Abstract
Mutations in BRCA1 are present in 45% of families that segregate with susceptibility for breast cancer and in 80–90% of families with both breast and ovarian cancer. Here we report that BRCA1 stimulates artificial and genomic promoter constructs containing p53-responsive elements. This activity of BRCA1 depends on the presence of wild-type p53, which was shown by using mouse fibroblasts expressing temperature-sensitive forms of p53, or p53(+/+) and p53(−/−) fibroblasts obtained from p53 knockout mice. Furthermore, mutant forms of BRCA1 lacking the C-terminal second BRCA1 C-terminal (BRCT) domain showed reduced p53-mediated transcriptional activation. Finally, we found that BRCA1 coimmunoprecipitates with p53, in vitro and in vivo. These findings suggest a function of BRCA1 as a p53 coactivator.
The tumor suppressor protein p53 is involved in a variety of human cancers (1). p53 protein consists of, at least, three functional domains: the N-terminal transcription activation domain, the central sequence-specific DNA binding domain, and the C-terminal oligomerization domain (2). The fact that most of the point mutations found in sporadic and familial cancers occur in the DNA binding domain suggests that transcription activity of p53 plays a crucial role in human carcinogenesis. Previous studies have shown that cellular proteins including mdm2 and CBP/p300 positively or negatively regulate this activity of p53, indicating that the p53-dependent gene regulation is tightly regulated (3–6). Furthermore, germline p53 mutations induce breast cancer in patients with Li-Fraumeni Syndrome supporting the notion that p53 plays an important role in breast cancer development (7).
BRCA1 was isolated as a candidate for the familial breast and ovarian cancer susceptibility gene (8), and encodes a 220-kDa nuclear phosphoprotein (9–12). Germ-line mutations of this gene are frequently found in kindreds with familial breast cancer (13, 14). BRCA1 acts as a tumor suppressor protein, because its overexpression leads to growth retardation of tumor cells in nude mice (15). Molecular features of the BRCA1 protein shows characteristic domains: the N-terminal RING-finger domain and two BRCT domains in the C terminus. BRCA1 C-terminal (BRCT) domains are also found in cell cycle checkpoint proteins including p53 binding protein 1 (p53BP1), the fission yeast replication checkpoint protein Rad4, DNA repair protein XRCC1 and the retinoblastoma family of proteins (16). We and others have shown that the C-terminal region has an intrinsic transactivation activity when fused with GAL4-DNA binding domain, suggesting a role of BRCA1 in the regulation of gene expression (17, 18).
In the present study, we tested the possible target of BRCA1 by using several reporter constructs, and found that BRCA1 can specifically stimulate p53-responsive elements. To investigate whether BRCA1 activity on p53-responsive element was dependent on the presence of p53, we used mouse fibroblasts expressing a temperature-sensitive form of p53 (19), and p53(+/+) or p53(−/−) primary fibroblasts obtained from p53 knockout mice (20). We also found that BRCA1 coimmunoprecipitates with p53. Our findings demonstrate that BRCA1 can enhance p53-dependent gene expression acting as a p53 coactivator.
MATERIALS AND METHODS
Luciferase Reporter Plasmids.
A 2.4-kb HindIII fragment of human p21Waf1 promoter from pWWPCAT (21), and a 1.0-kb BglII-HindIII fragment from COSX1CAT (22) were transferred to a luciferase reporter plasmid, pGL2 (Promega) to generate p21luc. For PGluc, a BglII-BamHI fragment containing two copies of p53 binding sequences and adenovirus major late TATA box from p50–2 (23) were cloned into the pGL2 vector. For p21(−)luc, p21luc was digested by SacI, and self-ligated. For mdm2smluc and mdm2pvluc, mdm2luc was digested by either SmaI or PvuII, and self-ligated, respectively. For E2Fluc, four copies of E2F binding sequences and TATA sequence from (E2F)4CAT (provided by E. Harlow, Harvard Medical School, Charlestown, MA) (24) were transferred to the pGL2 vector. For SREluc and NFκBluc, two copies of serum-responsive factor or NFκB consensus elements were cloned into pGL2 with the TATA box derived from c-fos promoter (provided from A. Chan, Mount Sinai Medical School, New York). Mycluc and GAL4luc were gifts from R. Eisenman (Fred Hutchinson Cancer Center, Seattle) or R. Davis (University of Massachusetts, Worcester, MA), respectively.
Cell Culture.
HBL100 cells (human breast epithelial cells) and 293T cells (human kidney cancer cells) were maintained in DMEM supplemented with 10% calf serum or fetal bovine serum in the presence of antibiotics. Mouse primary fibroblasts from p53 knockout mice (The Jackson Laboratory) and 10.1Val5 cells expressing a temperature sensitive form of p53 (a gift from A. Levine, Princeton University, Princeton, NJ) (19) were maintained in DMEM supplemented with 10% fetal bovine serum.
Luciferase Assay.
293T cells (2 × 105) were transiently transfected with 2 μg of DNA (0.5 μg of reporter plasmid and 1.5 μg of indicated plasmid) by a modified calcium phosphate method termed MBS (Stratagene). Cells were maintained below 80% confluence, and harvested 48 hr posttransfection. The luciferase assay was described (18). Values shown represent an average of three or four independent transfections normalized by the cotransfection of MFG-β-galactosidase plasmid (provided by R. Mulligan, Massachusetts Institute of Technology, Cambridge, MA). The procedure for transfection of primary fibroblasts or 10.1Val5 cells was basically the same as that of 293T cells. For 10.1Val5 cells, transfection was done at 39°C for 3 hr according to the protocol by the manufacturer, and then temperature was shifted to 32.5°C for further 48 hr before luciferase assay.
Effector Plasmids.
Full-length BRCA1 cDNA cloned in pcDNA3 (p385BRCA1) was a gift from M. Endos, National Institutes of Health, Bethesda, MD). All cDNAs used for these studies were cloned into pcDNA3.1 or pcDNA3.1/HisA (Invitrogen). For the C-terminal deletion mutant of BRCA1 (clone BRCA1–1769), the full-length BRCA1 cDNA cloned in pcDNA3.1/HisA was digested by ApaI and self-ligated to remove amino acids 1770 to 1863.
Immunoprecipitation and Western Blot.
Nuclear extracts of HBL100 cells were prepared as described (11). BRCA1 and p53 were immunoprecipitated from 300 μg of nuclear extract using anti-BRCA1 antibody (C-20) or anti-p53 antibody (DO-1) (Santa Cruz Biotechnology) in EBC buffer (0.5% Nonidet P-40, 50 mM Tris-HCl [pH 8.0], 120 mM NaCl, 100 mM NaF, 200 μM Na-orthovanadate, 10 μg/ml each of phenylmethylsulfonyl fluoride, aprotinin, and leupeptin). Samples were separated by SDS/PAGE, and transferred to Immobilon-P membrane (Millipore) followed by immunoblotting with the indicated antibodies.
Construction of Glutathione S-Transferase (GST)-Fusion Protein.
Segments of human p53 corresponding to amino acid 1 to 110 (primer A and B′), 115 to 263 (primer C and D), and 271 to 391 (primer E and F′) were PCR amplified using the following primers. Primer A: 5′-CCGGGTCGGATCCATGGAGG-3′, primer B′: 5′-TGTCCCAGGATCCTAGAAGCC-3′, primer C: 5′-CGTCTGGGGATCCTGCATTC-3′, primer D: 5′-GTTCCGTCCCTCGAGATTACC-3′, primer E: 5′-CTGGGACGGATCCGCTTTGAG-3′, and primer F′: 5′-AAGTGGGGATCCTCAGTCTGA-3′. Sequences of amplified fragments were confirmed by the dideoxy method, and cloned into pGEX vector (Pharmacia).
RESULTS
BRCA1 Stimulates p53-Responsive Elements.
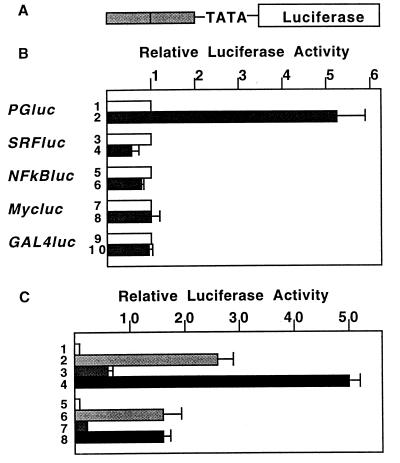
Because previous studies have suggested that BRCA1 is involved in the regulation of gene transcription (17, 18), a series of reporter plasmids containing consensus elements for various DNA binding proteins upstream of luciferase gene (Fig. 1A) were constructed and tested for activation by BRCA1. As shown in Fig. 1B, the reporter plasmid containing two p53-responsive elements (PGluc) showed 5-fold activation by BRCA1 (lane 2) while plasmids containing responsive sites for serum-responsive factor, NFκB, Myc and GAL4 showed no activation (lanes 4, 6, 8, and 10). Transfection of 293T cells with p53 caused an ≈25-fold activation of PGluc (Fig. 1C, lane 2), while coexpression of p53 and BRCA1 activated the reporter up to 50-fold (Fig. 1C, lane 4). A reporter containing only the TATA box, without p53-responsive elements, was not activated by either BRCA1, p53 or both together (not shown). To further determine whether the BRCA1 action was specific to transcriptional activation through a p53-responsive element, a similar reporter plasmid containing four of E2F1 recognition sites upstream of the TATA box was constructed. In cotransfected 293T cells, this E2Fluc reporter was activated ≈15-fold by E2F1/DP1, while ectopic BRCA1 had no effect itself and failed to cooperate with E2F1/DP1 in stimulating E2Fluc reporter activity (Fig. 1C, lanes 6–8). Neither BRCA1 nor E2F1/DP1 activated the control reporter containing only the TATA box upstream of the luciferase (not shown). All of these results suggested that BRCA1 is able to specifically activate p53-dependent gene expression, and is not a general partner for activation of transcription.
Figure 1.
BRCA1 targets p53-responsive elements. (A) Basic structure of the minimal reporter plasmid. Two (PGluc, SRFluc, NFκBluc, and E2Fluc) or four (Mycluc and GAL4luc) copies of DNA binding sequences were cloned upstream of c-fos promoter TATA sequence and luciferase genes. (B) BRCA1 activates p53-responsive elements. 293T cells were transfected with empty vector or BRCA1 expression plasmids together with several reporter plasmids indicated, and luciferase activity was measured. Each of the experiments was done in duplicate or triplicate, and normalized by β-galactosidase activity. Lanes: 1, 3, 5, 7, and 9, pcDNA3 vector; 2, 4, 6, 8, and 10, BRCA1/PCDNA3. (C) BRCA1 is not an general coactivator. 293T cells were cotransfected with p53, E2F1, and DP1 expression plasmids and/or BRCA1 to detect activation of PGluc or E2Fluc. Lanes: 1 and 5, pcDNA3 vector; 2, p53; 3 and 7, BRCA1; 4, p53 plus BRCA1; 6, E2F1 plus DP1; 8, E2F1, DP1 plus BRCA1.
BRCA1 Stimulates the mdm2 and p21Waf1 Promoter.
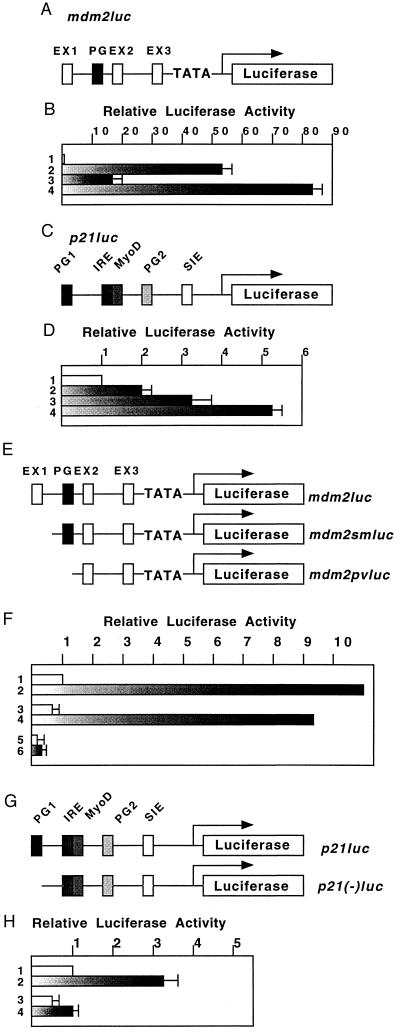
Because BRCA1 stimulated p53-dependent gene expression of PGluc (Fig. 1B), we analyzed its ability to activate natural genomic promoters (mdm2, p21Waf1) that have been shown (22, 23) to contain p53-responsive elements. 293T cells were cotransfected with reporter plasmids expressing luciferase under the control of a genomic fragment containing either a 1.0-kb fragment of the first intron of the murine mdm2 gene or a 2.4-kb fragment of the human p21Waf1 promoter region and vectors expressing p53 and/or BRCA1 (Fig. 2 A and C). The mdm2 promoter was activated ≈50-fold by p53 alone (Fig. 2B, lane 2), and ≈17-fold by BRCA1 alone (Fig. 2B, lane 3), whereas BRCA1 and p53 together stimulated the reporter >80-fold (Fig. 2B, lane 4). The p21Waf1 promoter was activated ≈2-fold by p53 alone (Fig. 2D, lane 2), ≈3-fold by BRCA1 (lane 3) and ≈5-fold by BRCA1 plus p53 (lane 4). The fact that the p21Waf1 promoter was activated more by ectopic BRCA1 alone than by p53 alone (Fig. 2D, lanes 2 and 3) probably reflects the presence of endogenous p53 (see Discussion). From these results, we conclude that BRCA1 is able to contribute to p53-mediated transcriptional activation through natural promoter elements.
Figure 2.
BRCA1 stimulates genomic promoters containing p53-responsive elements. (A) Genomic organization of the murine mdm2 promoter. The first intron of the 1.0-kb murine mdm2 and adenovirus major late TATA box/TdT initiation signal were cloned into pGL2 vector to generate mdm2luc. Exons 1–3 (EX1–EX3), and p53-responsive elements (PG) are indicated. (B and D) The procedure for luciferase analysis of mdm2luc (B) or p21luc (D) was basically the same with that of Fig. 1. Lanes: 1, pcDNA3 vector; 2, p53; 3, BRCA1; 4, p53 plus BRCA1. (C) Genomic organization of the human p21Waf1 promoter. Human p21Waf1 promoter (2.4 kb) was cloned upstream of luciferase gene of pGL2 vector to generate p21luc. p53-responsive elements (PG1 and 2), interferon-regulatory elements (IRE), MyoD-responsive elements (MyoD), and signal transducers and activators of transcription (STAT) serum-inducible elements (SIE) are indicated. (E) Structure of deletion mutants of mdm2luc. mdm2luc was digested with SmaI or PvuII to generate mdm2smluc or mdm2pvluv, respectively. (F) Activation of mutant mdm2 promoters by BRCA1. Reporter plasmids of mdm2luc (lane 1 and 2), mdm2smluc (lane 3 and 4) and mdm2pvluc (lane 5 and 6) were cotransfected with vector (lane 1, 3, and 5) or BRCA1 (lane 2, 4, and 6). (G) Structure of deletion mutant forms of p21Waf1 promoter. p21luc was digested with SacI to remove the first p53-responsive element (p21(−)luc). (H) Activation of p21luc or p21(−)luc by BRCA1. Lanes: 1 and 3, vector; 2 and 4, BRCA1.
The requirement for p53-responsive elements for BRCA1 function was further confirmed through the use of reporter plasmids with 5′ deletions in the mdm2 promoter (Fig. 2E). Reporters with deletion mutations obtained by SmaI digestion of the mdm2 promoter region retained the region containing the p53-responsive elements (mdm2smluc), and could still be activated to the same level as the wild-type promoter (Fig. 2F, lanes 2 and 4). In contrast, a reporter with a deletion mutant lacking this site (mdm2pvluc) could not be significantly activated by BRCA1 (Fig. 2F, lane 6).
It has been shown that, in the p21Waf1 promoter, there are two possible binding sequences for p53. The element, PG1, lying ≈2.4-kb upstream of the TATA box (Fig. 2G) is a primary target of p53 (21). Deletion of the p53-responsive element (PG1) markedly reduced activation of transcription by BRCA1 (Fig. 2H, lanes 2 and 4).
BRCA1 Requires Wild-type p53 to Stimulate p53-Responsive Elements.
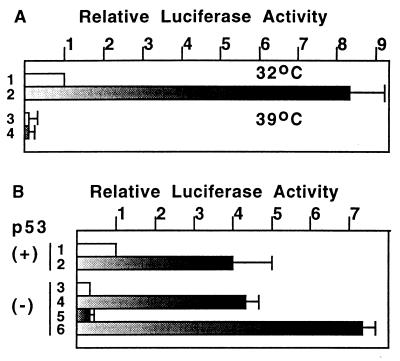
To examine whether BRCA1 requires wild-type p53 to activate transcription, we used two different cell types: mouse fibroblasts expressing a temperature-sensitive form of p53 and other primary fibroblasts derived from p53 knockout mice. Mouse fibroblasts 10.1Val5 express active p53 at the permissive temperature (32°C) but inactive p53 at the nonpermissive temperature (39°C). As shown in Fig. 3A, BRCA1 expression activated the PGluc reporter 8-fold at 32°C but not at all at 39°C. The control reporter containing only the TATA box did not show responsiveness to BRCA1 at any temperature tested (not shown). To rule out a general suppression of transcription at 39°C, cells were cotransfected with a vector expressing the GAL4 DNA binding domain fused to the transactivation domain of VP16 and a luciferase reporter under the control of four copies of the GAL4-responsive element. No significant difference in transcription activation at 39°C and at 32°C was found (not shown).
Figure 3.
BRCA1 requires wild-type p53 to activate p53-responsive elements. (A) 10.1Val5 mouse fibroblasts expressing temperature sensitive forms of murine p53 (wild type at 32°C and mutant at 39°C) were transfected with vector alone or BRCA1/pcDNA3 together with PGluc. Cells were transfected with the indicated DNA for 3 hr at 39°C, and were maintained at 32°C (lane 1 and 2) or 39°C (lane 3 and 4) for an additional 2 days before luciferase assay. Lanes: 1 and 3, pcDNA3 vector; 2 and 4, BRCA1/pcDNA3. (B) Primary fibroblasts prepared from p53 knockout mice were transfected and used for luciferase assays of PGluc. Approximately 2 × 106 p53 (+/+) cells (lane 1 and 2) or p53 (−/−) cells (lanes 3 to 6) were used for each of the assays. Lanes: 1 and 3, pcDNA3 vector; 2 and 5, BRCA1; 4, p53; 6, BRCA1 plus p53. Values shown here are the average of three or four independent experiments (A and B).
In the absence of ectopic factors, expression of PGluc was significantly lower in p53(−/−) primary cells than in p53(+/+) primary cells (Fig. 3B, lanes 1 and 3). Cotransfection of BRCA1 with the reporter resulted in a further 4-fold activation in p53(+/+) cells (Fig. 3B, lane 2), but had no detectable effect on expression of the reporter in p53 (−/−) cells (Fig. 3B, lanes 3 and 5). However, the reporter was significantly (4-fold) activated when p53 (−/−) cells were transfected with p53 alone, and this activation was further enhanced by cotransfection of BRCA1 (Fig. 3B, lane 4 and 6). All of these results demonstrate that BRCA1 requires wild-type p53 to stimulate p53-responsive elements.
The C-Terminal Region of BRCA1 Is Necessary for Transactivation of p53-Responsive Elements.
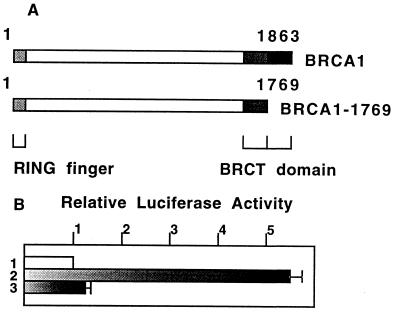
Mutations of BRCA1 gene are frequently found in kindreds with familial breast cancer. A variety of mutations can be observed in these patients and include point mutations of the RING-finger domain and truncational deletions of the C-terminal domain, suggesting that it is important to preserve the integrity of the molecule to suppress tumor development (13, 14). Because the C-terminal domain was shown to have an intrinsic transactivation activity (17, 18), we investigated the role of the BRCT domains in the regulation of p53-dependent gene expression. For this purpose, a C-terminal deletion form of BRCA1, BRCA1–1769, was generated and analyzed in a transcriptional activation assays (Fig. 4A). Whereas the wild-type BRCA1 activated the PGluc reporter 5- to 6-fold, deletion of amino acids 1770–1863 markedly reduced transcription activation (8% of wild-type activity) (Fig. 4B). These results show that the C-terminal domain of BRCA1 plays an important role in regulating p53-dependent gene expression, and support the concept that transcriptional activation is a physiological function of BRCA1.
Figure 4.
The second BRCT domain is required for the transactivation of PGluc reporter plasmid. (A) Molecular feature of the mutant form of BRCA1 lacking the second BRCT domain. BRCA1/pcDNA3 was digested by ApaI to generate BRCA1–1769 construct. (B) Relative luciferase activity of PGluc regulated by vector (lane 1), the wild-type (lane 2) or mutant BRCA1 (lane 3) was measured by transfection of 293T cells.
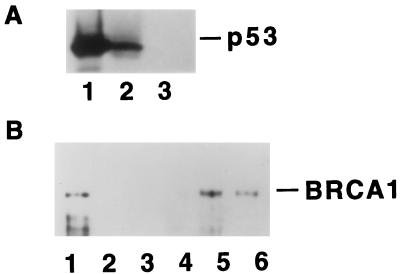
Detection of BRCA1/p53 Complex.
Our evidence that BRCA1 can collaborate with p53 led us to investigate whether a physical interaction between these proteins could be demonstrated. Nuclear extracts of HBL100, a human breast epithelial cell line, were subjected to immunoprecipitation with anti-BRCA1 polyclonal antibody C-20 and, as a negative control, normal rabbit IgG. Subsequent immunoblot analysis with anti-p53 mAb, DO-1, revealed p53 in the anti-BRCA1 immunoprecipitate (Fig. 5A, lane 2) and nuclear extract (lane 1), but not in the control immunoprecipitate (lane 3). To map the interaction site, segments of p53 were expressed as GST-fusion proteins, and incubated with lysates of Sf9 cells expressing BRCA1 from a baculovirus vector. The analysis in Fig. 5B shows that BRCA1 binds to intact p53 (lane 6) and to the C-terminal region (amino acisds 271–393) of p53 (lane 5) but not to the N-terminal region (lane 3) or the central region (lane 4). These results strongly suggest that BRCA1 forms a complex with p53 in vivo.
Figure 5.
Coimmunoprecipitation of BRCA1 and p53. (A) Endogenous BRCA1 protein was immunoprecipitated by anti-BRCA1 polyclonal antibody (C-20) from 300 μg of nuclear extract of human breast epithelial cell line HBL100. Samples were separated by SDS/10% polyacrylamide gel, and immunoblotted by anti-p53 mAb (DO-1). Lane 1, 10 μg of nuclear extract; lane 2, immunoprecipitation by C-20; lane 3, immunopreciptation by normal rabbit IgG. (B) BRCA1 binds to the C-terminal region of p53, in vitro. Lysates prepared from baculovirus-infected Sf9 cells expressing human BRCA1 were incubated with GST (lane 2), GST-p53(1–110) (lane 3), GST-p53(111–270) (lane 4), GST-p53(270–393) (lane 5) and GST-p53(1–393) (lane 6). Ten microliters of Sf9 lysates of baculoviral BRCA1 was loaded as a control of immunoblot (lane 1). Samples were separated by SDS/6% polyacrylamide gel, and blotted with C-20.
DISCUSSION
It becomes increasingly clear that BRCA1 is involved in transcriptional regulation (17, 18, 25, 26). The results presented here further support this notion. We show that, in cells expressing wild-type p53, ectopic BRCA1 activated promoters containing p53-responsive elements and also enhanced activation of the same promoters by exogenous p53 activation. Under the same conditions, BRCA1 exhibited no effect on several other responsive elements tested, suggesting at least some degree of specificity of BRCA1 function as a coactivator. The effects of BRCA1 were also reproduced in the context of two genomic promoters, further confirming that the major target for activation is the p53-responsive elements present in these promoters. Experiments using cells derived from p53 knockout mice or cells expressing temperature-sensitive p53 demonstrate the requirement for wild-type p53. Taken together, these findings suggest that at least one of the functions of BRCA1 in vivo is to act as a coactivator for p53. Further investigation will be needed to understand how BRCA1 activity is integrated with other proteins, such as CBP/p300 or Hdm2, known to regulate p53 function (3–6).
In the present study, BRCA1 activation of the p53-responsive element was found to require an intact BRCA1 C-terminal region. We and others have shown that the BRCA1 C-terminal region is able to act as a transactivation domain in the GAL4-fusion protein assay (17, 18). Moreover, the presence of cancer predisposing germ-line mutations (17, 18), but not benign polymorphisms (27), abolishes transcriptional activation. Therefore, the presence of an intact C-terminal region seems to be a requirement for transcriptional activity, a notion corroborated by the fact that a C-terminal truncation, Y1853X, fails to interact with the RNA polymerase II holoenzyme (26). It is tempting to speculate that the interaction of p53 with BRCA1 provides the complex with an additional activation domain. Alternatively, it also is possible that ability of BRCA1 to enhance p53 activity reflects a conformational change in p53 that increases its DNA binding capabilities and that this interaction depends on the presence of an intact BRCA1 C-terminal region. Interestingly, we defined the C terminus of p53 as the major target for BRCA1 interaction. The C terminus of p53 has been shown to be critical for the activation of p53 DNA binding activity by several means, including binding to a mAb, PAb421 (28), serine phosphorylation (29, 30), and acetylation (31). Therefore, it is conceivable that, besides contributing to an additional activation domain to the complex, BRCA1 may interact physically with the C-terminal domain of p53 to induce a conformational change that results in increased DNA binding.
While this manuscript was in preparation, Somasundaram et al. (25) recently reported that the genomic p21Waf1 promoter was activated by ectopic BRCA1 when cotransfected into SW480 cancer cells, which contain a functionally inactive p53 gene. These findings indicate that p21Waf1 promoter also can be activated by BRCA1 in the absence of wild-type p53, providing additional means by which BRCA1 can regulate p53-responsive genes. Although our present results argue against a generalized function of BRCA1 in gene transcription, we have obtained evidence that at least one other p21Waf1 promoter motif, a signal transducers and activators of transcription (STAT) binding serum-inducible element, can be activated in response to BRCA1 (T.O. and H.H., unpublished data). Therefore, BRCA1 may have different target motifs for activation. The choice of these motifs may vary according to the presence of transcription factors in different cell types. Somasundaram et al. (25) also demonstrated a critical role for BRCA1 C-terminal region, showing that germ-line mutations abolish activation of the p21Waf1 promoter.
Evidence that BRCA1 can be found in a complex with Rad51 has suggested a function of BRCA1 in maintaining genome integrity, possibly in a transcription-dependent manner (32). Rad51 also has been shown to interact with p53 (33). It is not known whether p53/Rad51/BRCA1 may form a ternary complex that is involved in sensing DNA damage. Of note is that the C-terminal region of p53, the interaction site with BRCA1, has been shown to recognize damaged DNA (34, 35). It is not known how such complexes containing BRCA1 and p53 might be organized in vivo but their identification and characterization may be instrumental in understanding how BRCA1 acts as a coactivator for p53.
Acknowledgments
We thank M. Ouchi and Q. Wang for technical assistance, R. G. Roeder for careful reading of the manuscript, W.-H. Lee and all members of the Hanafusa and the Aaronson Laboratories for critical advice and discussion. A.N.A.M. is a Pew Fellow in the Biomedical Sciences and on leave from the Institute of Chemistry, Federal University of Rio de Janeiro. A.A. is a recipient of a postdoctoral fellowship from the National Science Foundation. This work was supported by Grant CA44356 (H.H.) and SPORE in breast cancer 1P50CA68425 (S.A.A.) from the National Cancer Institute.
ABBREVIATIONS
- BRCT
BRCA1 C-terminal
- GST
glutathione S-transferase
References
- 1.Hollstein M, Sidransky D, Vogelstein B, Harris C C. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 2.Ko L J, Prives C. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 3.Momand J, Zambetti G P, Olson D C, George D, Levine A J. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 4.Thut C J, Goodrich J A, Tjian R. Genes Dev. 1997;11:1974–1986. doi: 10.1101/gad.11.15.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gu W, Shi X-L, Roeder R G. Nature (London) 1997;387:819–823. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- 6.Lill N L, Grossman S R, Ginsberg D, DeCaprio J, Livingston D M. Nature (London) 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 7.Malkin D, Li F P, Strong L C, Fraumeni J F, Jr, Nelson C E, Kim D H, Kassel J, Gryka M A, Bischoff F Z, Tainsky M A, et al. Science. 1990;250:1233–1238. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- 8.Miki Y, Swensen J, Shattuck-Eidens D, Futreal A P, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett L M, Ding W, et al. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 9.Chen Y, Farmer A A, Chen C-F, Jones D C, Chen P-L, Lee W-H. Cancer Res. 1996;56:3168–3172. [PubMed] [Google Scholar]
- 10.Ruffner H, Verma I M. Proc Natl Acad Sci USA. 1997;94:7138–7143. doi: 10.1073/pnas.94.14.7138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y, Chen C-F, Riley D, Allred D C, Chen P-L, Von Hoff D, Osborne K, Lee W-H. Science. 1995;270:789–791. doi: 10.1126/science.270.5237.789. [DOI] [PubMed] [Google Scholar]
- 12.Scully R, Ganesan S, Brown M, DeCaprio J A, Cannistra S A, Feunteun J, Schnitt S, Livingston D M. Science. 1996;272:123–125. doi: 10.1126/science.272.5258.123. [DOI] [PubMed] [Google Scholar]
- 13.Gayther S A, Warren W, Mazoyer S, Russell P A, Harrington P A, Chiano M, Seal S, Hamoudi R, Van Rensburg E J, Dunning A M, et al. Nat Genet. 1994;11:428–433. doi: 10.1038/ng1295-428. [DOI] [PubMed] [Google Scholar]
- 14.Futreal P A, Liu Q, Shattuck-Eiden D, Cochran C, Harshman K, Tavtigian S, Bennett L M, Haugen-Strano A, Swensen J, Miki Y, et al. Science. 1994;266:120–122. doi: 10.1126/science.7939630. [DOI] [PubMed] [Google Scholar]
- 15.Holt J T, Thompson M E, Szabo C, Robinson-Benion C, Arteaga C L, King M-C, Jensen R A. Nat Genet. 1996;12:298–302. doi: 10.1038/ng0396-298. [DOI] [PubMed] [Google Scholar]
- 16.Bork P, Hofmann K, Bucher P, Neuwald A F, Altschul S F, Koonin E V. FASEB J. 1997;11:68–76. [PubMed] [Google Scholar]
- 17.Chapman M, Verma I M. Nature (London) 1996;382:678–679. doi: 10.1038/382678a0. [DOI] [PubMed] [Google Scholar]
- 18.Monteiro A N A, August A, Hanafusa H. Proc Natl Acad Sci USA. 1996;93:13595–13599. doi: 10.1073/pnas.93.24.13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez J, Georgoff I, Martinez J, Levine A J. Genes Dev. 1991;5:151–159. doi: 10.1101/gad.5.2.151. [DOI] [PubMed] [Google Scholar]
- 20.Jacks T, Remington L, Williams B O, Schmitt E M, Halachmi S, Bronson R T, Weinberg R A. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 21.El-Deiry W S, Tokino T, Veculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 22.Wu X-W, Bayle J H, Olson D, Levine A J. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 23.Zambetti G, Bargonetti J, Walker K, Prives C, Levine A J. Genes Dev. 1992;6:1143–1152. doi: 10.1101/gad.6.7.1143. [DOI] [PubMed] [Google Scholar]
- 24.Helin K, Wu C-L, Fattaery A, Lees J A, Dynlacht B D, Ngwu C, Harlow E. Genes Dev. 1993;7:1850–1861. doi: 10.1101/gad.7.10.1850. [DOI] [PubMed] [Google Scholar]
- 25.Somasundaram K, Zang H, Zeng Y-X, Houvras Y, Peng Y, Zhang H, Wu G S, Licht J, Weber B L, El-Deiry W S. Nature (London) 1997;389:187–190. doi: 10.1038/38291. [DOI] [PubMed] [Google Scholar]
- 26.Scully R, Anderson S F, Chao D M, Wei W, Ye L, Young R A, Livingston D M, Parvin J D. Proc Natl Acad Sci USA. 1997;94:5605–5610. doi: 10.1073/pnas.94.11.5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monteiro A N A, August A, Hanafusa H. Am J Hum Genet. 1997;61:761–762. doi: 10.1086/515515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hupp T R, Sparko A, Lane D P. Cell. 1995;83:237–245. doi: 10.1016/0092-8674(95)90165-5. [DOI] [PubMed] [Google Scholar]
- 29.Hupp T R, Lane D P. Curr Biol. 1994;4:865–875. doi: 10.1016/s0960-9822(00)00195-0. [DOI] [PubMed] [Google Scholar]
- 30.Meek D. Cancer Biol. 1994;5:203–210. [PubMed] [Google Scholar]
- 31.Gu W, Roeder R G. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 32.Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, Ashley T, Livingston D M. Cell. 1997;88:265–275. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- 33.Sturzbecher H-W, Donzelmann B, Henning W, Knippschild U, Buchop S. EMBO J. 1996;15:1992–2002. [PMC free article] [PubMed] [Google Scholar]
- 34.Jayaraman L, Prives C. Cell. 1995;81:1021–1029. doi: 10.1016/s0092-8674(05)80007-8. [DOI] [PubMed] [Google Scholar]
- 35.Lee S, Elenbass B, Levine A, Griffith J. Cell. 1995;81:1013–1020. doi: 10.1016/s0092-8674(05)80006-6. [DOI] [PubMed] [Google Scholar]