Abstract
Members of the saframycin/safracin/ecteinascidin family of peptide natural products are potent antitumor agents currently under clinical development. Saframycin MX1, from Myxococcus xanthus, is synthesized by a nonribosomal peptide synthetase, SafAB, and an O-methyltransferase, SafC, although other proteins are likely involved in the pathway. SafC was overexpressed in Escherichia coli, purified to homogeneity, and assayed for its ability to methylate a variety of substrates. SafC was able to catalyze the O-methylation of catechol derivatives but not phenols. Among the substrates tested, the best substrate for SafC was l-dihydroxyphenylalanine (l-dopa), which was methylated specifically in the 4′-O position (kcat/Km = 5.5 × 103 M−1 s−1). SafC displayed less activity on other catechol derivatives, including catechol, dopamine, and caffeic acid. The more labile l-5′-methyldopa was an extremely poor substrate for SafC (kcat/Km = ∼2.8 × 10−5 M−1 s−1). l-Dopa thioester derivatives were also much less reactive than l-dopa. These results indicate that SafC-catalyzed 4′-O-methylation of l-dopa occurs prior to 5′-C-methylation, suggesting that 4′-O-methylation is likely the first committed step in the biosynthesis of saframycin MX1. SafC has biotechnological potential as a methyltransferase with unique regioselectivity.
Isoquinoline-containing natural products are widely used in biology and medicine because of their potent activities in a large number of biological assays (4, 7, 13, 21). Saframycins represent a class of isoquinoline alkaloids exhibiting antibiotic and antitumor activities (1). Since the original report of these molecules, a large number of analogs have been isolated, including saframycins and safracins from bacteria, renieramycins from sponges (phylum Porifera), and ecteinascidins from ascidians (phylum Chordata) (16, 19, 20, 29). The compounds interact with DNA, forming covalent bonds that are thought to be important in their cytotoxicity (37). Other mechanisms of action, either dependent upon or independent of DNA binding, have been reported (11). Interest in these molecules has been fueled by the advancement of ecteinascidin-743 to phase II/III clinical trials (12). Robust synthetic and semisynthetic approaches have been described to obtain ecteinascidin-743, which otherwise must be harvested by aquaculture or from wild stocks of the ascidian (9, 10). In order to improve the ability to synthesize analogs, a genetic approach has also been taken to modify the safracin biosynthetic pathway (33). Additional understanding of saframycin biosynthetic machinery and mechanisms would greatly aid in developing mutational approaches for the production of novel isoquinolines.
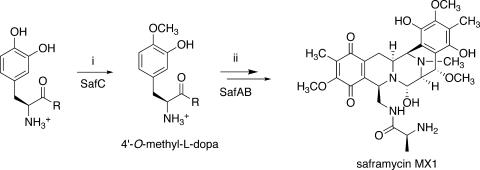
A number of precursor feeding studies with the saframycin family of metabolites have shown that the core structure is composed of amino acids (Fig. 1) (2). For example, safracins are composed of Ala, Gly, and two Tyr subunits, which are further elaborated by oxidative steps and by methylation with S-adenosylmethionine (33). Other biosynthetic steps, such as the folding of the tetrapeptide precursor into the final compound, remain obscure. Alternative possible routes to heterocycle formation include either Pictet-Spengler (P-S) or Bischler-Napieralski condensation (14).
FIG. 1.
Proposed biosynthetic route to saframycin MX1. (i) SAM methylation. (ii) NRPS-catalyzed formation of a tetrapeptide intermediate. This intermediate is folded by a Pictet-Spengler or Bischler-Napieralski condensation into a saframycin precursor. Further methylation and oxidation lead to the formation of saframycin MX1. This study defines intermediate 4′-O-methyl-l-dopa as the first committed intermediate; previously, the timing of C- versus O-methylation was unclear.
In 1995, Pospiech et al. reported the first biosynthetic gene cluster for isoquinoline biosynthesis, the saframycin MX1 (saf) cluster from Myxococcus xanthus (28). This cluster was described to consist of two nonribosomal peptide synthetase (NRPS) coding sequences, safA and safB, and a putative O-methyltransferase sequence, safC. When either the methyltransferase or safAB was knocked out, saframycin MX1 production was abolished in M. xanthus (27). However, when NRPS and methyltransferase mutants were cocultivated, antibacterial activity was restored. This result was interpreted to indicate that the SafC O-methyltransferase acts on a precursor molecule, which is then incorporated by SafAB into the saframycin structure. More recently, a homologous pathway to safracins was reported by Velasco et al. (33). Despite the similarity of NRPS proteins, the predicted modifying enzymes, including methyltransferases, are not homologous to those of saframycin MX1.
SafC is homologous to a variety of O-methyltransferases, particularly those involved in the methylation of catechols. Catechol O-methyltransferases (COMTs) are widely important in biology, agriculture, and medicine. For example, they are involved in the degradation of dopamine and estrogen as well as in the synthesis of caffeic acid and important plant natural products (5, 6, 15, 38). The characterized COMTs are primarily 3′-O-methyltransferases, while some produce a mixture of both 3′- and 4′-O-methylated products (24). Such 4′-O-methylation seems to play a role in the metabolism of anthocyanins and catechins (18, 23). There are two examples of methyltransferases selective for the para-hydroxyl group in benzylisoquinoline alkaloids in plants, although this methylation takes place after the Pictet-Spengler folding step (25, 39). Because l-dihydroxyphenylalanine (l-dopa) is O-methylated at the 4′ position in saframycins, SafC was potentially a new catalyst that could find use in the study of catechol metabolism and natural product biosynthesis. As part of our biochemical analysis of the saf pathway, we report the kinetic characterization of the SafC O-methyltransferase.
MATERIALS AND METHODS
General.
Chemicals were purchased from Sigma-Aldrich, except for derivatives of l-dopa, which were synthesized as described below or in a previous work (32). For all catechol derivatives with the exception of thioesters, authentic unmethylated, 3-O-methylated, and 4-O-methylated standards were available and used for regioselectivity studies. S-[methyl-3H]Adenosyl-l-methionine was purchased from PerkinElmer. High-pressure liquid chromatography (HPLC) was performed using a Hitachi L6200 intelligent pump and L3000 photo diode array detector with either a 100-Å Microsorb MV C18 column (Varian) or a Phenomenex Gemini 5-μm C18 column (250 by 4.6 mm). Thin-layer chromatography (TLC) analysis was performed using aluminum-backed silica plates with 13:5:2 butanol-water-acetic acid as the mobile phase and ninhydrin-based detection. M. xanthus Mx ×48 was provided by H. Reichenbach.
N-(tert-Butyloxycarbonyl)-3,4-dihydroxy-l-phenyalanine thiophenolate.
l-Dopa (300 mg, 1.5 mmol) was added to aqueous NaOH (1 M, 1.5 ml) in 1,4-dioxane (3 ml). The resulting solution was stirred for 10 min at 4°C, and then di-tert-butyldicarbonate (330 mg, 1.5 mmol) was added. The reaction was stirred overnight at 25°C. A rotary evaporator was used to remove dioxane, and the remaining solution was acidified with 1 ml of 1 N HCl and extracted with ethyl acetate. The crude product was dried under Ar and then dissolved in N,N-dimethylformamide (200 μl). A solution of (benzotriazol-1-yloxy)tripyrrolidinophosphonium hexafluorophosphate (210 mg, 0.40 mmol) and benzenethiol (50 μl, 0.41 mmol) in N,N-dimethylformamide (200 μl) was then added, and the resulting mixture was stirred for 1 h at 25°C. K2CO3 (27.6 mg, 0.20 mmol) was added, and the reaction was stirred for 2 h at 25°C. Phosphate buffer (1 ml, 10 mM, pH 6) was added, and the mixture was extracted with ethyl acetate. Following column chromatography (silica; 30 to 50% ethyl acetate in hexane gradient), Boc-l-dopa thiophenolate (35.0 mg, 90 μmol, 6% yield over two steps) was obtained. 1H nuclear magnetic resonance (NMR), (CDCl3, 400 MHz) δ 7.3 to 7.4 (5H, m), 6.78 (1H, d, J = 8 Hz), 6.67 (1H, d, J = 2 Hz), 6.57 (1H, dd, J = 2, 8 Hz), 5.12 Hz (1H, d, J = 8.8 Hz), 4.66 (1H, m), 3.00 (2H, m), 1.41 (9H, s); 13C NMR (CDCl3, 125 MHz) δ 200.1, 155.5, 144.1, 143.2, 134.6, 129.5, 129.2, 127.7, 127.0, 121.6, 116.2, 115.4, 81.0, 60.6, 37.8, 28.3; high-resolution fast atom bombardment mass spectrum, m/z = 390.1372 [M+H]+, C20H24NO5S requires m/z = 390.1375 (Δ = −0.3 ppm).
Synthesis of l-dopa-CoA.
Boc-l-dopa-thiophenolate (4.8 mg, 0.01 mmol) was suspended in 1:1 dichloromethane-trifluoroacetic acid (0.2 ml) and stirred for 30 min at ambient temperature. Following the removal of solvent under a stream of Ar, the crude product was dissolved in a solution of coenzyme A (CoA) sodium salt (8.2 mg, 0.01 mmol) in aqueous phosphate buffer (200 μl, 10 mM, pH 8). The resulting mixture was stirred for 2.5 h at 25°C and then injected onto a Phenomenex C18 analytical HPLC column with a 1-ml/min flow rate. The mobile phase consisted of 10 mM aqueous ammonium acetate and acetonitrile. The major compound at 280 nm was collected, lyophilized to yield l-dopa-CoA as an acetate salt (3.9 mg, 0.004 mmol, 35%), and confirmed by 1H NMR and mass spectrometry analyses: 1H NMR, (CD3OD, 400 MHz) δ 8.56 (1H, s), 8.18 (1H, s), 6.75 (1H, s), 6.67 (1H, d, J = 8.4 Hz), 6.53 (1H, d, J = 7 Hz), 6.11 (1H, d, J = 6.4 Hz), 4.79 (1H, m), 4.47 (1H, br), 4.27 (2H, br), 4.18 (1H, m), 4.08 (1H, s), 3.99 (1H, dd, J = 5.2, 8.4 Hz), 3.60 (1H, m), 3.54 (1H, m), 3.39 (1H, s), 3.25 (1H, m), 3.14 (1H, m), 2.97 (3H, m), 2.41 (2H, m), 1.91 (3H, s), 1.28 (1H, t, J = 7.2 Hz), 1.17 (1H, t, J = 7.2 Hz), 1.06 (3H, s), 0.84 (3H, s); matrix-assisted laser desorption ionization-time of flight mass spectrum, m/z = 947.36 [M+H]+, C30H46N8O19P3S requires m/z = 947.17.
Bacterial strains, plasmids, and culture conditions.
Escherichia coli strain DH5α was used for routine subcloning and was transformed as described by Sambrook and Russell (31). For protein expression, E. coli BL21(DE3) competent cells were used with standard procedures (Novagen). To construct the vector harboring the SafC gene, PCR was performed with Pfu DNA polymerase (Stratagene), using primers SafCF (5′-GGAATTCCATATGATCCACCACGTCGAATTGACACAG-3′, NdeI site underlined) and SafCR (5′-GGGAAGCTTGCGCTTGCGAGCGAGGGT-3′, HindIII site underlined). Amplification was achieved with 30 cycles of denaturation at 94°C for 30 s, annealing at 53°C for 30 s, and extension at 72°C for 5 min. The 663-bp PCR product was recovered by 1% agarose gel electrophoresis. After digestion with NdeI and HindIII, the fragment was inserted into pET20b to generate pSafC with a C-terminal hexahistidine tag.
Protein expression and purification.
E. coli BL21(DE3) harboring pSafC was grown at 37°C in a shake flask in LB medium in the presence of 50 μg/ml of ampicillin. Overnight cultures were diluted to 1:1,000 in fresh medium and grown to an optical density at 600 nm of 0.6 until the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) at a final concentration of 1 mM. Induction was continued for 2 h, and the cells were harvested.
The cell pellet was resuspended in 10 ml lysis buffer (50 mM Tris-HCl, 500 mM NaCl, and 10 mM imidazole, pH 8.0). Cells were disrupted by sonication, and the lysate was clarified by centrifugation (45 min at 14,400 × g). The protein was purified on Ni-nitrilotriacetic acid resin (QIAGEN). SafC was eluted with a linear gradient of imidazole from 20 to 250 mM and then analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Fractions containing pure protein were dialyzed against buffer (50 mM Tris-HCl, 0.2 mM MgCl2, 2 mM dithiothreitol [DTT], and 10% glycerol, pH 7.5). After overnight dialysis, the samples were quantified by the Bradford protein dye assay, aliquoted, and stored frozen at −80°C until use.
Large-scale SafC reaction monitored by 1H NMR.
l-Dopa-CoA was used in a large-scale reaction with SafC in D2O in an NMR tube. SafC (7.5 μg) was added to 10 mM (pH 8.0) phosphate buffer (450 μl) with S-adenosylmethionine (SAM; 1 mg) and l-dopa-CoA (1 mg). The reaction was monitored by 1H NMR (D2O, 400 MHz).
HPLC characterization of SafC regioselectivity.
SafC (1.6 μM) and either l-dopa (500 μM) or l-dopa-CoA (200 μM) were added to HEPES (10 mM, pH 7.2), MgCl2 (200 μM), DTT (2 mM), and SAM (200 μM) in 50-μl reaction volumes. Enzyme reaction mixtures were incubated overnight at 37°C. Reaction quenching and thioester hydrolysis were accomplished by adding 5.5 μl of 1 M Na2HPO4 and boiling the reaction mix for 15 min. The reaction mixtures were then frozen at −80°C until analyzed by HPLC. Appropriate standards and the products of SafC-l-dopa-CoA and SafC-l-dopa reactions were resolved using a 100-Å Microsorb MV C18 column (Varian) in an aqueous solvent consisting of 80.3 ml of methanol, 75 ml of 1 M NaH2PO4, 22.5 ml of 1 M EDTA, and 695.3 ml of H2O. Additional l-dopa reactions and standards were resolved using a Phenomenex Gemini 5-μm C18 column (250 by 4.6 mm) in an aqueous buffer: 960 ml of 40 mM MgCl2, pH 3.0, and 40 ml of acetonitrile.
l-Tyr reactivity was assessed in overnight reactions as with l-dopa, except that reactions were not quenched prior to HPLC analysis. Similar reactions of SafC with dopamine and caffeic acid were also performed, and the products were resolved and analyzed by HPLC: caffeic acid, isocratic in 125 ml of acetonitrile and 375 ml of 10.42 mM MgCl2, pH 3.0; dopamine, gradient run in acetonitrile and 10.42 mM MgCl2, pH 3.0.
SafC kinetics.
4′-O-Methyl-l-dopa, 5′-methyl-l-dopa, and 4′-O-methyl-5′-methyl-l-dopa were previously synthesized (32). SafC (0.78 μM) and various concentrations of substrate were incubated with HEPES (10 mM, pH 7.2), MgCl2 (100 μM), DTT (2 mM), and SAM (400 μM) at 37°C. The optimum buffer composition and pH were determined by various parameters to achieve maximum reaction rates with l-dopa. Optimum SAM concentrations were determined by varying the amount of SAM under conditions of saturating l-dopa (507 μM) to measure an approximate Vmax and Km. Since the Vmax was achieved at a SAM concentration of ∼300 μM, a saturating amount of SAM was used in all reactions. Following incubation, reaction vials were frozen in liquid nitrogen. Reaction products were resolved using a Phenomenex Gemini 5-μm C18 column (250 by 4.6 mm). D-6000 HPLC Manager software (Hitachi) was used for the integration of product peaks, and KaleidaGraph 4.0 (Synergy Software) was used to graph and fit data by using the Michaelis-Menten approximation.
Triplicate l-dopa reactions were run for 20 min at concentrations of 25 μM, 50 μM, 75 μM, 190 μM, 300 μM, 370 μM, and 480 μM. HPLC product resolution of l-dopa reactions was accomplished in a pH 3 aqueous solvent (960 ml of 40 mM MgCl2, pH 3.0, and 40 ml of acetonitrile). Reactions with catechol were run similarly; however, triplicate reactions were each run for a period of 50 min at substrate concentrations of 36 μM, 270 μM, 900 μM, 2,200 μM, and 3,100 μM. Product resolution of catechol reactions was performed in an aqueous buffer consisting of 125 ml of acetonitrile and 375 ml of 40 mM MgCl2 at pH 3.0.
Approximate kinetics for SafC reacting with 5′-methyl-l-dopa were obtained using single, 60-min reactions at substrate concentrations of 1 mM, 2 mM, 5 mM, 10 mM, 18 mM, and 24 mM. The HPLC solvent for resolving these later reaction products was 187 ml of 40 mM MgCl2 at pH 3 combined with 17 ml of acetonitrile. Standard curves for reactions with all three substrates were produced using 4′-O-methyl-l-dopa, 4′-O-methyl-5′-methyl-l-dopa, and guaiacol.
For caffeic acid and dopamine, similar reactions were run under various substrate concentrations of 25 μM, 50 μM, 75 μM, 190 μM, 300 μM, 370 μM, 480 μM, and 800 μM. Reactions were analyzed by HPLC in 125 ml of acetonitrile and 375 ml of 10.42 mM MgCl2, pH 3.0. The dopamine reactions were similarly analyzed but with an acetonitrile gradient.
l-Dopa-CoA reactivity.
Kinetic runs were performed using l-dopa-CoA, following the method used for l-dopa, except that reactions were quenched by boiling in various bases prior to HPLC analysis. Additionally, a series of runs was performed in which l-dopa-CoA hydrolysis to l-dopa and CoA was measured using Ellman's reagent at 25°C. A 1-ml solution containing 100 μM MgCl2, 10 mM HEPES, pH 7.2, 30 μl of ethanol, and 2 mM Ellman's reagent was used to zero a UV spectrophotometer. To this solution was added 10 μl of dopa-CoA (1 mg ml−1). Absorbance at 412 nm was monitored every 1 min for 30 min and then after 60 min and 120 min. A standard curve was prepared using the same buffer containing free CoA.
l-Dopa-CoA kinetics.
SafC was incubated in 100 μM MgCl2, 2 mM DTT, 400 μM SAM, 10 mM HEPES, pH 7.2, 8 fmol (0.5 μCi) [3H]SAM, and different concentrations of either l-dopa or l-dopa-CoA in a total volume of 50 μl at 37°C for 20 min. After incubation, the entire reaction volume was spotted onto a C8 RP-8 F254S TLC plate (Merck). The TLC plates were developed in a solvent of n-butanol-water-acetic acid (13:5:2). Following drying, the plate was sprayed with En3Hance (PerkinElmer) and exposed to X-Omat AR film (Kodak) for 17 h. Radiolabeled spots were scraped and quantified using a liquid scintillation counter (Packard).
RESULTS
Sequence analysis.
BLASTP searches (http://www.ncbi.nlm.nih.gov) revealed that SafC is closely related to COMTs (∼58% identity at the amino acid level) and to several methyltransferases from natural product biosynthetic pathways (∼55% identity at the amino acid level). Using a neighbor-joining tree algorithm available in the ClustalX 1.81 software package (National Center for Biotechnology Information), a phylogenetic tree was generated for SafC and relatives. SafC does not branch with either the COMTs or the natural product genes but instead appears to occupy a distinct branch within this group of proteins.
Related COMTs have been kinetically characterized and shown to specifically 3′-O-methylate caffeoyl-CoA derivatives (35, 36). Hoffmann et al. developed a threading model for the Nicotiana COMT protein, and specific residues were proposed to be responsible for catechol versus CoA binding (17). Those authors then tested their predictions by systematically mutating residues and feeding truncated caffeoyl-CoA analogs to mutant proteins. Importantly, this COMT could not accept unmodified caffeic acid as a substrate. Taken together, the data indicated that specific, predictable residues may be responsible for binding of the CoA motif.
In contrast to the COMTs, much less is known about the related natural product O-methyltransferases found in bacteria. The closest bacterial relatives seem to be involved in the methylation of hydroxymalonyl-acyl carrier protein or hydroxymalonyl-CoA. Indeed, the residues identified by Hoffmann et al. (17) as being important for CoA binding appear to be universal within this group of enzymes, and thus, we predicted that SafC should methylate l-dopa-thioester derivatives specifically. It was predicted that l-dopa-CoA or l-dopa-peptidyl carrier protein (PCP) derivates should be specifically methylated. Since NRPS knockouts in M. xanthus were reported to methylate a substrate (27), the CoA derivative or a PCP thioester from an unclustered part of the pathway seemed likely, since thiolation by SafAB was no longer a possibility.
Synthesis of thioesters.
To mimic the putative CoA- or PCP-bound l-dopa, l-dopa-CoA thioester was synthesized. A variety of methods were attempted to directly thioesterify l-dopa, but they were unsuccessful. The method of Vitali et al. (34) was adapted for synthesis via a thiophenolate intermediate, leading to l-dopa-CoA via a transthiolation reaction. This product could be characterized with proton NMR and mass spectrometry, but its relative instability was problematic for additional characterization and long-term storage. Fresh l-dopa-CoA was therefore prepared immediately prior to assays. The N-acetylcysteamine (SNAC) thioester of l-dopa was prepared in a similar manner (data not shown).
Expression of SafC.
SafC with a C-terminal His6 tag was readily expressed to high levels in E. coli. The protein was also readily visualized by sodium dodecyl sulfate-polyacrylamide gel electrophoresis after transformation into Streptomyces lividans TK24 by using the pEM4-XH vector. Ni-nitrilotriacetic acid-purified SafC could be stored for several months at −80°C without a noticeable loss in activity. Stored aliquots of SafC were used in a series of experiments to determine substrate selectivity.
Regioselectivity of SafC.
l-Dopa-CoA was used in a large-scale reaction with SafC and SAM, and the reaction was monitored by 1H NMR. Peak evolution in the aromatic region was observed that corresponded to the production of 3-(3-hydroxy-4-methoxyphenyl)-l-alanine: δ 6.83 (1H, d, J = 8.4 Hz, C6-H) and δ 6.64 (1H, s, C2-H) (32). Peaks at δ 6.63 (1H, d, J = 8.4 Hz, C5-H) overlapped with a decreasing peak from the l-dopa-CoA substrate. Shift assignments were based upon comparison with authentic standards of l-dopa derivatives prepared during previous work by our lab (32), and thus, we could readily distinguish between methylation at oxygen and carbon, as well as regioselectivity, by this method. Further analysis described below revealed that SafC was likely acting directly on the hydrolyzed product, l-dopa, rather than on l-dopa-CoA itself.
HPLC methods were used to assess the products of SafC with either l-dopa or l-dopa-CoA. 4′-O-methylated dihydroxyphenylalanine products were observed from both substrates. No 3′-O-methyl or 5′-methyl products were observed from SafC acting on either substrate. In subsequent kinetics experiments with 5′-methyl-l-dopa, only 4′-O-methyltransferase activity was observed. However, when similar methods were used to test l-tyrosine as a substrate for SafC, no 4′-O-methyltransferase activity was detected.
SafC kinetics and substrate selectivity.
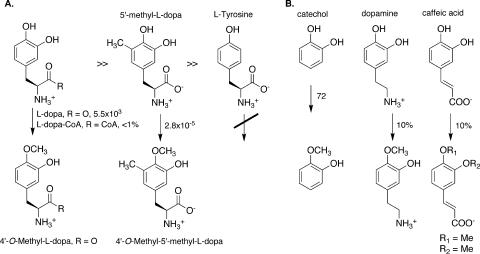
l-Dopa, l-dopa-CoA, catechol, and 5′-methyl-l-dopa were used to assess substrate selectivity (Table 1 and Fig. 2). To our surprise, l-dopa itself was the best substrate, with a kcat/Km of 5.5 × 103 M−1 s−1. Catechol incubated with SafC exhibited a kcat/Km of 72 M−1 s−1.
TABLE 1.
Kinetic constants for SafC
| Substrate | kcat (s−1) | Km (μM) | kcat/Km (M−1 s−1) |
|---|---|---|---|
| l-Dopa | 0.48 | 87 | 5.5 × 103 |
| 5′-Methyl-l-dopa | 1.2 × 10−7 | 4.4 × 103 | 2.8 × 10−5 |
| Catechol | 0.032 | 440 | 72 |
FIG. 2.
Substrate selectivity of SafC. (A) l-Dopa is a better substrate than methyldopa, and only catechols are substrates. (B) Catechol, dopamine, and caffeic acid are also substrates, although caffeic acid lacks the strict regioselectivity of the other substrates. The associated plain text numbers indicate kinetic constants (kcat/Km [M−1 s−1]) or relative percent activities in comparison to that of l-dopa (see the text). Me, methyl.
Numerous attempts to obtain kinetic constants for l-dopa-CoA by HPLC assay failed to yield consistent numbers. l-Dopa-CoA was unstable, completely hydrolyzing to l-dopa and coenzyme A in overnight reactions. The hydrolysis rate was difficult to precisely quantify because it was highly dependent upon substrate concentration and initial conditions, but at an 11 mM dopa-CoA concentration, ∼0.1% of the thioester decayed to free thiol per min at 25°C in the reaction buffer. Because of this hydrolysis problem and because we had no authentic standard of 4′-O-methyl-l-dopa-CoA, we used a comparative radiographic TLC method with l-dopa as a standard. Under these conditions, spots with approximately the same Rf as l-dopa, but also with Rfs quite close to the Rf of l-dopa-CoA, were detected by autoradiography and quantified by scintillation counting. By comparison with the same reaction applied to l-dopa, these spots indicated an approximate kcat/Km for l-dopa-CoA that was 100 times less than that observed for l-dopa. Because this rate was comparable in magnitude to estimates of the hydrolysis rate, it is probable that l-dopa-CoA does not serve as a substrate for SafC, but in any case, it is much slower than l-dopa. In trial runs with the SNAC derivative of l-dopa (data not shown), we were unable to observe 4′-O-methylation. Thioester derivatives are apparently inefficient substrates of SafC.
Authentic, enantiopure 5′-methyl-l-dopa was previously synthesized in our lab (32). Although this compound was much more light and base sensitive than l-dopa, it was sufficiently stable for approximate kinetic analysis. The C-methylated dopa derivative was a very poor substrate in comparison to l-dopa, with a kcat/Km approximately 2 × 108-fold lower than that for l-dopa. This decrease was due mainly to a greatly lowered kcat in comparison to that for the native substrate, although Km was also slightly increased. It is likely that steric interactions with the active site decrease the reaction rate, since the electronics of C-methyl-dopa still greatly favor methylation. The 4′-O-methyl product was the only regioisomer detected in the reaction.
Caffeic acid and dopamine were also substrates for SafC, but full kinetic data were not obtained (Fig. 2). Initial kinetic runs indicated that the reaction of dopamine and caffeic acid with SafC is non-Michaelis-Menten, possibly due to substrate inhibition. However, under optimal conditions, both substrates exhibited approximately 10% of the reaction rate of l-dopa at identical concentrations. For example, at 190 μM, dopamine was converted to 4′-O-methyldopamine at a velocity of 30 nM s−1 under the same conditions as those used for l-dopa. At 370 μM, caffeic acid was methylated at 40 nM s−1. For dopamine, only the 4′-O-methylated product was observed, but for caffeic acid, both the 4′- and 3′-O-methyl derivatives were produced at a 3:1 ratio, respectively.
DISCUSSION
Although sequence analysis indicates homology between SafC and the group of CoA/pantetheine-dependent methyltransferases, thiolation is not required, and l-dopa itself is the best substrate. The enzyme accepts a variety of catechol substrates, but simple phenol (l-Tyr) is not a substrate. SafC exhibits a regioselectivity different from that of most known catechol methyltransferases, since l-dopa is methylated exclusively at the 4′ oxygen. Steric interaction with the catechol moiety appears to be the most important factor for substrate selectivity. The replacement of a single proton with methyl at the 5′ carbon of the catechol greatly decreases the turnover number.
Because SafC operates most efficiently on l-dopa, at least in vitro, it is likely that free l-dopa is the natural in vivo substrate of the enzyme. l-Dopa-CoA is probably not a substrate and is at best an extremely poor substrate, and SNAC derivatives were inactive under these conditions. The removal of portions of the alaninyl side chain, as in catechol, caffeic acid, and dopamine, resulted in a loss of ≥90% of activity, substantiating l-dopa as the enzyme substrate. Further evidence for the intermediacy of l-dopa comes from the knockout experiments of Pospiech et al., who definitively showed that SafC does not act on an intermediate that is covalently bound to the SafAB NRPS (27). Because SafC reacts with l-dopa and not with simple phenols or 5′-methyldopa and because SafC is required for saframycin MX1 production, SafC 4′-O-methylation of l-dopa is likely the first committed step in the biosynthesis of saframycin MX1.
Velasco et al. reported the gene cluster for the production of safracins in Pseudomonas fluorescens (33). Three NRPS genes (sacABC) are homologous in sequence and domain order with safAB. However, there are seven other clustered sequences (sacDEFGHIJ) involved in precursor synthesis and tetrapeptide modification; in contrast, only one modifying gene (safC) is known to be involved in saframycin MX1 production. The two methyltransferases in the sac cluster are not homologous to SafC, and knockout experiments strongly suggest that they do not methylate l-dopa. Instead, they putatively C- and O-methylate free tyrosine, which is later oxidized to the dopa derivative. These differences indicate that the biosyntheses of safracins and saframycins do not follow identical routes. The related metabolite, naphthyridinomycin, contains a similar Tyr-derived unit. Feeding studies demonstrated that either 3′-methyltyrosine or 5′-methyldopa could be incorporated into naphthyridinomycin, indicating that multiple routes to bacterial isoquinoline alkaloids are possible (26).
The ability of SafC to 4′-O-methylate a variety of catechols has potential biotechnological applicability not just in the modification of the saframycin pathway but also in other areas of medicinal and biological research. In particular, dopa and dopamine methylations control the levels of these signaling molecules and are important in neurochemistry (3, 6). The ability of SafC to regioselectively methylate dopamine in a relatively efficient manner indicates that SafC may be a general catalyst of biotechnological importance. 4′-O-methylated dopa and dopamine have been proposed to be important toxins with possible involvement in neurological conditions such as Parkinson's disease (8, 22, 30). The availability of a recombinant, catechol 4′-O-methyltransferase could allow hypotheses about the importance of these molecules to be readily explored.
Acknowledgments
This work was supported by UU Startup funds to E.W.S. and an NIH Training Grant in Biological Chemistry awarded to J.T.N.
We are grateful to H. Reichenbach for the gift of M. xanthus.
Footnotes
Published ahead of print on 20 April 2007.
REFERENCES
- 1.Arai, T., K. Takahashi, K. Ishiguro, and Y. Mikami. 1980. Some chemotherapeutic properties of two new antitumor antibiotics, saframycins A and C. Gann 71:790-796. [PubMed] [Google Scholar]
- 2.Arai, T., K. Yazawa, K. Takahashi, A. Maeda, and Y. Mikami. 1985. Directed biosynthesis of new saframycin derivatives with resting cells of Streptomyces lavendulae. Antimicrob. Agents. Chemother. 28:5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Assicot, M., and C. Bohuon. 1970. Purification and studies of catechol-O-methyltransferase of rat liver. Eur. J. Biochem. 12:490-495. [DOI] [PubMed] [Google Scholar]
- 4.Aune, G. J., T. Furuta, and Y. Pommier. 2002. Ecteinascidin 743: a novel anticancer drug with a unique mechanism of action. Anticancer Drugs 13:545-555. [DOI] [PubMed] [Google Scholar]
- 5.Axelrod, J. 1966. Methylation reactions in the formation and metabolism of catecholamines and other biogenic amines. Pharmacol. Rev. 18:95-113. [PubMed] [Google Scholar]
- 6.Axelrod, J., and R. Tomchick. 1958. Enzymatic O-methylation of epinephrine and other catechols. J. Biol. Chem. 233:702-705. [PubMed] [Google Scholar]
- 7.Bentley, K. W. 2006. β-Phenylethylamines and the isoquinoline alkaloids. Nat. Prod. Rep. 23:444-463. [DOI] [PubMed] [Google Scholar]
- 8.Bonifati, V., and G. Meco. 1999. New, selective catechol-O-methyltransferase inhibitors as therapeutic agents in Parkinson's disease. Pharmacol. Ther. 81:1-36. [DOI] [PubMed] [Google Scholar]
- 9.Chen, J., X. Chen, M. Bois-Choussy, and J. Zhu. 2006. Total synthesis of ecteinascidin 743. J. Am. Chem. Soc. 128:87-89. [DOI] [PubMed] [Google Scholar]
- 10.Chen, X., J. Chen, M. De Paolis, and J. Zhu. 2005. Synthetic studies toward ecteinascidin 743. J. Org. Chem. 70:4397-4408. [DOI] [PubMed] [Google Scholar]
- 11.Cuevas, C., M. Perez, M. J. Martin, J. L. Chicharro, C. Fernandez-Rivas, M. Flores, A. Francesch, P. Gallego, M. Zarzuelo, F. de la Calle, J. Garcia, C. Polanco, I. Rodriguez, and I. Manzanares. 2000. Synthesis of ecteinascidin Et-743 and phthalascidin Pt-650 from cyanosafracin B. Org. Lett. 2:2545-2548. [DOI] [PubMed] [Google Scholar]
- 12.Fayette, J., I. R. Coquard, L. Alberti, H. Boyle, P. Meeus, A. V. Decouvelaere, P. Thiesse, M. P. Sunyach, D. Ranchere, and J. Y. Blay. 2006. Et-743: a novel agent with activity in soft-tissue sarcomas. Curr. Opin. Oncol. 18:347-353. [DOI] [PubMed] [Google Scholar]
- 13.Fischera, D. C. H., N. C. A. Gualda, D. Bachiega, C. S. Carvalho, F. N. Lupo, S. V. Bonotto, M. O. Alves, A. Yogi, S. M. Di Santi, P. E. Avila, K. Kirchgatter, and P. R. H. Moreno. 2004. In vitro screening for antiplasmodial activity of isoquinoline alkaloids from Brazilian plant species. Acta Trop. 92:261-266. [DOI] [PubMed] [Google Scholar]
- 14.Fodor, G., and S. Nagubandi. 1980. Correlation of the von Braun, Ritter, Bischler-Napieralski, Beckmann and Schmidt reactions via nitrilium salt intermediates. Tetrahedron 36:1279-1300. [Google Scholar]
- 15.Guldberg, H. C., and C. A. Marsden. 1975. Catechol-O-methyl transferase: pharmacological aspects and physiological role. Pharmacol. Rev. 27:135-206. [PubMed] [Google Scholar]
- 16.He, H. Y., and D. J. Faulkner. 1989. Renieramycin E and renieramycin F from the sponge Reniera sp. Reassignment of the stereochemistry of the renieramycins. J. Org. Chem. 54:5822-5824. [Google Scholar]
- 17.Hoffmann, L., S. Maury, M. Bergdoll, L. Thion, M. Erard, and M. Legrand. 2001. Identification of the enzymatic active site of tobacco caffeoyl-coenzyme A O-methyltransferase by site-directed mutagenesis. J. Biol. Chem. 276:36831-36838. [DOI] [PubMed] [Google Scholar]
- 18.Ichiyanagi, T., M. M. Rahman, Y. Kashiwada, Y. Ikeshiro, Y. Shida, Y. Hatano, H. Matsumoto, M. Hirayama, and T. Konishi. 2004. Absorption and metabolism of delphinidin 3-O-β-d-glucoside in rats. Biofactors 21:411-413. [DOI] [PubMed] [Google Scholar]
- 19.Ikeda, Y., H. Idemoto, F. Hirayama, K. Yamamoto, K. Iwao, T. Asao, and T. Munakata. 1983. Safracins, new antitumor antibiotics. I. Producing organism, fermentation and isolation. J. Antibiot. (Tokyo) 36:1279-1283. [DOI] [PubMed] [Google Scholar]
- 20.Irschik, H., W. Trowitzsch-Kienast, K. Gerth, G. Hofle, and H. Reichenbach. 1988. Saframycin Mx1, a new natural saframycin isolated from a myxobacterium. J. Antibiot. (Tokyo) 41:993-998. [DOI] [PubMed] [Google Scholar]
- 21.Jiang, B., K. Cao, and R. Wang. 2004. Inhibitory effect of protopine on KATP channel subunits expressed in HEK-293 cells. Eur. J. Pharmacol. 506:93-100. [DOI] [PubMed] [Google Scholar]
- 22.Kaakkola, S. 2000. Clinical pharmacology, therapeutic use and potential of COMT inhibitors in Parkinson's disease. Drugs 59:1233-1250. [DOI] [PubMed] [Google Scholar]
- 23.Lu, H., X. Meng, and C. S. Yang. 2003. Enzymology of methylation of tea catechins and inhibition of catechol-O-methyltransferase by (−)-epigallocatechin gallate. Drug Metab. Dispos. 31:572-579. [DOI] [PubMed] [Google Scholar]
- 24.Männistö, P. T., and S. Kaakkola. 1999. Catechol-O-methyltransferase (COMT): biochemistry, molecular biology, pharmacology, and clinical efficacy of the new selective COMT inhibitors. Pharmacol. Rev. 51:593-628. [PubMed] [Google Scholar]
- 25.Morishige, T., T. Tsujita, Y. Yamada, and F. Sato. 2000. Molecular characterization of the S-adenosyl-l-methionine:3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase involved in isoquinoline alkaloid biosynthesis in Coptis japonica. J. Biol. Chem. 275:23398-23405. [DOI] [PubMed] [Google Scholar]
- 26.Palaniswamy, V. A., and S. J. Gould. 1986. Incorporation of 3′-methyltyrosine and 5′-methyl-DOPA into naphthyridinomycin. J. Am. Chem. Soc. 108:5651-5652. [Google Scholar]
- 27.Pospiech, A., J. Bietenhader, and T. Schupp. 1996. Two multifunctional peptide synthetases and an O-methyltransferase are involved in the biosynthesis of the DNA-binding antibiotic and antitumour agent saframycin Mx1 from Myxococcus xanthus. Microbiology 142:741-746. [DOI] [PubMed] [Google Scholar]
- 28.Pospiech, A., B. Cluzel, J. Bietenhader, and T. Schupp. 1995. A new Myxococcus xanthus gene cluster for the biosynthesis of the antibiotic saframycin Mx1 encoding a peptide synthetase. Microbiology 141:1793-1803. [DOI] [PubMed] [Google Scholar]
- 29.Rinehart, K. L., T. G. Holt, N. L. Fregeau, J. G. Stroh, P. A. Keifer, F. Sun, L. H. Li, and D. G. Martin. 1990. Ecteinascidin-729, ecteinascidin-743, ecteinascidin-745, ecteinascidin-759a, ecteinascidin-759b, and ecteinascidin-770. Potent antitumor agents from the Caribbean tunicate Ecteinascidia turbinata. J. Org. Chem. 55:4512-4515. [Google Scholar]
- 30.Ruottinen, H. M., and U. K. Rinne. 1998. COMT inhibition in the treatment of Parkinson's disease. J. Neurol. 245:P25-P34. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 32.Schmidt, E. W., J. T. Nelson, and J. P. Fillmore. 2004. Synthesis of tyrosine derivatives for saframycin MX1 biosynthetic studies. Tetrahedron Lett. 45:3921-3924. [Google Scholar]
- 33.Velasco, A., P. Acebo, A. Gomez, C. Schleissner, P. Rodríguez, T. Aparicio, S. Conde, R. Muñoz, F de la Calle, J. L. Garcia, and J. M. Sánchez-Puelles. 2005. Molecular characterization of the safracin biosynthetic pathway from Pseudomonas fluorescens A2-2: designing new cytotoxic compounds. Mol. Microbiol. 56:144-154. [DOI] [PubMed] [Google Scholar]
- 34.Vitali, F., K. Zerbe, and J. A. Robinson. 2003. Production of the vancomycin aglycone conjugated to a peptide carrier domain derived from a biosynthetic non-ribosomal peptide synthetase. Chem. Commun. 21:2718-2719. [DOI] [PubMed] [Google Scholar]
- 35.Ye, Z. H., R. E. Kneusel, U. Matern, and J. E. Varner. 1994. An alternative methylation pathway in lignin biosynthesis in Zinnia. Plant Cell 6:1427-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ye, Z. H., and J. E. Varner. 1995. Differential expression of two O-methyltransferases in lignin biosynthesis in Zinnia elegans. Plant Physiol. 108:459-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zewail-Foote, M., V.-S. Li, H. Kohn, D. Bearss, M. Guzman, and L. H. Hurley. 2001. The inefficiency of incisions of ecteinascidin 743-DNA adducts by the UvrABC nuclease and the unique structural feature of the DNA adducts can be used to explain the repair-dependent toxicities of this antitumor agent. Chem. Biol. 8:1033-1049. [DOI] [PubMed] [Google Scholar]
- 38.Zhu, B. T., and A. H. Conney. 1998. Is 2-methoxyestradiol an endogenous estrogen metabolite that inhibits mammary carcinogenesis? Cancer Res. 58:2269-2277. [PubMed] [Google Scholar]
- 39.Ziegler, J., M. L. Diaz-Chavez, R. Kramell, C. Ammer, and T. M. Kutchan. 2005. Comparative macroarray analysis of morphine containing Papaver somniferum and eight morphine free Papaver species identifies an O-methyltransferase involved in benzylisoquinoline biosynthesis. Planta 222:458-471. [DOI] [PubMed] [Google Scholar]