Abstract
To identify secreted virulence factors involved in bacterial wilt disease caused by the phytopathogen Ralstonia solanacearum, we mutated tatC, a key component of the twin-arginine translocation (Tat) secretion system. The R. solanacearum tatC mutation was pleiotropic; its phenotypes included defects in cell division, nitrate utilization, polygalacturonase activity, membrane stability, and growth in plant tissue. Bioinformatic analysis of the R. solanacearum strain GMI1000 genome predicted that this pathogen secretes 70 proteins via the Tat system. The R. solanacearum tatC strain was severely attenuated in its ability to cause disease, killing just over 50% of tomato plants in a naturalistic soil soak assay where the wild-type parent killed 100% of the plants. This result suggested that elements of the Tat secretome may be novel bacterial wilt virulence factors. To identify contributors to R. solanacearum virulence, we cloned and mutated three genes whose products are predicted to be secreted by the Tat system: RSp1521, encoding a predicted AcvB-like protein, and two genes, RSc1651 and RSp1575, that were identified as upregulated in planta by an in vivo expression technology screen. The RSc1651 mutant had wild-type virulence on tomato plants. However, mutants lacking either RSp1521, which appears to be involved in acid tolerance, or RSp1575, which encodes a possible amino acid binding protein, were significantly reduced in virulence on tomato plants. Additional bacterial wilt virulence factors may be found in the Tat secretome.
Extracellular proteins secreted by plant pathogenic bacteria form a molecular arsenal that facilitates niche adaptation, host invasion, and evasion of plant defenses. The consortium of plant cell wall-degrading enzymes produced by the soft rot erwiniae, which are secreted via the type II secretion system (23), offers an example of how bacterial secreted proteins work together during plant pathogenesis. Agrobacterium tumefaciens uses a specialized type IV secretion system to deliver Ti-DNA that transforms plant host cells (8). Many plant pathogenic bacteria use host contact-mediated type III secretion systems to optimize the host environment and suppress plant defense responses (9). Cooperatively secreted proteins are critical for the pathogenicity and virulence of bacterial plant pathogens (31).
Ralstonia solanacearum, a widely distributed soilborne plant pathogen, causes bacterial wilt disease, which affects such economically important crops as potato, tomato, and banana (18). Bacterial wilt pathogenesis is incompletely understood. However, it is known that contributing factors are controlled by interlocking environmentally responsive regulatory cascades and that disease development depends on the action of several proteins secreted by the type III and type II secretion systems (35). Type II-secreted factors include the plant cell wall-degrading pectinases (PehA, PehB, PehC, and Pme), an endoglucanase (Egl), and extracellular polysaccharide (EPS); these individually contribute quantitatively to pathogen success in colonization and subsequent disease development (1, 11, 15, 33). However, several additional as-yet-unidentified virulence factors must be secreted by R. solanacearum in a type II-dependent manner because a gspK type II secretion mutant is significantly less virulent than a pyramided mutant lacking six of the previously identified type II-dependent virulence factors (24, 26). Analysis of the R. solanacearum strain GMI1000 genome sequence identified over 200 candidate virulence proteins; we hypothesize that among these are many secreted proteins that contribute to virulence (34).
In gram-negative bacteria, extracellular proteins must first be secreted through the cytoplasmic membrane into the periplasm prior to their secretion out of the bacterial cell by the type II system. Proteins move into the periplasm via one of two parallel secretion systems: Sec and twin-arginine protein translocation (Tat) (3, 32). Tat secretion is a Sec-independent, ATP-independent mechanism of protein translocation. Proteins secreted by Tat are characterized by a signature twin-arginine motif (S-R-R-x-F-L-K) located at the N terminus (37). Relative to their Sec-secreted counterparts, Tat-secreted proteins have a longer signal peptide (14 amino acids longer on average), decreased hydrophobicity in the hydrophobic central domain (h-region), and a basic or uncharged, but seldom acidic, polar carboxy-terminal region (c-region). A genomic survey of available bacterial genomes found putative Tat system genes in pathogens of both plants (Xanthomonas campestris, X. axonopodis, and Xyllela fastidiosa) and animals (Salmonella enterica serovar Typhi, Yersinia pestis, Neisseria meningitidis, and Vibrio cholerae) (12). A functional Tat system is essential for establishing relationships with eukaryotic hosts both pathogenic (Pseudomonas aeruginosa, P. syringae, Escherichia coli O157:H7, and A. tumefaciens) and symbiotic (Rhizobium leguminosarum) (5, 7, 13, 28, 29). These results suggested that that the Tat system might also play a role in R. solanacearum virulence. To test this hypothesis we mutated the tatC locus of R. solanacearum, which is predicted to encode an indispensable component of the Tat secretion system.
We used a 2003 genomic survey of putative Tat-secreted proteins in published bacterial genomes (12) to identify potential virulence factors among R. solanacearum candidate Tat-secreted proteins. Several R. solanacearum proteins predicted to be secreted by Tat are virulence factors in other host-pathogen systems. In P. aeruginosa, phospholipases PlcH and PlcN, which degrade host cellular lipids, are secreted by Tat and contribute significantly to virulence (28, 42). In E. coli O157:H7, Shiga toxin Stx1 synthesis is severely affected in a ΔtatABC mutant (29). Thus, we further hypothesized that additional virulence factors await identification among the R. solanacearum Tat-secreted proteins. In this report we characterize an R. solanacearum tatC mutant and describe the individual contributions to bacterial wilt virulence of two novel predicted Tat-secreted R. solanacearum proteins.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Strains and plasmids used in this research
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| Escherichia coli strain | ||
| DH5α | F−endA1 relA φ80lacZΔ supE44 thi-1 recA1 gyrA96 | 16 |
| Ralstonia solanacearum strains | ||
| GMI1000 | Wild-type phylotype I, biovar 3 | 4 |
| K60 | Wild-type phylotype II, biovar 1 | 25 |
| K60-409 | pehC::aphA-3; Kmr | 15 |
| K60TAT | tatC::pTatC(s); Kmr | This study |
| K1575 | RSp1575::aacC1; Gmr | This study |
| K1651 | RSc1651::aacC1; Gmr | This study |
| K1521 | RSp1521::aacC1; Gmr | This study |
| Plasmids | ||
| pSTblue-1 | Kmr Apr | Novagen |
| pLAFR3 | Tcr | 38 |
| pUCGM | Gmr Apr | 36 |
| pCR-1 | Kmr Zeor | Epicenter |
| pTatC(s) | 533-bp internal tatC fragment cloned into pCR-1; Kmr Zeor | This study |
| pSTTAT | 1.8-kb insert containing tatABC in pSTblue-1; Kmr Apr | |
| pLTat | 1.8-kb insert containing tatABC in pLAFR3; Tetr | This study |
| pST2131 | 701-bp internal RSp1575 fragment cloned into pSTblue-1; Kmr Apr | This study |
| pST2131Gm | pST2131 with 850-bp Gmr cassette insertion from pUCGM; Kmr Apr Gmr | This study |
| pST4028 | 1,185-bp insert containing RSc1651 in pSTblue-1; Kmr Apr Gmr | This study |
| pST4048Gm | pST4028 with 850-bp Gmr cassette insertion from pUCGM; Kmr Apr Gmr | This study |
| pST4798 | 1,809-bp insert containing RSp1521 in pSTblue-1; Kmr Apr | This study |
| pST4798Gm | pST4798 with 850-bp Gmr cassette insertion from pUCGM; Km Kmr Apr Gmr | This study |
| pLPehCRRKK | 3.2-kb insert in pLAFR3 containing R25K and R26K mutations PehC; Tetr | This study |
| pPehC11 | 3.8-kb BalI-Alw44I fragment containing pehC-exuT in pLAFR3; Tcr | 15 |
Ap, ampicillin; Tc, tetracycline; Km, kanamycin; Gm, gentamicin; Zeo, zeocin.
Cultivation of E. coli and R. solanacearum strains.
E. coli strains were grown in LB medium (27) at 37°C. R. solanacearum strains were grown at 28°C in either CPG broth (20) or CPG solid medium amended with 0.05% (wt/vol) tetrazolium chloride (25). Antibiotics were added to cultures when needed at the following concentrations: ampicillin, 50 μg/ml; kanamycin, 25 μg/ml; tetracycline, 15 μg/ml; and gentamicin, 12.5 μg/ml. To test for auxotrophic phenotypes and carbon source utilization, we used BMM (4) amended with 0.2% (wt/vol) glucose, 0.2% (wt/vol) polygalacturonate, or 4 mM of various amino acids. BMM amended with 0.2% (wt/vol) citrate and 0.1% (wt/vol) yeast extract was used to grow cells for extracellular exopolygalacturonase activity assays. Motility was assayed in plates containing 0.325% Noble agar and BMM plus 0.1% (wt/vol) citrate.
To measure the generation times of strains K1521 and K60, cultures were grown in BMM plus 0.2% glucose at pH 7.0 or pH 5.5 for 6 hours. Populations were enumerated by dilution plating at time zero and at 6 h, and the generation time was calculated. Bacterial sensitivity to detergents was measured by exposing equivalent-density CPG broth cultures of strains to a series of sodium dodecyl sulfide (SDS) concentrations (0, 0.01, 0.015, 0.02, and 0.025% [wt/vol]) overnight. Relative sensitivity was calculated as the optical density (A600) of the strain grown in the presence of SDS divided by the optical density (A600) of the same strain grown in the absence of SDS.
In planta growth of R. solanacearum was measured as multiplication in the apoplast of tobacco (Nicotiana tabacum cv. “Bottom Special”) as previously described; growth of this pathogen in tobacco leaf tissue is correlated with multiplication in tomato stems (40). We measured growth of R. solanacearum strains K1521, K1651, and K1575 in either CPG or BMM supplemented with different carbon sources as previously described (39). We measured growth of R. solanacearum wild-type K60 and K60TAT under oxygen-limiting conditions supplemented with 0.2% (wt/vol) KNO3 as described previously (19).
DNA manipulations.
All DNA manipulations, including cloning, Southern transfer, restriction mapping, sequencing, and PCR, were performed using standard procedures (2). R. solanacearum and E. coli electrocompetent cells were used as previously described (1). Unless otherwise noted, chemical reagents were purchased from Sigma-Aldrich (St. Louis, MO) and molecular biology reagents and kits were purchased from Promega (Madison, WI).
Oligonucleotides and sequence analysis.
DNA sequences were assembled using the Sequencher program (Gene Codes Corporation, Ann Arbor, MI). Oligonucleotides for PCR and sequencing were ordered from Sigma-Genosys (The Woodlands, TX), Integrated DNA Technologies (Coralville, IA), or the University of Wisconsin Biotechnology Center (Madison, WI). All DNA sequencing was performed at the University of Wisconsin Biotechnology Center (Madison, WI). Sequences of the oligonucleotide primers used to amplify tatC, tatABC, RSp1521, RSc1651, and RSp1575 are listed in Table 2.
TABLE 2.
Primers used in PCR amplification of Tat secretion system genes and putative Tat-secreted genes
| Primer | Target | Sequence |
|---|---|---|
| tatCshortL | Internal tatC | 5′-CTTCAACCTGTTCTCGCGTC-3′ |
| tatCshortR | 5′-CAACAGCTGCGACATCACAT-3′ | |
| TATORFL | tatABC | 5′-GCTGCCTTTTCTTTTGGATG-3′ |
| TATORFR | 5′-TATCTCAACGACAGCAAGCC-3′ | |
| Rs04798L | RSp1521 | 5′-TACATTGACTGATGTCGCGG-3′ |
| Rs04798R | 5′-GTAATCGCCGTCGAAGTGA-3′ | |
| Rs02131L | RSp1575 | 5′-ATTGCCGATATCAAGAAGCG-3′ |
| Rs02131R | 5′-AACCACTTTGTCTGCAGCG-3′ | |
| Rs04028L | RSc1651 | 5′-GTCGGAGGACATCATGGAAC-3′ |
| Rs04028R | 5′-CTACGCAAGCTGTGGCTG-3′ | |
| PehCKRKKFor | pehC | 5′-GCTCGCATTCGCCAGCGAAGAAGGCATTCGTCGTCTGGTC-3′ |
| PehCKRKKRev | 5′-GACCAGACGACGAATGCCTTCTTCGCTGGCGAATGCGAGC-3′ |
Construction of R. solanacearum tatC mutant K60TAT.
PCR primers were designed from the R. solanacearum GMI1000 sequence to amplify a 533-bp internal portion of the tatC gene from R. solanacearum strain K60 (Table 2). The resulting PCR product was cloned into pCR-1, creating the R. solanacearum suicide plasmid pTatC(s), which was introduced into R. solanacearum K60 via electroporation. A single exchange gene interruption mutant was selected, and its insertion into tatC was verified by Southern blot analysis.
Complementation of R. solanacearum K60TAT.
PCR primers were designed to amplify the entire tatABC operon from R. solanacearum GMI1000. The resulting 1.8-kb PCR product was cloned into pSTBlue-1, creating pSTTAT. The insert was sequenced and subcloned into broad-host-range vector pLAFR3, creating pLTAT. For trans complementation studies, plasmid pLTAT was introduced into both R. solanacearum wild-type and tatC mutant K60TAT cells by electroporation.
Construction of R. solanacearum mutant strains K1521, K1575, and K1652.
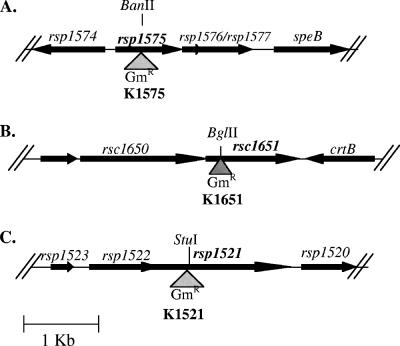
Open reading frames (ORFs) of RSp1521, RSp1575, and RSc1651 were PCR amplified from R. solanacearum K60 genomic DNA by use of primers listed in Table 2. The resulting PCR products were cloned into pSTBlue-1 and sequenced to confirm the identity of the cloned fragment. Each ORF was interrupted by inserting the gentamicin resistance cassette aacC1 from pUCGM, and the resulting mutagenesis constructs were individually introduced into R. solanacearum K60 via allelic replacement, as shown in Fig. 1. The resulting RSp1521, RSp1575, and RSc1651 mutants were called K1521, K1575, and K1651, respectively. Insertion location for all mutant strains was verified by Southern blot analysis.
FIG. 1.
Genomic context and mutagenesis strategies for R. solanacearum strains K1575, K1651, and K1521. (A) Gene map of RSp1575 mutant K1575. RSp1575 is directly upstream of RSp1576 and RSp1577, which are predicted to encode an ABC-type transporter. Further downstream is speB, which is predicted to encode an arginase. (B) Gene map of RSc1651 mutant K1651. RSc1651 is upstream of crtB, a gene predicted to encode a phytoene synthase; the genes are oriented in opposite directions. (C) Gene map of RSp1521 mutant K1521. RSp1521 is preceded by lpiA-like gene RSp1522; the two genes are overlapping and may therefore form an operon. RSp1575, RSp1651, and RSp1521 were mutated by the insertion of an aacC1 gentamicin resistance cassette (shaded triangle) into a unique restriction site in the gene of interest. Correct allelic replacements were confirmed by Southern blot analysis.
Microscopy.
To study cell morphology, a 10-microliter droplet containing R. solanacearum liquid cultures of K60 or K60TAT grown to mid-log phase was viewed under ×400 magnification using an Olympus BX60F-3 phase microscope. Images were collected using an Olympus digital camera (Olympus America, Inc.).
Site-directed mutagenesis of the PehC twin-arginine domain from RR to KK.
We used the QuikChange approach (Stratagene) to mutate the twin-arginine domain of PehC to R25K/R26K using primers PehCKRKKFor and PehCKRKKRev (Table 2). The mutated gene was sequenced to confirm the site changes and subcloned into pLAFR3, creating pLPehCRRKK for use in trans complementation studies.
PehC activity assay.
Release of galacturonic acid from polygalacturonate by the exopolygalacturonase PehC was measured as previously described by the separation and visualization of reaction products on thin-layer chromatography plates (21).
Virulence assays.
To compare virulence levels of mutant and wild-type R. solanacearum strains on the tomato host plant, we used a naturalistic soil soak inoculation as previously described (39). Fourteen- to 15-day-old plants (susceptible cv. Bonny Best) were inoculated to a soil density of approximately 1.56 × 107 to 3.12 × 107 CFU/g soil. Plants were monitored for disease progress over a 14-day period, and symptoms were scored on a 0-to-4 disease index by a rater blind to inoculation to generate a disease progress curve, where 0 indicates no disease, 1 indicates 1 to 25% of leaves wilted, 2 indicates 25 to 50% of leaves wilted, 3 indicates 51 to 75% of leaves wilted, and 4 indicates 76 to 100% of leaves wilted. Each experiment contained a minimum of 16 plants per treatment, and each assay included a minimum of three trials. Strain virulence levels were analyzed using a repeated-measures analysis of variance (40).
RESULTS
R. solanacearum GMI1000 has Tat secretion genes.
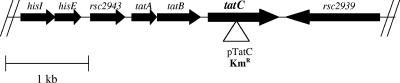
To determine if R. solanacearum contains homologs of the core genes encoding a Tat secretion system, we searched the R. solanacearum GMI1000 database (http://bioinfo.genopole-toulouse.prd.fr/annotation/iANT/bacteria/ralsto/) for genes similar to the well-characterized tatABC genes from P. aeruginosa and E. coli. On the 3.1-Mb circular chromosome we found a tatABC gene cluster encoding proteins with 46%, 37%, and 43% identity, respectively, to the TatA, TatB, and TatC proteins of P. aeruginosa PAO1 (Fig. 2).
FIG. 2.
R. solanacearum Tat gene organization (GMI1000 genome). To create a Tat secretion mutant of R. solanacearum, a kanamycin resistance gene cassette (open triangle) was inserted into tatC, a gene predicted to encode an essential component of the Tat secretion system. R. solanacearum tatC is predicted to encode a 267-amino-acid protein with a molecular mass of 29.7 kDa. The entire tatABC gene cluster was used for all trans complementation studies of the R. solanacearum tatC mutant, K60TAT.
A search of the GMI1000 genome sequence using the algorithm TATFIND1.2 revealed 70 putative Tat-secreted proteins, identified by the presence of the signature twin-arginine domain within the first 35 amino acids followed by a stretch of at least 13 uncharged amino acids (12). The putative Tat secretome of GMI1000 contained proteins required for nitrate metabolism (NasF, NosL, and NosZ), plant cell wall degradation (PehC), and cell membrane function (AmiC, MltB, and LpxK), to name a few. A complete list of putative R. solanacearum GMI1000 Tat-secreted proteins is given elsewhere (see Table S1 in the supplemental material).
R. solanacearum tatC mutant K60TAT has a pleiotropic phenotype.
To investigate the contribution of Tat-secreted proteins to R. solanacearum virulence, we mutated tatC to create Tat-defective strain K60TAT (Fig. 2). tatC encodes an indispensable component of the Tat secretion system (3). In P. aeruginosa and A. tumefaciens, tatC mutants have a pleiotropic phenotype that features defects in swimming motility, extracellular enzyme production, and virulence (13, 28). We tested R. solanacearum K60TAT for a subset of the phenotypes observed in these systems.
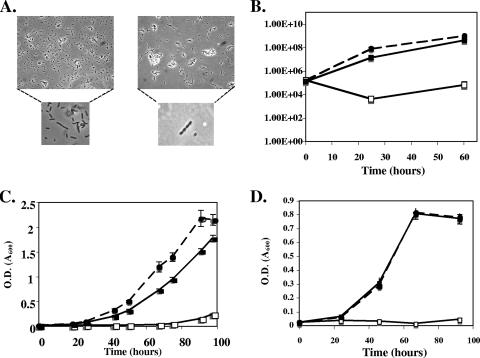
K60TAT had a distinctive colony morphology; colonies were smaller and surrounded by flocculent EPS different from those of the wild-type colonies, which produced opaque, milky EPS. K60TAT also overexpressed the reddish-brown pigment melanin on agar plates. Mid-log-phase cultures of R. solanacearum K60TAT viewed under the phase microscope contained chains of approximately four to five cells, in contrast to the single or doublet cells formed by the wild-type parent under the same growth conditions (Fig. 3A). In addition, cells of K60TAT in culture were smaller and rounder than those of the wild type (Fig. 3A). The tatC mutant also had reduced swimming motility on low-agar motility plates (data not shown). Although there was a slight early growth lag, the tatC mutant ultimately grew to wild-type levels in minimal medium broth supplemented with 0.2% (wt/vol) glucose and in CPG rich medium (data not shown). To test for membrane defects, we exposed wild-type and R. solanacearum K60TAT cells to detergent stress; R. solanacearum K60TAT was significantly more sensitive to SDS treatment than wild-type strain K60. In the presence of 0.02% SDS, K60 cultures grew a density fivefold greater than that of mutant strain K60TAT (data not shown).
FIG. 3.
Phenotypes of R. solanacearum tatC mutant K60TAT. (A) Cells of wild-type strain K60 and tatC mutant K60TAT, viewed by phase-contrast microscopy at ×400 magnification. (B) Growth in tobacco leaf tissue of R. solanacearum wild type (K60; closed circles), tatC mutant (K60TAT; open squares), and tatC mutant complemented [K60TAT(pLTAT); closed squares]. (C and D) Growth curves of R. solanacearum wild type (K60; closed circles), tatC mutant (K60TAT; open squares), and tatC mutant complemented [K60TAT(pLTAT); closed squares] in rich medium supplemented with 0.2% (wt/vol) KNO3 under O2-limiting conditions (panel C) or in minimal medium supplemented with 0.2% (wt/vol) polygalacturonic acid (panel D). All growth curves were performed in triplicate; bars indicate standard error.
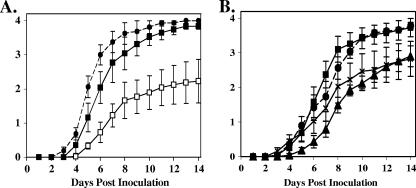
To cause disease, R. solanacearum must proliferate in plants, and bacterial populations normally reach 108 to 109 CFU/g of stem tissue-infected hosts (22). To measure the ability of the mutant to multiply in planta, we infused tobacco leaves with equivalent populations of wild-type R. solanacearum K60, tatC strain K60TAT, and the tatC strain carrying the tatABC complementation construct. K60TAT grew more poorly in tobacco leaf apoplast than its wild-type parent did, but K60TAT complemented with tatABC in trans reached wild-type population sizes (Fig. 3B). Tobacco leaf tissue became necrotic and collapsed 48 h after infusion with wild-type K60, but no necrosis was apparent 48 h after infusion with K60TAT (data not shown). These results suggested that R. solanacearum K60TAT would not be as virulent on tomato host plants as its wild-type parent. To test this hypothesis we inoculated susceptible heirloom tomato cultivar “Bonny Best” by pouring a bacterial suspension into pots of unwounded 2-week-old plants. K60TAT was significantly less virulent than the wild type, reaching a final disease index of only 2.2 of a possible 4 (P = 0.026) (Fig. 4A). However, the presence of tatABC in trans restored the virulence of K60TAT to wild-type levels (P = 0.479) (Fig. 4A).
FIG. 4.
Disease progress curves of R. solanacearum strains on tomato plants. (A) Equivalent bacterial cell suspensions of R. solanacearum wild type (K60; closed circles), tatC mutant (K60TAT; open squares), and complemented tatC mutant [K60TAT(pLTAT); solid squares] were poured onto the crowns of young susceptible tomato seedling plants, which were monitored for disease over the course of 14 days on a disease index scale where 0 indicated healthy and 4 indicated 100% wilted. (B) Similar assay using R. solanacearum wild type (K60; closed circles), RSc1651 mutant (K1651; closed squares), RSp1575 mutant (K1575; X's), and RSp1521 mutant (K1521, closed triangles). All assays had a minimum of 16 plants per treatment per assay. The graph in panel A represents the mean of three assays and the graph in panel B represents four assays; bars indicate standard error.
Several proteins involved in nitrate metabolism are predicted Tat substrates in R. solanacearum: NasF, the nitrate transporter; NosZ, nitrous oxide reductase; and NosL, a nitrous oxide reductase co-cofactor insertion protein. We hypothesized that an R. solanacearum tatC mutant would grow poorly under oxygen-limiting conditions with 0.2% (wt/vol) KNO3 as an energy source. Indeed, R. solanacearum K60TAT did not grow at all under these conditions, but near-wild-type growth was restored if the tatABC operon was provided in trans (Fig. 3C).
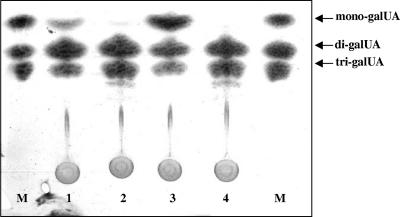
One of R. solanacearum's well-characterized plant cell wall-degrading exopolygalacturonases, PehC, is a predicted Tat substrate (15). This suggested that like a pehC mutant, the tatC mutant would not be able to use polygalacturonate as the sole carbon source. Indeed, K60TAT did not grow in minimal medium supplemented with 0.2% (wt/vol) polygalacturonate, although the growth of the mutant on this carbon source was restored by the wild-type tatABC gene cluster in trans (Fig. 3D). To more directly determine if R. solanacearum PehC is a Tat-secreted protein, we destroyed the twin-arginine motif in the N terminus of PehC by introducing R25K and R26K mutations by site-directed mutagenesis. If the twin-arginine motifs in Tat-dependent proteins are changed to twin lysines they are no longer translocated by the Tat secretion system (10). When heterologously expressed in E. coli, the resulting mutant twin-lysine PehC protein still had polygalacturonase activity, degrading polygalacturonate into galacturonate monomers like the wild-type enzyme (data not shown). However, the twin-lysine mutant protein could not restore extracellular PehC enzyme activity to strain K60-409 (pehC negative), indicating that even though it retained its enzyme activity, without its signature twin arginines the mutant PehC protein was not secreted (Fig. 5). Collectively, these data demonstrated the presence of a fully functional Tat secretion system in R. solanacearum K60 and that PehC is secreted by this system.
FIG. 5.
Thin-layer chromatography analysis of secreted polygalacturonase products from trans complementation analyses of R. solanacearum pehC mutant K60-409. Culture supernatants of R. solanacearum strains were incubated with 0.4% (wt/vol) polygalacturonic acid in 50 mM MES [2-(N-morpholino)ethanesulfonic acid], pH 5.7, for 1 h at 45°C and separated on a K5 silica gel plate in a 5:3:2 n-butanol:water:glacial acetic acid buffer system. Wild-type strain K60 (lane 1), pehC mutant K60-409 (lane 2), pehC mutant K60-409(pPehC11) (wild-type pehC) (lane 3), and pehC mutant K60-409(pLPehCRRKK) (R25K/R26K PehC) (lane 4). Four micrograms of pure monogalacturonate (mono-galUA), di-galUA, and tri-galUA standards were spotted for reference (lanes M). Monogalacturonate is the product of PehC activity.
Searching for more R. solanacearum virulence factors in the Tat secretome.
The significantly reduced virulence of the tatC mutant suggested that one or more R. solanacearum Tat-secreted proteins contribute to bacterial wilt virulence. To begin assessing the contributions of individual Tat-secreted proteins to virulence, we mutated three genes, RSc1651, RSp1575, and RSp1521, whose products are predicted Tat substrates (12). Genes RSc1651 and RSp1575 were chosen because they were previously identified in an in vivo expression technology (IVET) screen as upregulated during growth in tomato plants, suggesting a possible role in bacterial wilt virulence (6). RSc1651 is predicted to encode a conserved hypothetical protein most similar to a hypothetical protein from Caulobacter crescentus (NCBI NC_00296.2; E = 2e−34). We mutated RSc1651 by marker-mediated allelic replacement (Fig. 1A) and assayed the virulence of the resulting mutant, called K1651, on tomato using the naturalistic soil soak inoculation method. The virulence of strain K1651 was indistinguishable from that of its wild-type parent (Fig. 4B).
Another IVET-identified gene, RSp1575, is predicted to encode a protein resembling a periplasmic amino acid binding protein from P. aeruginosa (NCBI AE004920.1; E = 2e−22) and is upregulated approximately 17-fold when R. solanacearum grows in tomato plants (6). We also mutated this gene by marker-mediated allelic replacement mutagenesis to create R. solanacearum K1575 (Fig. 1B); this mutant grew like the wild type in culture and in planta (data not shown). This mutant had significantly reduced virulence (P = 0.0009), killing only just over half of the inoculated plants under conditions in which the wild-type parent killed all inoculated plants (Fig. 4B).
The protein encoded by RSp1575 is a predicted amino acid binding protein because it contains a bacterial periplasmic binding protein (PBPb) domain from amino acids 42 to 266, so we hypothesized that the mutant K1575 would be unable to grow in media containing one or several specific amino acids as the sole carbon source. However, when we tested the growths of the wild type and of mutant K1575 in 20 different BMM aliquots individually supplemented with 4 mM of 1 of the 20 amino acids as a sole carbon source, the growth pattern of K1575 was indistinguishable from that of the wild-type parent (data not shown).
The third Tat-dependent gene, RSp1521, resembled virulence-related proteins in several plant-associated bacteria, including X. fastidiosa, X. campestris, Rhizobium tropici, and A. tumefaciens, and Rsp1521 is 1 of 200 proteins previously identified as potential R. solanacearum virulence factors (34). RSp1521 is predicted to encode a protein most similar to AcvB from A. tumefaciens (E = 1e−38) and AtvA from R. tropici (E = 2e−36) over the whole length of the protein, apart from the N terminus; however, AcvB and AtvA are not predicted Tat-secreted proteins. An A. tumefaciens acvB mutant is attenuated in virulence, and an R. tropici atvA mutant cannot tolerate changes in extracellular pH level (41, 43). We cloned RSp1521 by amplifying the ORF from R. solanacearum K60 and mutated it to create R. solanacearum K1521 (Fig. 1C); this mutant grew as well as the wild type both in culture and in planta (data not shown). Mutant K1521 was significantly reduced in virulence on tomato plants, reaching a disease index of only 2.9 out of a possible 4 (P = 0.0018) (Fig. 4B). Because the R. tropici atvA mutant has reduced acid tolerance, we measured growth of K1521 in minimal medium at pH 5.5, the pH of healthy tomato plant xylem fluid. Under these relatively low pH conditions, the doubling time of K1521 was significantly longer than that of its wild-type parent, about 1.3 times longer (P = 0.05, Student's t test; data not shown). However, the two strains grew equally well at pH 7.0. This is similar to what was reported for an R. tropici atvA mutant. However, for R. solanacearum this pH-dependent growth phenotype varied with the carbon source, suggesting that Rsp1521 function involves several environmental variables and that the protein plays a complex role in the physiology of the bacterium.
DISCUSSION
The accumulating genomic data from plant-associated bacteria is impressive, but we are only beginning to use these genomes to understand how plant-associated bacteria form intimate relationships with their host plant(s) and other microbial partners in the phyllo- or rhizosphere. We used a postgenomic approach to look for new virulence determinants in the bacterial plant pathogen R. solanacearum, focusing on secreted potential virulence factors. We disrupted the tatC gene with the expectation that this would eliminate the Tat system. An R. solanacearum tatC mutant, like tatC mutants from P. aeruginosa, A. tumefaciens, and E. coli O157:H7, is pleiotropic with defects in cell division, colony morphology, swimming motility, extracellular enzyme production, growth in planta, and virulence. The importance of the Tat system for R. solanacearum's fitness in the host plant is demonstrated by the fact that a pLAFR3-derived plasmid carrying that tatABC locus was highly stable in K60TAT mutant background and was retained over weeks during growth in planta and pathogenesis. pLAFR3 plasmids are usually moderately unstable in planta (C. Allen, unpublished data), but restoration of the Tat secretion system clearly conferred a strong selective advantage on the complemented strains.
Collectively, Tat-secreted proteins contributed to R. solanacearum virulence; however, the pleiotropic nature of the tatC mutation made it difficult to identify the specific mechanisms of virulence involved. We therefore mutated several individual putative Tat-secreted proteins to determine their roles in virulence. Among the 70 putative Tat-secreted protein genes were four, RSc0532, RSc1651, RSp0881, and RSp1575, that were also identified as highly expressed in planta by an independent IVET screen (6). We cloned and mutated two of the four IVET Tat genes, RSp1575 and RSc1651.
An RSc1651 mutant was fully virulent on the susceptible tomato cultivar “Bonny Best.” Although this gene is expressed in planta, its product is apparently not required for virulence. It is possible that this gene encodes a redundant virulence function. In planta competition assays with wild-type K60 in tomato could determine if the mutant has reduced fitness under these conditions.
An RSp1575 mutant which lacks a gene predicted to encode an amino acid binding protein was severely attenuated in virulence on tomato, killing just over 50% of the inoculated plants under conditions when the wild-type parent killed all inoculated plants. Expression of RSp1575 is induced 17-fold when the pathogen grows in the tomato plant host rather than in rich broth, suggesting that this protein plays a role in plant-microbe interactions (6). Although this gene is predicted to encode an amino acid binding protein, mutant K1575 had an amino acid utilization pattern indistinguishable from that of the wild type, indicating either that amino acids are not the preferred substrate for Rsp1575 or that alternate redundant binding proteins can compensate if this protein is absent. We speculate that Rsp1575 may play a role in R. solanacearum taxis, the ability of bacteria to move towards more favorable conditions. Taxis is required for full virulence of this pathogen and is regulated by the motility master regulatory protein complex FlhDC (44; J. Yao, A. Milling, and C. Allen, unpublished data). Solute-binding proteins are often involved in chemotaxis, and we previously found that RSp1575 is negatively regulated by FlhDC (6). In preliminary experiments, K1575 had reduced swimming motility on low-agar motility plates. Further detailed chemotaxis and motility studies are necessary to explore the connection between RSp1575 and bacterial taxis.
We chose to mutate RSp1521 based on its similarity to genes required for normal interactions with plant interaction by other bacteria, namely, acvB from A. tumefaciens and atvA from R. tropici. Although the RSp1521 mutant grew normally in tobacco leaf apoplast, the strain was significantly attenuated in virulence on tomatoes. The function of Rsp1521 remains unclear, however.
Bioinformatic and in vitro growth experiments suggested that RSp1521 contributes to the ability to grow at low pH in R. solanacearum, and we speculate that poor acid tolerance may explain the reduced virulence of strain K1521. K1521 grows like the wild type in tobacco leaves; however, the leaf apoplast is not R. solanacearum's primary habitat inside plants. Quantitative analyses of bacterial growth in tomato xylem fluid could clarify the role of Rsp1521 in this trait. The pH of tomato plant xylem fluid during bacterial wilt pathogenesis is unknown, but R. solanacearum grows faster in xylem fluid of diseased plants than in any laboratory medium tested to date (Allen, unpublished data). Acidic and/or toxic by-products caused by the rapid growth of R. solanacearum in xylem fluid could contribute to an inhibitory change in pH in this environment, and the degradation of the reactive oxygen species produced during plant defense can also lower xylem pH significantly. Preliminary in vitro studies of R. solanacearum K152 found that this strain has wild-type levels of hydrogen peroxide tolerance; however, it is the active degradation of peroxides in planta that can be inhibitory. Measuring the growth or virulence of this strain in cultivars of tobacco that overproduce reactive oxygen species could test this hypothesis (17).
Of six genes encoding putative Tat-secreted proteins (pehC, RSp1521, RSp1575, RSc1651, nosZ, and plcN), four had a virulence defect when mutated, suggesting that there are additional contributors to virulence among the 70 putative Tat-secreted proteins. A nonhemolytic phospholipase C, PlcN, and a nitrous oxide reductase, NosZ, are putative Tat-secreted proteins in R. solanacearum GMI1000, and these proteins have been shown to be Tat dependent in other bacteria (19, 28). In separate experiments, we found that nosZ and plcN mutants of R. solanacearum GMI1000 are reduced in virulence (E. Gonzalez, J. Zarate, and C. Allen, unpublished data). However, the genes encoding NosZ and PlcN are not present in the genome of R. solanacearum K60, the strain used in the experiments presented in this paper. NosZ and PlcN may thus be strain-specific virulence factors for GMI1000. Further analysis of these proteins and their role in virulence is forthcoming.
The draft sequence of another R. solanacearum strain, phylotype II biovar 2 (“race 3”) strain UW551 (http://vision.biotech.ufl.edu/mycap/jsp/project/description.jsp?projectID = 1), suggests that this strain carries a somewhat different repertoire of Tat-secreted proteins (14). Interestingly, 15 proteins that are predicted Tat-secreted substrates in GMI1000 are present in the UW551 genome but lack the signature N-terminal twin-arginine motif, suggesting that although these proteins are conserved across strains, their secretion pathways are not. There may be adaptive value in secreting virulence factors via different pathways under various environmental conditions. These differences in secretory strategy also reinforce the idea that genomes of strains in the R. solanacearum species complex are fluid and significantly functionally divergent.
It is hypothesized that the Tat system could be a potential target for antimicrobial agents. However, mounting data suggest that in addition to virulence factors, Tat secretes numerous proteins necessary for basic metabolic and physiological bacterial processes. Tat proteins are well conserved among bacteria both beneficial (e.g., Rhizobium spp.) and pathogenic (12). Therefore, it could be unwise to use such a broad antimicrobial strategy against bacterial plant pathogens that occupy the same niche as benevolent bacteria. Rather, a comparative genomic analysis of predicted Tat-secreted proteins from plant pathogenic bacteria may shed light on other virulence-related or host adaptation proteins common to pathogens that share the same habitat (i.e., plant xylem tissue). These proteins would be good candidates for further mutagenesis and study using R. solanacearum and might offer useful targets for antimicrobial control strategies.
Supplementary Material
Acknowledgments
We acknowledge J. Clifford, J. Cramer, and Z. Flores for technical assistance, J. Yao for useful discussions, and L. Yakabe for statistical assistance.
E.T.G. was supported by an NIH Biotechnology Training Grant (NIGMS 5 T32 GM08349) and a Howard Hughes Medical Institute Teaching Fellowship. This research was supported by National Science Foundation grant IBN 0090692 and by the University of Wisconsin—Madison College of Agricultural and Life Sciences.
Footnotes
Published ahead of print on 27 April 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Allen, C., Y. Huang, and L. Sequeira. 1991. Cloning of genes affecting polygalacturonase production in Pseudomonas solanacearum. Mol. Plant-Microbe Interact. 4:147-154. [Google Scholar]
- 2.Ausubel, F., R. Brent, R. Kingston, D. Moore, J. Seidman, J. Smith, and K. Struhl. 1995. Short protocols in molecular biology, 3rd ed. John Wiley and Sons, New York, NY.
- 3.Berks, B. C., T. Palmer, and F. Sargent. 2003. The TAT protein translocation pathway and its role in microbial physiology. Adv. Microb. Physiol. 47:187-254. [DOI] [PubMed] [Google Scholar]
- 4.Boucher, C., P. Barberis, A. Trigalet, and D. Demery. 1985. Transposon mutagenesis of Pseudomonas solanacearum: isolation of Tn5-induced avirulent mutants. J. Gen. Microbiol. 131:2449-2457. [Google Scholar]
- 5.Bronstein, P. A., M. Marrichi, S. Cartinhour, D. J. Schneider, and M. P. DeLisa. 2005. Identification of a twin-arginine translocation system in Pseudomonas syringae pv. tomato DC3000 and its contribution to pathogenicity and fitness. J. Bacteriol. 187:8450-8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, D. G., and C. Allen. 2004. Ralstonia solanacearum genes induced during growth in tomato: an inside view of bacterial wilt. Mol. Microbiol. 53:1641-1660. [DOI] [PubMed] [Google Scholar]
- 7.Caldelari, I., S. Mann, C. Crooks, and T. Palmer. 2006. The Tat pathway of the plant pathogen Pseudomonas syringae is required for virulence. Mol. Plant-Microbe Interact. 19:200-212. [DOI] [PubMed] [Google Scholar]
- 8.Christie, P., K. Atmakuri, V. Krishnamoorthy, S. Jakubowski, and E. Cascales. 2005. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 59:451-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cornelis, G. R., and F. VanGijsegem. 2000. Assembly and function of type III secretion systems. Annu. Rev. Microbiol. 54:735-774. [DOI] [PubMed] [Google Scholar]
- 10.Cristobal, S., J.-W. de Gier, H. Nielsen, and G. von Heijne. 1999. Competition between Sec- and TAT-dependent protein translocation in Escherichia coli. EMBO J. 18:2982-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denny, T. P., and S. R. Baek. 1991. Genetic evidence that extracellular polysaccharide is a virulence factor of Pseudomonas solanacearum. Mol. Plant-Microbe Interact. 4:198-206. [Google Scholar]
- 12.Dilks, K., R. W. Rose, E. Hartmann, and M. Pohlschroder. 2003. Prokaryotic utilization of the twin-arginine translocation pathway: a genomic survey. J. Bacteriol. 185:1478-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding, Z., and P. J. Christie. 2003. Agrobacterium tumefaciens twin-arginine-dependent translocation is important for virulence, flagellation, and chemotaxis but not type IV secretion. J. Bacteriol. 185:760-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gabriel, D. W., C. Allen, M. Schell, T. P. Denny, J. T. Greenberg, Y. P. Duan, Z. Flores-Cruz, Q. Huang, J. M. Clifford, G. Presting, E. T. González, J. Reddy, J. Elphinstone, J. Swanson, J. Yao, V. Mulholland, L. Liu, W. Farmerie, M. Patnaikuni, B. Balogh, D. Norman, A. Alvarez, J. A. Castillo, J. Jones, G. Saddler, T. Walunas, A. Zhukov, and N. Mikhailova. 2006. Identification of open reading frames unique to a select agent: Ralstonia solanacearum race 3 biovar 2. Mol. Plant-Microbe Interact. 19:69-79. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez, E. T., and C. Allen. 2003. Characterization of a Ralstonia solanacearum operon required for polygalacturonate degradation and uptake of galacturonic acid. Mol. Plant-Microbe Interact. 16:536-544. [DOI] [PubMed] [Google Scholar]
- 16.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 17.Harding, S. A., O. Suk-Heung, and D. M. Roberts. 1997. Transgenic tobacco expressing a foreign calmodulin gene shows an enhanced production of active oxygen species. EMBO J. 16:1137-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayward, A. C. 1991. Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu. Rev. Phytopathol. 29:65-87. [DOI] [PubMed] [Google Scholar]
- 19.Heikkila, M. P., U. Honsisch, P. Wunsch, and W. G. Zumft. 2001. Role of the Tat transport system in nitrous oxide reductase translocation and cytochrome cd1 biosynthesis in Pseudomonas stutzeri. J. Bacteriol. 183:1663-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hendrick, C., and L. Sequeira. 1984. Lipopolysaccharide-defective mutants of the wilt pathogen Pseudomonas solanacearum. Appl. Environ. Microbiol. 48:94-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang, Q., and C. Allen. 1997. An exo-poly-α-d-galacturonosidase, PehB, is required for wild-type virulence of Ralstonia solanacearum. J. Bacteriol. 179:7369-7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang, Q., and C. Allen. 2000. Polygalacturonases are required for rapid colonization and full virulence of Ralstonia solanacearum on tomato plants. Physiol. Mol. Plant Pathol. 57:77-83. [Google Scholar]
- 23.Hugouvieux-Cotte-Pattat, N., G. Condemine, W. Nasser, and S. Reverchon. 1996. Regulation of pectinolysis in Erwinia chrysanthemi. Annu. Rev. Microbiol. 50:213-257. [DOI] [PubMed] [Google Scholar]
- 24.Kang, Y., J. Huang, G. Mao, L. He, and M. A. Schell. 1994. Dramatically reduced virulence of mutants of Pseudomonas solanacearum defective in export of extracellular proteins across the outer membrane. Mol. Plant-Microbe Interact. 7:370-377. [Google Scholar]
- 25.Kelman, A. 1954. The relationship of pathogenicity of Pseudomonas solanacearum to colony appearance in a tetrazolium medium. Phytopathology 44:693-695. [Google Scholar]
- 26.Liu, H., S. Zhang, M. A. Schell, and T. P. Denny. 2005. Pyramiding unmarked deletions in Ralstonia solanacearum shows that secreted proteins in addition to plant cell-wall-degrading enzymes contribute to virulence. Mol. Plant-Microbe Interact. 18:1296-1305. [DOI] [PubMed] [Google Scholar]
- 27.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Press, Cold Spring Harbor, NY.
- 28.Ochsner, U. A., A. Snyder, A. I. Vasil, and M. L. Vasil. 2002. Effects of the twin-arginine translocase on secretion of virulence factors, stress response, and pathogenesis. Proc. Natl. Acad. Sci. USA 99:8312-8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pradel, N., C. Ye, V. Livrelli, J. Xu, B. Joly, and L.-F. Wu. 2003. Contribution of the twin arginine translocation system to the virulence of enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 71:4908-4916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reference deleted.
- 31.Preston, G., D. J. Studholme, and I. Calelari. 2005. Profiling the secretomes of plant pathogenic proteobacteria. FEMS Microbiol. Rev. 29:331-360. [DOI] [PubMed] [Google Scholar]
- 32.Pugsley, A. P., O. Francetic, A. J. Driessen, and V. deLorenzo. 2004. Getting out: protein traffic in prokaryotes. Mol. Microbiol. 52:3-11. [DOI] [PubMed] [Google Scholar]
- 33.Roberts, D. P., T. P. Denny, and M. Schell. 1988. Cloning of the egl gene of Pseudomonas solanacearum and analysis of its role in phytopathogenicity. J. Bacteriol. 170:1445-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salanoubat, M., S. Genin, F. Artiguenave, J. Gouzy, S. Mangenot, M. Arlat, A. Billault, P. Brottier, J. Camus, L. Cattolico, M. Chandler, N. Choisne, C. Claudel-Renard, S. Cunnac, N. Demange, C. Gaspin, M. Lavie, A. Moisan, C. Robert, W. Saurin, T. Schiex, P. Siguier, P. Thebault, M. Whalen, P. Wincker, M. Levy, J. Weissenbach, and C. A. Boucher. 2002. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415:497-502. [DOI] [PubMed] [Google Scholar]
- 35.Schell, M. A. 2000. Control of virulence and pathogenicity genes of Ralstonia solanacearum by an elaborate sensory array. Annu. Rev. Phytopathol. 38:263-292. [DOI] [PubMed] [Google Scholar]
- 36.Schweizer, H. P. 1993. Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques 15:831-833. [PubMed] [Google Scholar]
- 37.Stanley, N. R., T. Palmer, and B. C. Berks. 2000. The twin-arginine consensus motif of Tat signal peptides is involved in Sec-independent protein targeting in Escherichia coli. J. Biol. Chem. 275:11591-11596. [DOI] [PubMed] [Google Scholar]
- 38.Staskawicz, B., D. Dahlbeck, N. Keen, and C. Napoli. 1987. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J. Bacteriol. 169:5789-5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tans-Kersten, J., Y. Guan, and C. Allen. 1998. Ralstonia solanacearum pectin methylesterase is required for growth on methylated pectin but not for bacterial wilt virulence. Appl. Environ. Microbiol. 64:4918-4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tans-Kersten, J., H. Huang, and C. Allen. 2001. Ralstonia solanacearum needs motility for invasive virulence on tomato. J. Bacteriol. 183:3597-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vinuesa, P., F. Newumann-Silkow, C. Pacios-Bras, H. P. Spaink, E. Martinez-Romero, and D. Werner. 2003. Genetic analysis of a pH-regulated operon from Rhizobium tropici CIAT899 involved in acid tolerance and nodulation competitiveness. Mol. Plant-Microbe Interact. 16:159-168. [DOI] [PubMed] [Google Scholar]
- 42.Voulhoux, R., G. Ball, B. Ize, M. L. Vasil, A. Lazdunski, L.-F. Wu, and A. Filloux. 2001. Involvement of the twin-arginine translocation system in protein secretion via the type II pathway. EMBO J. 20:6735-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wirawan, I. G. P., H. W. Kang, and M. Kojima. 1993. Isolation and characterization of a new chromosomal virulence gene of Agrobacterium tumefaciens. J. Bacteriol. 175:3208-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yao, J., and C. Allen. 2006. Chemotaxis is required for virulence and competitive fitness in the bacterial wilt pathogen Ralstonia solanacearum. J. Bacteriol. 188:3697-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.