not been experimentally verified yet. The flanking housekeeping genes have homology with genes in a single region of the C. violaceum ATCC 12472 genome (CV_3375 through CV_3386) (4). Interestingly, five ATCC 12472 genes (CV_3379 through CV_3383) are seemingly replaced by the dep gene cluster, suggesting that a lateral gene transfer event occurred (34). Further evidence that supports this notion comes from a G+C content analysis. The flanking housekeeping genes have an average G+C content of 62.9%, while the dep gene cluster has a G+C content of 69.0%. C. violaceum no. 968 could have acquired the dep gene cluster from an organism with a higher-G+C genome at the expense of a five-gene deletion of its own.
Cotranscription is common among related genes in bacteria. In the dep gene cluster and flanking regions, orf1 through orf3, orf18 through orf21, depABCDEFGH, and depIJ are very likely organized as operons, because genes within each putative operon have overlapping stop and start codons. In contrast, the depK, depL, depM, and depN genes are separated by variable lengths of intergenic DNA. This analysis facilitated the prediction that depJ is the downstream boundary of the dep gene cluster because orf18 through orf21 are housekeeping genes in a single putative operon.
Model for FK228 biosynthesis by a hybrid NRPS-PKS-NRPS assembly line.
Many natural products are often biosynthesized by modular NPRSs, PKSs, or hybrid NRPS-PKS or PKS-NRPS assembly lines in a colinearity model in which the substrate specificity and the number and order of modules dictate the chemical makeup of the products (for recent comprehensive reviews, see references 15, 16, and 22); meanwhile, variations from the canonical model, including colinearity violation, iterative polymerization (iteration), missing or misplaced domains, module skipping or stuttering, stand-alone domains, alternative chain termination, the presence of unique domains, or trans-acting enzymes, have all been documented in individual biosynthetic pathways (for recent comprehensive reviews, see references 16, 44, and 55). Based on extensive bioinformatic analyses of the domain and module organization of biosynthetic enzymes encoded by the dep gene cluster, a model for FK228 biosynthesis by a hybrid NRPS-PKS-NRPS assembly line is proposed (Fig. 2B), and this model should serve as a general guideline for future studies and experimental validation. The proposed pathway includes nine proteins (DepA, DepB, DepC, DepD, DepE, DepF, DepH, and DepM, as well as DepJ [not drawn in the model]) that constitute five NRPS modules, two PKS modules, and accessory activities; each module is responsible for the incorporation of one contributing building block.
Based on the model, FK228 biosynthesis starts with the activation of a Cys by the A domain in module 1 to form a cysteinyl-S-PCP intermediate. DepM (an aminotransferase) is proposed to act in trans to remove an amino group from the intermediate to form 4-mercaptobutanyl-S-PCP. Aminotransferase domains have been found to be an integral part of the PKSs in the biosynthesis of mycosubtilin (12) and iturin A (50), adding an amino group; no such domain, however, has been found to remove an amino group in a reverse reaction. The C domain in module 1 appears to be nonfunctional because of a lack of a critical catalytic motif, HHXXXDG; a nonfunctional C domain disconnects the possible chemical interaction between the upstream acyl coenzyme A ligase domain and the downstream A domain. Next, PKS modules 2 and 3 sequentially extend the growing chain with two C2 units from MCoA. However, module 2 contains only a remnant nonfunctional AT domain that lacks essential motifs (e.g., GHSXG and A[FS]HS), and module 3 lacks an AT domain. The dehydratase (DH) domains in modules 2 and 3 also appear to be nonfunctional because of a lack of a conserved active site motif, HXXXGXXXXP. An unknown stand-alone AT-DH didomain protein (or, alternatively, discrete AT and DH proteins) is proposed to act in trans to compensate the modules in the PKS mode of biosynthesis. Furthermore, since no gene encoding a stand-alone AT-DH didomain is present in the dep gene cluster, it must exist in another region of the genome. Stand-alone AT domains or AT-X didomains (where X is any domain) have been identified in recent years in the biosynthetic pathways of natural products, including leinamycin (9), pederin (37), and many other compounds; a recent molecular cellular study of the bacillaene biosynthetic enzyme complex revealed an amazing interaction between a stand-alone AT-X didomain and the rest of a mega-PKS complex in Bacillus subtilis (48). In addition, DepF, an FadE2-like acyl coenzyme A dehydrogenase, has been proposed to act in trans on module 2 to generate a double bond on the β-hydroxyl-5-mercaptopentanoyl-S-ACP intermediate to form the β-5-mercaptopent-2-enoyl-S-ACP intermediate. If this is true, DepF would be functionally equivalent to an enoylreductase. KR domains in modules 2 and 3, although intact, are proposed to be inactive, probably due to a lack of proper interaction with the putative trans-acting AT-DH didomain. Modules 4, 5, and 6 extend the growing intermediate chain with activated d-Val, d-Cys, and 2,3-dehydro-2-aminobutanoic acid (2,3-dehydrothreonine) sequentially in the canonical model of the NRPS mode of biosynthesis. Module 7 is expected to incorporate a Val, but an A domain is completely missing in this module. It is proposed that the A domain in module 4, which specifies Val, acts in trans to aminoacylate the PCP domain in module 7. This phenomenon has been observed in the biosynthetic pathways of viomycin (49), yersiniabactin (19), and other compounds. Finally, terminal thioesterase (TE) on DepE should catalyze the formation of an ester linkage between a hydroxyl group originating from MCoA and a β-keto group from Val to form a 16-membered macrolactone ring, and a flavin adenine dinucleotide-dependent pyridine nucleotide-disulfide oxidoreductase encoded by depH is proposed to bring the free sulfhydryl groups from two Cys residues close to form an intramolecular disulfide bond. Disulfide bond formation hallmarks the formation of a 17-membered ring structure and brings the FK228 biosynthesis to completion. DepJ, a discrete type II TE, is not drawn into the model, and type II TEs are generally believed to have a proofreading function during chain elongation to ensure smooth biosynthesis by selectively removing misprimed thioesters or shunt intermediates (21). It is necessary to point out that, in the model described above, several unique features that include the trans-acting DepM, DepF, an unknown stand-alone AT-DH didomain, and a trans-acting A domain are highly speculative and require experimental validation.
Other genes in the dep gene cluster.
There are two apparent resistance genes in the dep gene cluster. An esterase/lipase, encoded by depI, is proposed to hydrolyze the ester linkage and/or the disulfide bond in FK228 to prevent the accumulation of an excess concentration of FK228 in cells where FK228 may become toxic. A phosphotransferase, encoded by depG, is proposed to further mask and quench the hydrolyzed FK228 by adding a phosphate group to the freed hydroxyl and/or sulfhydroxyl group(s). Surprisingly, no gene encoding exportation machinery is found in the dep gene cluster. The depL gene encodes a typical transcriptional regulator that contains a helix-turn-helix motif, indicating its DNA-binding activity. The depK gene encodes a conserved functionally unknown protein. Finally, depN encodes a nonfunctional PCP remnant without a critical serine residue in a conserved motif, GX(HD)S, necessary for phosphopantetheinylation and covalent substrate aminoacylation.
In conclusion, FK228, a promising anticancer agent, is moving fast towards clinical uses. Its unique structural characteristics and mode of action warrant efforts to generate analogs that can be tested as additional and hopefully even better anticancer drug candidates. The adaptation of an efficient genetic system for C. violaceum no. 968, in combination with cloning of the dep gene cluster, provides an excellent platform for further investigations of the FK228 biosynthetic pathway and makes future genetic manipulations of the biosynthetic pathway for generating novel FK228 analogs by engineered biosynthesis possible. In addition, the Flp-FRT recombination system validated in this study could be applicable to other strains of C. violaceum.
Acknowledgments
We thank Herbert Schweizer, Colorado State University, for providing components of the Flp-FRT recombination system; Dale Noel, Marquette University, for providing E. coli strain MT607(pRK600); and Patrick Anderson, Great Lakes WATER Institute at the University of Wisconsin-Milwaukee, for assistance with LC-MS analysis. We also thank Kenneth Chan and Mark McBride for critical reading of the manuscript.
This work was supported by funds from the University of Wisconsin-Milwaukee under the UWM Biotechnology Initiative.
Footnotes
Published ahead of print on 30 March 2007.
REFERENCES
- 1.Acharya, M. R., A. Sparreboom, J. Venitz, and W. D. Figg. 2005. Rational development of histone deacetylase inhibitors as anticancer agents: a review. Mol. Pharmacol. 68:917-932. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Ansari, M. Z., G. Yadav, R. S. Gokhale, and D. Mohanty. 2004. NRPS-PKS: a knowledge-based resource for analysis of NRPS/PKS megasynthases. Nucleic Acids Res. 32:W405-W413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brazilian National Genome Project Consortium. 2003. The complete genome sequence of Chromobacterium violaceum reveals remarkable and exploitable bacterial adaptability. Proc. Natl. Acad. Sci. USA 100:11660-11665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Byrd, J. C., G. Marcucci, M. R. Parthun, J. J. Xiao, R. B. Klisovic, M. Moran, T. S. Lin, S. Liu, A. R. Sklenar, M. E. Davis, D. M. Lucas, B. Fischer, R. Shank, S. L. Tejaswi, P. Binkley, J. Wright, K. K. Chan, and M. R. Grever. 2005. A phase 1 and pharmacodynamic study of depsipeptide (FK228) in chronic lymphocytic leukemia and acute myeloid leukemia. Blood 105:959-967. [DOI] [PubMed] [Google Scholar]
- 6.Byrd, J. C., C. Shinn, R. Ravi, C. R. Willis, J. K. Waselenko, I. W. Flinn, N. A. Dawson, and M. R. Grever. 1999. Depsipeptide (FR901228): a novel therapeutic agent with selective, in vitro activity against human B-cell chronic lymphocytic leukemia cells. Blood 94:1401-1408. [PubMed] [Google Scholar]
- 7.Challis, G. L., J. Ravel, and C. A. Townsend. 2000. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem. Biol. 7:211-224. [DOI] [PubMed] [Google Scholar]
- 8.Cheng, Y. Q. 2006. Deciphering the biosynthetic codes for the potent anti-SARS-CoV cyclodepsipeptide valinomycin in Streptomyces tsusimaensis ATCC 15141. ChemBioChem 7:471-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng, Y. Q., G. L. Tang, and B. Shen. 2003. Type I polyketide synthase requiring a discrete acyltransferase for polyketide biosynthesis. Proc. Natl. Acad. Sci. USA 100:3149-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dokmanovic, M., and P. A. Marks. 2005. Prospects: histone deacetylase inhibitors. J. Cell. Biochem. 96:293-304. [DOI] [PubMed] [Google Scholar]
- 11.Du, L., Y. Q. Cheng, G. Ingenhorst, G. L. Tang, Y. Huang, and B. Shen. 2003. Hybrid peptide-polyketide natural products: biosynthesis and prospects towards engineering novel molecules. Genet. Eng. 25:227-267. [DOI] [PubMed] [Google Scholar]
- 12.Duitman, E. H., L. W. Hamoen, M. Rembold, G. Venema, H. Seitz, W. Saenger, F. Bernhard, R. Reinhardt, M. Schmidt, C. Ullrich, T. Stein, F. Leenders, and J. Vater. 1999. The mycosubtilin synthetase of Bacillus subtilis ATCC6633: a multifunctional hybrid between a peptide synthetase, an amino transferase, and a fatty acid synthase. Proc. Natl. Acad. Sci. USA 96:13294-13299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duran, N., and C. F. Menck. 2001. Chromobacterium violaceum: a review of pharmacological and industrial perspectives. Crit. Rev. Microbiol. 27:201-222. [DOI] [PubMed] [Google Scholar]
- 14.Finan, T. M., B. Kunkel, G. F. De Vos, and E. R. Signer. 1986. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J. Bacteriol. 167:66-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finking, R., and M. A. Marahiel. 2004. Biosynthesis of nonribosomal peptides1. Annu. Rev. Microbiol. 58:453-488. [DOI] [PubMed] [Google Scholar]
- 16.Fischbach, M. A., and C. T. Walsh. 2006. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: logic, machinery, and mechanisms. Chem. Rev. 106:3468-3496. [DOI] [PubMed] [Google Scholar]
- 17.Furumai, R., A. Matsuyama, N. Kobashi, K. H. Lee, M. Nishiyama, H. Nakajima, A. Tanaka, Y. Komatsu, N. Nishino, M. Yoshida, and S. Horinouchi. 2002. FK228 (depsipeptide) as a natural prodrug that inhibits class I histone deacetylases. Cancer Res. 62:4916-4921. [PubMed] [Google Scholar]
- 18.Garber, K. 2007. HDAC inhibitors overcome first hurdle. Nat. Biotechnol. 25:17-19. [DOI] [PubMed] [Google Scholar]
- 19.Gehring, A. M., E. DeMoll, J. D. Fetherston, I. Mori, G. F. Mayhew, F. R. Blattner, C. T. Walsh, and R. D. Perry. 1998. Iron acquisition in plague: modular logic in enzymatic biogenesis of yersiniabactin by Yersinia pestis. Chem. Biol. 5:573-586. [DOI] [PubMed] [Google Scholar]
- 20.Grunewald, J., and M. A. Marahiel. 2006. Chemoenzymatic and template-directed synthesis of bioactive macrocyclic peptides. Microbiol. Mol. Biol. Rev. 70:121-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heathcote, M. L., J. Staunton, and P. F. Leadlay. 2001. Role of type II thioesterases: evidence for removal of short acyl chains produced by aberrant decarboxylation of chain extender units. Chem. Biol. 8:207-220. [DOI] [PubMed] [Google Scholar]
- 22.Hill, A. M. 2006. The biosynthesis, molecular genetics and enzymology of the polyketide-derived metabolites. Nat. Prod. Rep. 23:256-320. [DOI] [PubMed] [Google Scholar]
- 23.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 24.Johnstone, R. W. 2002. Histone-deacetylase inhibitors: novel drugs for the treatment of cancer. Nat. Rev. Drug Discov. 1:287-299. [DOI] [PubMed] [Google Scholar]
- 25.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. John Innes Foundation, Norwich, England.
- 26.Lee, C., H. Gorisch, H. Kleinkauf, and R. Zocher. 1992. A highly specific d-hydroxyisovalerate dehydrogenase from the enniatin producer Fusarium sambucinum. J. Biol. Chem. 267:11741-11744. [PubMed] [Google Scholar]
- 27.Li, K. W., J. Wu, W. Xing, and J. A. Simon. 1996. Total synthesis of the antitumor depsipeptide FR-901,228. J. Am. Chem. Soc. 118:7237-7238. [Google Scholar]
- 28.MacNeil, D. J., K. M. Gewain, C. L. Ruby, G. Dezeny, P. H. Gibbons, and T. MacNeil. 1992. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 111:61-68. [DOI] [PubMed] [Google Scholar]
- 29.Magarvey, N. A., M. Ehling-Schulz, and C. T. Walsh. 2006. Characterization of the cereulide NRPS alpha-hydroxy acid specifying modules: activation of alpha-keto acids and chiral reduction on the assembly line. J. Am. Chem. Soc. 128:10698-10699. [DOI] [PubMed] [Google Scholar]
- 30.Marshall, J. L., N. Rizvi, J. Kauh, W. Dahut, M. Figuera, M. H. Kang, W. D. Figg, I. Wainer, C. Chaissang, M. Z. Li, and M. J. Hawkins. 2002. A phase I trial of depsipeptide (FR901228) in patients with advanced cancer. J. Exp. Ther. Oncol. 2:325-332. [DOI] [PubMed] [Google Scholar]
- 31.Monneret, C. 2005. Histone deacetylase inhibitors. Eur. J. Med. Chem. 40:1-13. [DOI] [PubMed] [Google Scholar]
- 32.Mootz, H. D., D. Schwarzer, and M. A. Marahiel. 2002. Ways of assembling complex natural products on modular nonribosomal peptide synthetases. Chembiochem 3:490-504. [DOI] [PubMed] [Google Scholar]
- 33.Nakajima, H., Y. B. Kim, H. Terano, M. Yoshida, and S. Horinouchi. 1998. FR901228, a potent antitumor antibiotic, is a novel histone deacetylase inhibitor. Exp. Cell Res. 241:126-133. [DOI] [PubMed] [Google Scholar]
- 34.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 35.Piekarz, R., and S. Bates. 2004. A review of depsipeptide and other histone deacetylase inhibitors in clinical trials. Curr. Pharm. Des. 10:2289-2298. [DOI] [PubMed] [Google Scholar]
- 36.Piekarz, R. L., R. Robey, V. Sandor, S. Bakke, W. H. Wilson, L. Dahmoush, D. M. Kingma, M. L. Turner, R. Altemus, and S. E. Bates. 2001. Inhibitor of histone deacetylation, depsipeptide (FR901228), in the treatment of peripheral and cutaneous T-cell lymphoma: a case report. Blood 98:2865-2868. [DOI] [PubMed] [Google Scholar]
- 37.Piel, J. 2002. A polyketide synthase-peptide synthetase gene cluster from an uncultured bacterial symbiont of Paederus beetles. Proc. Natl. Acad. Sci. USA 99:14002-14007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rajgolikar, G., K. K. Chan, and H. C. Wang. 1998. Effects of a novel antitumor depsipeptide, FR901228, on human breast cancer cells. Breast Cancer Res. Treat. 51:29-38. [DOI] [PubMed] [Google Scholar]
- 39.Reeves, C. D., S. Murli, G. W. Ashley, M. Piagentini, C. R. Hutchinson, and R. McDaniel. 2001. Alteration of the substrate specificity of a modular polyketide synthase acyltransferase domain through site-specific mutations. Biochemistry 40:15464-15470. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook, J., and D. W. Russell. 2000. Molecular cloning: a laboratory manual, 3d ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 41.Sandor, V., S. Bakke, R. W. Robey, M. H. Kang, M. V. Blagosklonny, J. Bender, R. Brooks, R. L. Piekarz, E. Tucker, W. D. Figg, K. K. Chan, B. Goldspiel, A. T. Fojo, S. P. Balcerzak, and S. E. Bates. 2002. Phase I trial of the histone deacetylase inhibitor, depsipeptide (FR901228, NSC 630176), in patients with refractory neoplasms. Clin. Cancer Res. 8:718-728. [PubMed] [Google Scholar]
- 42.Sandor, V., A. R. Robbins, R. Robey, T. Myers, E. Sausville, S. E. Bates, and D. L. Sackett. 2000. FR901228 causes mitotic arrest but does not alter microtubule polymerization. Anticancer Drugs 11:445-454. [DOI] [PubMed] [Google Scholar]
- 43.Sandor, V., A. Senderowicz, S. Mertins, D. Sackett, E. Sausville, M. V. Blagosklonny, and S. E. Bates. 2000. P21-dependent G1 arrest with downregulation of cyclin D1 and upregulation of cyclin E by the histone deacetylase inhibitor FR901228. Br. J. Cancer. 83:817-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen, B. 2003. Polyketide biosynthesis beyond the type I, II and III polyketide synthase paradigms. Curr. Opin. Chem. Biol. 7:285-295. [DOI] [PubMed] [Google Scholar]
- 45.Shigematsu, N., H. Ueda, S. Takase, H. Tanaka, K. Yamamoto, and T. Tada. 1994. FR901228, a novel antitumor bicyclic depsipeptide produced by Chromobacterium violaceum No. 968. II. Structure determination. J. Antibiot. (Tokyo) 47:311-314. [DOI] [PubMed] [Google Scholar]
- 46.Simon, R., U. Priefer, and A. Puehler. 1983. A broad host range mobilisation system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 47.Stachelhaus, T., H. D. Mootz, and M. A. Marahiel. 1999. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 6:493-505. [DOI] [PubMed] [Google Scholar]
- 48.Straight, P. D., M. A. Fischbach, C. T. Walsh, D. Z. Rudner, and R. Kolter. 2007. A singular enzymatic megacomplex from Bacillus subtilis. Proc. Natl. Acad. Sci. USA 104:305-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomas, M. G., Y. A. Chan, and S. G. Ozanick. 2003. Deciphering tuberactinomycin biosynthesis: isolation, sequencing, and annotation of the viomycin biosynthetic gene cluster. Antimicrob. Agents Chemother. 47:2823-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsuge, K., T. Akiyama, and M. Shoda. 2001. Cloning, sequencing, and characterization of the iturin A operon. J. Bacteriol. 183:6265-6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ueda, H., H. Nakajima, Y. Hori, T. Fujita, M. Nishimura, T. Goto, and M. Okuhara. 1994. FR901228, a novel antitumor bicyclic depsipeptide produced by Chromobacterium violaceum No. 968. I. Taxonomy, fermentation, isolation, physico-chemical and biological properties, and antitumor activity. J. Antibiot. (Tokyo) 47:301-310. [DOI] [PubMed] [Google Scholar]
- 52.Ueda, H., H. Nakajima, Y. Hori, T. Goto, and M. Okuhara. 1994. Action of FR901228, a novel antitumor bicyclic depsipeptide produced by Chromobacterium violaceum no. 968, on Ha-ras transformed NIH3T3 cells. Biosci. Biotechnol. Biochem. 58:1579-1583. [DOI] [PubMed] [Google Scholar]
- 53.Vigushin, D. M. 2002. FR-901228 Fujisawa/National Cancer Institute. Curr. Opin. Investig. Drugs 3:1396-1402. [PubMed] [Google Scholar]
- 54.Walsh, C. T. 2002. Combinatorial biosynthesis of antibiotics: challenges and opportunities. Chembiochem 3:125-134. [DOI] [PubMed] [Google Scholar]
- 55.Wenzel, S. C., and R. Muller. 2005. Formation of novel secondary metabolites by bacterial multimodular assembly lines: deviations from textbook biosynthetic logic. Curr. Opin. Chem. Biol. 9:447-458. [DOI] [PubMed] [Google Scholar]
- 56.Xiao, J. J., J. Byrd, G. Marcucci, M. Grever, and K. K. Chan. 2003. Identification of thiols and glutathione conjugates of depsipeptide FK228 (FR901228), a novel histone protein deacetylase inhibitor, in the blood. Rapid Commun. Mass Spectrom. 17:757-766. [DOI] [PubMed] [Google Scholar]
- 57.Yoo, C. B., and P. A. Jones. 2006. Epigenetic therapy of cancer: past, present and future. Nat. Rev. Drug Discov. 5:37-50. [DOI] [PubMed] [Google Scholar]
- 58.Zazopoulos, E., K. Huang, A. Staffa, W. Liu, B. O. Bachmann, K. Nonaka, J. Ahlert, J. S. Thorson, B. Shen, and C. M. Farnet. 2003. A genomics-guided approach for discovering and expressing cryptic metabolic pathways. Nat. Biotechnol. 21:187-190. [DOI] [PubMed] [Google Scholar]