Abstract
Artificial designer minicellulosomes comprise a chimeric scaffoldin that displays an optional cellulose-binding module (CBM) and bacterial cohesins from divergent species which bind strongly to enzymes engineered to bear complementary dockerins. Incorporation of cellulosomal cellulases from Clostridium cellulolyticum into minicellulosomes leads to artificial complexes with enhanced activity on crystalline cellulose, due to enzyme proximity and substrate targeting induced by the scaffoldin-borne CBM. In the present study, a bacterial dockerin was appended to the family 6 fungal cellulase Cel6A, produced by Neocallimastix patriciarum, for subsequent incorporation into minicellulosomes in combination with various cellulosomal cellulases from C. cellulolyticum. The binding of the fungal Cel6A with a bacterial family 5 endoglucanase onto chimeric miniscaffoldins had no impact on their activity toward crystalline cellulose. Replacement of the bacterial family 5 enzyme with homologous endoglucanase Cel5D from N. patriciarum bearing a clostridial dockerin gave similar results. In contrast, enzyme pairs comprising the fungal Cel6A and bacterial family 9 endoglucanases were substantially stimulated (up to 2.6-fold) by complexation on chimeric scaffoldins, compared to the free-enzyme system. Incorporation of enzyme pairs including Cel6A and a processive bacterial cellulase generally induced lower stimulation levels. Enhanced activity on crystalline cellulose appeared to result from either proximity or CBM effects alone but never from both simultaneously, unlike minicellulosomes composed exclusively of bacterial cellulases. The present study is the first demonstration that viable designer minicellulosomes can be produced that include (i) free (noncellulosomal) enzymes, (ii) fungal enzymes combined with bacterial enzymes, and (iii) a type (family 6) of cellulase never known to occur in natural cellulosomes.
The conversion of biomass into biofuels such as ethanol has become a major industrial challenge over the past decades. The main issue in such a process concerns the degradation in fermentable sugars of the main plant cell wall polysaccharide, cellulose, with the most efficient (and cost-effective) enzymatic cocktail. Microbial cellulolysis naturally occurs in the aerobic and anaerobic biotopes where cellulose accumulates. The cellulolytic fungi and bacteria that occupy these biotopes have developed different strategies to degrade this biopolymer and the related plant cell wall polysaccharides. Thus, aerobic microorganisms usually secrete copious amounts of free cellulases which act synergistically to degrade cellulose. For instance, the main cellulases of the aerobic fungus Trichoderma reesei are secreted at yields of >10 g/liter, and these enzymes are often used to saccharify the biomass in bioethanol production processes. On the other hand, anaerobes produce large extracellular complexes called cellulosomes. Typical bacterial cellulosomes are composed of a scaffolding protein, termed scaffoldin, that contains a powerful cellulose-binding module (CBM) and a number of cohesin modules which tightly bind to the complementary dockerin modules borne by the catalytic subunits (1). Within a given species, the cohesin-dockerin interaction is usually nonspecific; i.e., one enzyme-borne dockerin can bind to any of the cohesins of the scaffoldin with similar affinity (25, 32). The cohesin-dockerin interaction was, however, shown to be species specific among three different bacterial species (13, 24). This observation was exploited to build chimeric minicellulosomes, or designer cellulosomes, composed of a chimeric scaffoldin which displays an optional CBM and cohesins of different bacterial species which bind tightly and selectively to the cellulases with matching dockerins of the corresponding species appended (10, 12, 13). Thus, in the artificial designer minicellulosomes, each enzyme was bound at a specific location onto the chimeric scaffoldin, contrary to the heterogeneous nature of “natural” cellulosomes. This strategy was applied to C. cellulolyticum cellulosomal cellulases which were engineered for incorporation into chimeric cellulosomes. The resulting complexes displayed enhanced activity on crystalline cellulose and hatched straw substrates, compared to the corresponding free-enzyme systems (10, 13). The activity enhancement was essentially due to additional synergy induced by enzyme proximity within the complex and the targeting effect of the CBM, borne by the chimeric scaffoldin, which anchors the whole complex at the substrate surface. The cellulolytic complexes produced by anaerobic fungi have been less well characterized, but the presence of a “scaffolding” protein(s) has been reported in various species (15, 29). Furthermore, the “cellulosomal” enzymes were shown to contain a short 40-residue module(s) that functions as a bacterial dockerin, though the fungal modules display no homology with their prokaryote counterparts (15, 27, 29). The cellulosomes are believed to play a pivotal role during anaerobic cellulolysis, but both cellulosome-producing bacteria and fungi also secrete free (dockerin-lacking) cellulases and hemicellulases (5, 9, 19).
The cellulases (EC 3.2.1.4 and EC 3.2.1.91) discovered to date belong to approximately a dozen different families of glycoside hydrolases (GH), classified on the basis of sequence similarities. Interestingly, some cellulase types, like GH-5 cellulases, are extensively distributed in nature and participate in the cellulolytic systems of fungal and bacterial microorganisms that occupy both aerobic and anaerobic biotopes. On the contrary, GH-7 cellulases seem to be exclusively produced by aerobic fungi. In this respect, the distribution of GH-6 cellulases among cellulolytic microorganisms is even more intriguing. These enzymes are widespread in aerobic fungi (3, 23) and bacteria (8, 20), and several species of anaerobic fungi have been shown to produce GH-6 cellulases as part of their putative cellulosome (18) or as a free (dockerin-lacking) enzyme (9). Nevertheless, GH-6 cellulases have not been selected by cellulosome-producing bacteria since, to date, such an enzyme has not been discovered in anaerobic prokaryotes, neither as a free cellulase nor as a cellulosomal subunit.
This observation prompted us to engineer and incorporate a fungal GH-6 cellulase into bacterial chimeric minicellulosomes with cellulosomal C. cellulolyticum cellulases. Our purpose was to determine whether the technology of designer cellulosomes may be extended to noncellulosomal enzymes and used to create new artificial cellulosomes with superior cellulolytic performance. The selected enzyme, termed Cel6A (9), is secreted as a free enzyme by the anaerobic rumen fungus Neocallimastix patriciarum since it is devoid of a fungal dockerin. This cellulase was chosen because its pH and temperature optima are compatible to those of C. cellulolyticum cellulases. The fungal cellulase has previously been successfully produced in an active form in two different bacterial hosts (9, 22), thus indicating that glycosylation is not absolutely required for Cel6A activity. In light of its rather high specific activity on carboxymethyl cellulose (CMC), Cel6A can be considered an endoglucanase, but the enzyme also displays substantial activity on crystalline cellulose (9).
A C. thermocellum dockerin was appended to the fungal cellulase, and it was incorporated together with various C. cellulolyticum cellulases into bi- or trifunctional chimeric cellulosomes. The selected clostridial enzymes include three endoglucanases (Cel5A, Cel9G, and Cel9M) and two endoprocessive cellulases (Cel48F and Ce9E). The resulting complexes were assayed on crystalline cellulose, and the data obtained prompted us to also engineer and incorporate into the complexes a GH-5 cellulase from N. patriciarum (31).
MATERIALS AND METHODS
Plasmids and strains.
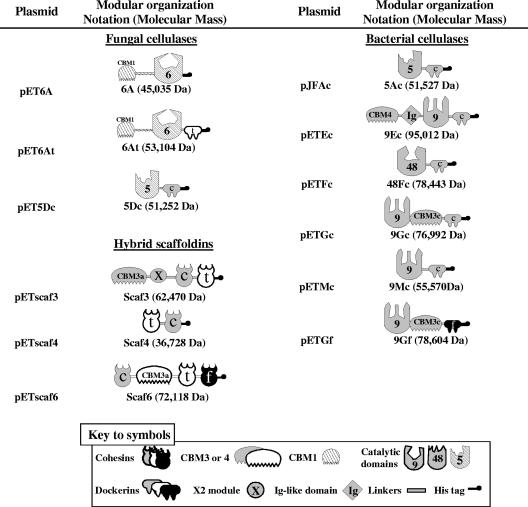
The plasmids and encoded proteins used in this study are summarized in Fig. 1. The construction of pETscaf3 (12), pETscaf6 (13), pJFAc (13), pETEc (17), pETGc (16), pETGf (13), pETFc (28), pETFt (12), and pETMc (4) was described previously.
FIG. 1.
Schematic representation of the recombinant proteins used. Gray (C. cellulolyticum), white (C. thermocellum), black (R. flavefaciens), and hatched (N. patriciarum) denote the sources of the respective modules (see key to symbols). In the shorthand notation for the engineered enzymes, numbers refer to the corresponding GH family of the catalytic module; A, D, E, F, G, and M refer to the original names of the enzymes (CelA and CelD from N. patriciarum and CelA, CelE, CelF, CelG, and CelM from C. cellulolyticum); c, f, and t indicate the sources of the dockerin modules of C. cellulolyticum, R. flavefaciens, and C. thermocellum, respectively. Note that, unlike CBM1 of the fungal enzyme and CBM3a of the scaffoldins, the CBMs (CBM3c and CBM4) associated with the C. cellulolyticum enzymes 9Gf, 9Gc, and 9Ec are not CBMs per se but serve as ancillary modules that modify the activity of the catalytic modules of the parent enzyme. The X2 module in Scaf3 is a 100-residue-long module that the scaffoldin CipC contains and whose functions remain unknown. Ig, immunoglobulin.
Construction of plasmid pET6A encoding the mature form of Cel6A of N. patriciarum with a C-terminal His tag appended.
The DNA encoding Cel6A was amplified from pWUR3 (22) with forward primer 6AmatF (5′-AAAACATATGGACGCGTGTGGTGGTGCC-3′; the introduced NdeI site is in bold) and reverse primer 6AmatR (5′-AAACTCGAGGAATGATGGTCTAGCGTTTCTTACTAATTGAGCG-3′; the introduced XhoI site is in bold). The fragment was cloned into NdeI-XhoI-linearized pET22b(+) (Novagen, Madison, WI), thereby generating pET6A.
Construction of plasmid pET6At encoding the engineered Cel6A protein of N. patriciarum that contains a C-terminal C. thermocellum dockerin module and a His tag.
Plasmid pET6At was obtained by the overlap extension PCR method. The DNA encoding the catalytic module of Cel6A was amplified from pET6A with forward primer 6AmatF and reverse primer 6AtR (5′-TGGGCTAGGGCTTGTCTTGGCAAATGATGGTCTAGCGTTTCTTAC-3′; the sequence matching the 3′ end of the DNA encoding Cel6A is underlined). The DNA encoding the C. thermocellum dockerin module was amplified from pJFAt (12) with forward primer 6AtF (5′-GTAAGAAACGCTAGACCATCATTTGCCAAGACAAGCCCTAGCCCA-3′; the sequence matching the 5′ end of the DNA encoding the C. thermocellum dockerin module is underlined) and reverse primer tagXhoR (5′-AAACTCGAGTTAGTGGTGGTGGTGGTGGTG-3′; the introduced XhoI site is in bold). The two resulting overlapping fragments (the overlapping region is in italics) were mixed, and a combined fragment was synthesized with external primers 6AmatF and tagXhoR. The fragment was cloned into NdeI-XhoI-linearized pET22b(+), thereby generating pET6At.
Construction of plasmid pETD5c encoding an engineered form of Cel5D from N. patriciarum containing a C-terminal C. cellulolyticum dockerin module and a His tag.
Plasmid pETD5c was obtained by the overlap extension PCR method. The DNA encoding the catalytic module of Cel5D was amplified from pWUR4 (22) with forward primer 5DmatF (5′-AAAACATATGACTAATCCAGAAGAACCAACCGG-3′; the introduced NdeI site is in bold) and reverse primer 5DcR (5′-GGGTCAGGATCTGTCTTGGCTTCGAATTTTTTTTCAACAGCATGAACAAC-3′; the sequence matching the 3′ end of the DNA encoding Cel5D is underlined). The DNA encoding a C. cellulolyticum dockerin module was amplified from pJFAc with forward primer 5DcF (5′-GTTGTTCATGCTGTTGAAAAAAAATTCGAAGCCAAGACAGATCCTGACCC-3′; the sequence matching the 5′ end of the DNA encoding the C. cellulolyticum dockerin module is underlined) and reverse primer tagXhoR. The two resulting overlapping fragments (the overlapping region is in italics) were mixed, and a combined fragment was synthesized with external primers 5DmatF and tagXhoR. Digestion of this amplified DNA with NdeI and XhoI generated two fragments, a 0.66-kb NdeI-NdeI fragment which encodes the N-terminal part of Cel5D and a 0.72-kb NdeI-XhoI fragment which encodes the C-terminal part of Cel5D with the C. cellulolyticum dockerin module. The 0.72-kb fragment was initially cloned into NdeI-XhoI-linearized pET22(+), generating pETtrunc5Dc. The 0.66-kb fragment was cloned into NdeI-linearized pETtrunc5Dc, thereby generating pET5Dc.
Positive clones were verified by DNA sequencing. Escherichia coli strain BL21 (DE3) (Novagen) was used as the production host.
Production and purification of recombinant proteins.
The production and purification of Scaf3, Scaf4, Scaf6, Cel5Ac (5Ac), Cel9Gc (9Gc), Cel9Gf (9Gf), Cel9Ec (9Ec), Cel48Fc (48Fc), and Cel9Mc (9Mc) were performed as previously described (13).
Cel5D-overproducing strain BL21 (DE3), with a C. cellulolyticum dockerin (5Dc) appended, was grown at 37°C to an A600 of 1.5 in Luria-Bertani medium supplemented with 1.2% (wt/vol) glycerol and the appropriate antibiotic. The culture was then cooled to 20°C, and isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 100 μmol/liter. After 16 h, the cells were centrifuged (10 min, 3,000 × g), resuspended in 30 mmol/liter Tris-HCl (pH 8)-1 mmol/liter CaCl2-0.1 mg/ml DNase I (Roche, Mannheim, Germany), and broken in a French press. The modified Cel5D protein was purified on nickel-nitrilotriacetic acid resin (QIAGEN, Vanloo, The Netherlands). This purification was achieved on Q-Sepharose fastflow resin (GE Healthcare, Uppsala, Sweden) equilibrated in 30 mmol/liter Tris-HCl (pH 8.0)-1 mmol/liter CaCl2. The cellulase was eluted by a linear gradient of 0 to 500 mmol/liter NaCl in 30 mmol/liter Tris-HCl (pH 8.0)-1 mmol/liter CaCl2. The purified protein was dialyzed and concentrated by ultrafiltration against 10 mmol/liter Tris-HCl (pH 8)-1 mmol/liter CaCl2 and stored at −80°C.
The strains overproducing the wild-type and engineered forms of Cel6A were grown at 37°C to an A600 of 1.5 in Luria-Bertani medium supplemented with 1.2% (wt/vol) glycerol and the appropriate antibiotic. IPTG (250 μmol/liter) was then added, and the culture was grown again at 37°C. After 3 h 30 min, the cells were harvested by centrifugation (10 min, 3,000 × g), resuspended in 30 mmol/liter Tris-HCl (pH 8.0)-1 mmol/liter CaCl2-0.1 mg/ml DNase I (Roche), and broken in a French press. The crude extract was centrifuged (10 min, 10,000 × g), and the pellet which contains the inclusion bodies of Cel6A was incubated in 50 ml of 30 mmol/liter Tris-HCl (pH 8.5)-1 mmol/liter CaCl2-10 mmol/liter 2-mercaptoethanol-8 mol/liter urea at room temperature with mild shaking until complete dissolution of the inclusion bodies (approximately 1 h). The sample was then loaded onto nickel-nitrilotriacetic acid resin equilibrated in the same buffer. The cellulase was eluted with 50 mmol/liter sodium acetate (pH 4.8)-1 mmol/liter CaCl2-10 mmol/liter 2-mercaptoethanol-8 mol/liter urea. The enzyme fraction (approximately 15 ml) was then slowly diluted 10-fold (flow rate, 0.5 ml/min) in 50 mmol/liter sodium acetate (pH 4.8)-1 mmol/liter CaCl2 at room temperature. The diluted sample was dialyzed three times at 4°C against 3 liters of 25 mmol/liter Tris-HCl (pH 7.5)-1 mmol/liter CaCl2. The enzyme was then concentrated by ultrafiltration and stored at −80°C.
The concentration of the purified proteins was estimated by determining absorbance (280 nm) in 6 mol/liter guanidine hydrochloride-25 mmol/liter sodium phosphate, pH 6.5, with the program ProtParam tool (www.expasy.org/tools/protparam.html).
Nondenaturing PAGE.
Samples (10-μmol/liter final concentration) were mixed at room temperature in 20 mmol/liter Tris-maleate (pH 6.0)-1 mmol/liter CaCl2, and 4 μl was subjected to nondenaturing polyacrylamide gel electrophoresis (PAGE; 4 to 15% gradient) with a Phastsystem apparatus (GE Healthcare).
Gel filtration analyses.
Gel filtration experiments were carried out with an Ákta fast protein liquid chromatography system (GE Healthcare). Samples (100 μl, 10 μmol/liter) were injected onto a Superdex 200 (10 by 300 mm) gas-liquid analysis grade column (GE Healthcare) equilibrated in 50 mmol/liter potassium phosphate (pH 7.0)-150 mmol/liter NaCl. A flow rate of 0.5 ml/min was used during chromatography.
Enzyme activity.
Kinetics on CMC (medium viscosity; Sigma, St. Louis, MO) were determined by incubating, at 40 or 37°C, aliquots (40 μl) of the protein samples (2 μmol/liter in 20 mmol/liter Tris-maleate [pH 6.0]-1 mmol/liter CaCl2) with 4 ml of CMC at 4 or 8 g/liter in 20 mmol/liter Tris-maleate (pH 6.0)-1 mmol/liter CaCl2-0.01% (wt/vol) NaN3. The final enzyme concentration during the determination of kinetics on CMC was 20 nmol/liter. Aliquots (500 μl) were extracted at 0, 3, 6, 9, 12, 15, and 18 min and examined for soluble reducing sugars by the method of Park and Johnson (26) with glucose as the standard.
Kinetics on microcrystalline cellulose Avicel PH101 (Fluka, Buchs, Switzerland) were determined at 37°C as follows. Aliquots (40 μl) of the protein samples (10 μmol/liter in 20 mmol/liter Tris-maleate [pH 6.0]-1 mmol/liter CaCl2) were mixed with 4 ml of Avicel at 3.5 g/liter in 20 mmol/liter Tris-maleate (pH 6.0)-1 mmol/liter CaCl2-0.01% (wt/vol) NaN3. Thus, the final enzyme concentration during the determination of kinetics was 0.1 μmol/liter. Aliquots (900 μl) were extracted at 0, 1, 6, and 24 h, centrifuged, and examined for soluble reducing sugars by the method of Park and Johnson (26) with glucose as the standard.
RESULTS
Heterologous production, purification, and characterization of wild-type and engineered Cel6A from N. patriciarum.
The cellulase produced by the anaerobic fungus N. patriciarum contains a C-terminal family 6 catalytic module and an N-terminal family 1 CBM characteristic of fungal enzymes. The catalytic and substrate-targeting modules are connected by an unusual linker almost exclusively composed of Asn residues (33 Asn residues out of 36 amino acids). The DNA encoding the mature form of the fungal cellulase was modified to introduce six His codons at the C terminus of the enzyme (termed 6A, Fig. 1). In a second genetic construction, the DNA encoding a C. thermocellum dockerin and six His codons were introduced at the 3′ end of the gene cel6A. The resulting engineered cellulase, called 6At (Fig. 1), was designed for subsequent incorporation into cellulosome chimeras.
Overexpression of both engineered genes in E. coli systematically induced the formation of inclusion bodies composed of the heterologous enzymes. The inclusion bodies of wild-type or engineered Cel6A were totally inactive on CMC, and neither reduced concentrations of the inducer nor lower temperatures during induction prevented the aggregation of the recombinant cellulases. Thus, direct purification of the cellulases from the inclusion bodies with an elevated concentration of urea (8 mol/liter) in the presence of a reducing agent (2-mercaptoethanol) was finally selected. The purified inactive enzymes were afterwards renatured by slow dilution in mild acidic buffer (pH 4.8), followed by extensive dialyses at 4°C against pH 7.5 (Tris-HCl) buffer. This procedure led to purified enzymes with CMCase specific activities of 1,252 and 1,057 IU/μmol for 6A and 6At, respectively. These values are higher than the specific activity previously determined for the recombinant Cel6A protein (approximately 700 IU/μmol) produced in the periplasm of E. coli (9). The specific activities were estimated under similar experimental conditions (40°C, substrate at 4 g/liter), but medium-viscosity CMC was used in the present study, whereas low-viscosity CMC was previously used (9).
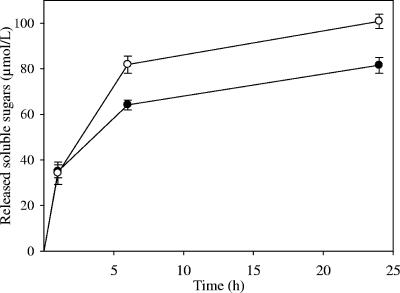
As indicated above, grafting a C. thermocellum dockerin to 6A reduced its specific activity on CMC by 20%. Kinetics determined on Avicel (Fig. 2) indicated that 6At is also approximately 20% less active that the wild-type GH-6 cellulase on crystalline cellulose. Thus, introduction of a dockerin at the C terminus of Cel6A slightly affected its activity. Nevertheless, the engineered fungal cellulase remained twofold more active on Avicel than the any of the C. cellulolyticum cellulases discovered to date (16).
FIG. 2.
Kinetics of crystalline cellulose degradation by 6A and 6At. Curves are labeled as follows: wild-type 6A, ○; engineered 6At, •. The final protein concentration in this and the subsequent figures was 0.1 μmol/liter. Released soluble sugars were assayed by the method of Park and Johnson (26). The data are the means of three independent experiments, and the bars indicate the standard deviations.
The avicelase activity of 6At was also assayed in the presence of different endoglucanases (5Ac, 9Mc, and 9Gc) or processive endocellulases (9Ec and 48Fc) from C. cellulolyticum in the free state. In these kinetic experiments, the fungal and bacterial cellulases were maintained at the same concentration. The soluble sugars released by the mixture were in the same range as the theoretical sum of individual activities (data not shown). This indicates that, in the free state, 6At neither acts synergistically nor competes with any of the C. cellulolyticum cellulases tested for the degradation of the crystalline substrate.
Incorporation of the GH-6 cellulase into bifunctional chimeric minicellulosomes.
Cellulosome chimeras containing the fungal cellulase to which the C. thermocellum dockerin was appended were obtained by mixing, in the presence of calcium, stoichiometric amounts of chimeric scaffoldin Scaf3 or Scaf4 (Fig. 1) with 6At and one of the five selected C. cellulolyticum cellulases bearing their native dockerin. As shown in previous studies (10, 12), both Scaf3 and Scaf4 contain two cohesins, derived from the native scaffoldins of C. thermocellum and C. cellulolyticum, that exhibit divergent specificities; i.e., each recognizes and binds tightly to its own dockerin but fails to interact with that of the other species. The functional difference between the two chimeric scaffoldins is that Scaf3 includes an N-terminal CBM designed for substrate targeting, whereas Scaf4 lacks a CBM. The dockerin of the 6At chimeric enzyme would thus bind selectively to the C. thermocellum cohesin in either Scaf3 or Scaf4, and the native C. cellulolyticum cellulases would be incorporated into the chimeric scaffoldins via their C. cellulolyticum cohesin.
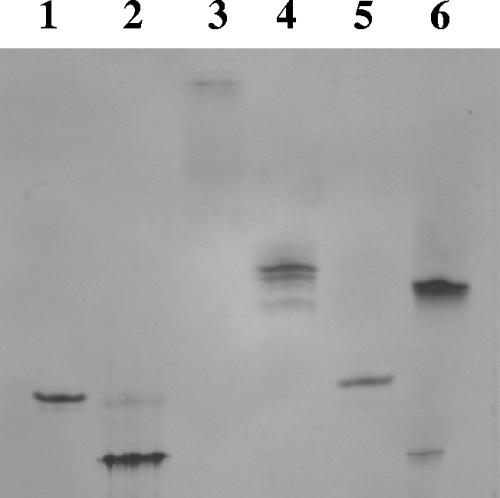
The complexation of the various partners was routinely checked by gel filtration analyses (data not shown) and nondenaturing PAGE, as shown in Fig. 3. The complexation was found to be total or nearly total in all cases, thus indicating that the C. thermocellum dockerin which has been grafted to Cel6A is fully operational.
FIG. 3.
Electrophoretic mobility of components and assembled trifunctional complexes on nondenaturing gel. Lane 1, Scaf3 alone; lane 2, 9Gc alone; lane 3, 6At alone; lane 4, chimeric cellulosome containing Scaf3, 6At, and 9Gc; lane 5, Scaf4 alone; lane 6, chimeric cellulosome containing Scaf4, 6At, and 9Gc. In each lane, equimolar concentrations (10 μmol/liter) of the indicated proteins were used. Similar-quality gels were obtained for all of the complexes used in this study.
Avicelase activity of the bifunctional chimeric minicellulosomes.
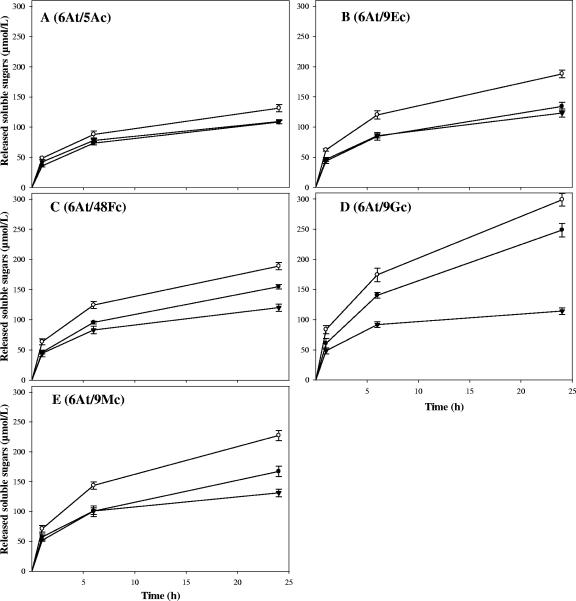
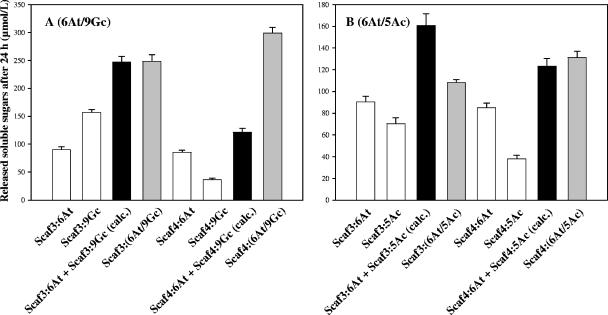
The kinetics obtained for Scaf3- and Scaf4-based complexes and the corresponding free-enzyme systems are reported in Fig. 4. The assembled minicellulosomes behaved very differently, depending on the C. cellulolyticum cellulase incorporated together with the fungal enzyme hybrid 6At and the type of chimeric scaffoldin used. Contrary to previous studies on bacterial enzyme pairs (10, 12), Scaf4-based complexes displayed superior activity compared to the corresponding Scaf3-based minicellulosomes. Furthermore, the Scaf3-based complexes exhibited similar levels of avicelase activity compared to the corresponding free-enzyme system for enzyme pairs composed of 6At and the bacterial cellulase 9Ec or 5Ac. The enzyme pair stimulated the most by complexation onto chimeric scaffoldins was 6At plus 9Gc (Fig. 4D). The binding of these two cellulases onto Scaf4 induced a 2.6-fold increase in activity compared to free 6At and 9Gc. Incorporation of 9Gc and 6At into Scaf3 also generated a complex 2.1-fold more active than the corresponding free-enzyme system. The other enzyme pairs were more moderately stimulated by complexation onto the chimeric scaffoldins, with activity enhancements in the case of the Scaf4-based complexes varying from 75% for 9Mc and 6At (Fig. 4E) to no significant stimulation for 6At and 5Ac (Fig. 4A). The avicelase activity of the latter enzyme pair was indeed the least stimulated by complexation with either Scaf4 or Scaf3.
FIG. 4.
Kinetic studies of Avicel hydrolysis by enzyme pairs in the free state or complexed with Scaf3 or Scaf4. The proteins were mixed in stoichiometric amounts, and the status of the protein mixture was verified by nondenaturing PAGE (data not shown) prior to addition of the substrate. The panels indicate the selected enzyme pair as follows: A, 6At and 5Ac; B, 6At and 9Ec; C, 6At and 48Fc; D, 6At and 9Gc; and E, 6At and 9Mc. Curves are labeled as follows: free enzyme pair, ▾; enzyme pair complexed to Scaf3, •; enzyme pair complexed to Scaf4, ○. The data are the means of four independent experiments, and the bars indicate the standard deviations.
To further investigate the avicelase activities of the assembled complexes, additional kinetics were determined with the various enzyme pairs. Each enzyme was bound alone to Scaf3 or Scaf4, and the avicelase activity of the resulting single-enzyme complexes was measured individually and compared to that of Scaf3- and Scaf4-based bifunctional complexes. In Fig. 5 are reported the data obtained with 6At plus 9Gc (Fig. 5A) and 6At plus 5Ac (Fig. 5B), for which complexation had a distinctive impact on activity. For the cellulase pair 6At plus 9Gc, the amount of released soluble sugars by the Scaf3-based complex matches the theoretical sum of the single-enzyme complexes. This indicates that the enhanced activity of the complex Scaf3-6At/9Gc seems exclusively due to the CBM effect of the scaffoldin. This effect mainly applies to 9Gc, since the binding of 9Gc to Scaf3 induces a fourfold increase in the avicelase activity of 9Gc, whereas the Scaf3-6At complex is barely 10% more active than free 6At. Nevertheless, this important CBM effect on 9Gc is maintained in Scaf3-6At/9Gc. In contrast, the minicellulosome Scaf4-6At/9Gc is approximately 2.5-fold more active than the corresponding cellulases bound separately onto Scaf4, thus reflecting a dramatic “proximity” effect occurring in the bifunctional complex. Similar data were obtained with the enzyme pairs 6At plus 48Fc, 6At plus 9Ec, and 6At plus 9Mc, except that the proximity effect observed in Scaf4-based complexes (+50 to 75%) and the CBM effect induced by Scaf3 on the bacterial cellulase (+10 to 30%) were more modest (data not shown).
FIG. 5.
Avicelase activities of 6At plus 9Gc (A) and 6At plus 5Ac (B) bound jointly or individually to Scaf3 and Scaf4. The designated proteins were mixed in stoichiometric amounts, and the status of the protein mixture was verified by nondenaturing PAGE (data not shown) prior to addition of the substrate. White bars, soluble sugars released by single-enzyme complexes; gray bars, soluble sugars released by Scaf3- and Scaf4-based complexes; black bars, calculated (calc.) sum of soluble sugars released by single-enzyme complexes individually. The data are the means of four independent experiments, and the bars indicate the standard deviations.
The enzyme pair 6At plus 5Ac (Fig. 5B) displayed a very different pattern. Binding the two cellulases together or separately to Scaf4 failed to have any significant impact on their avicelase activity, which remains basically in the same range compared to that of the corresponding free-enzyme system. This result indicates that no major proximity effect seems to occur between this particular pair of cellulases in this context. In fact, the activity of the minicellulosome Scaf3-6At/5Ac was found to be significantly (approximately 35%) lower than the cumulative activity of the single-enzyme complexes. As in the case of 9Gc, the binding of 5Ac alone to Scaf3 induced a significant 2.3-enhancement of the avicelase activity of the bacterial cellulase but this stimulation was apparently completely lost when 5Ac was bound together with 6At to Scaf3.
Incorporation of the GH-5 catalytic module of Cel5D from N. patriciarum.
The unexpected data obtained with the enzyme pair 6At plus 5Ac prompted us to assay 6At in combination with another GH-5 cellulase. The cellulase we selected for these experiments was the central GH-5 catalytic module of the cellulosomal enzyme Cel5D from N. patriciarum (31). This module has already been successfully produced in an active form in C. beijerinckii (22), but the recombinant cellulase has not been purified. Since the catalytic module of Cel5D is derived from the same fungal species as 6At, the effects of their combined incorporation into a minicellulosome would help explain whether reduced synergy results from an inherent incompatibility in coupling this particular family 6 cellulase with a family 5 cellulase.
A C. cellulolyticum dockerin and a His tag were thus grafted to the C terminus of the fungal GH-5 catalytic module, and the engineered enzyme, termed 5Dc (Fig. 1), was overproduced in E. coli. Contrary to 6A and 6At, 5Dc did not form inclusion bodies and the recombinant enzyme was easily purified in a soluble form. The specific activity (2,477 IU/μmol) of the engineered cellulase at 37°C on 8 g/liter CMC was found to be in the same range as that of homologous bacterial cellulase 5Ac (11), indicating that the enzyme is an endoglucanase.
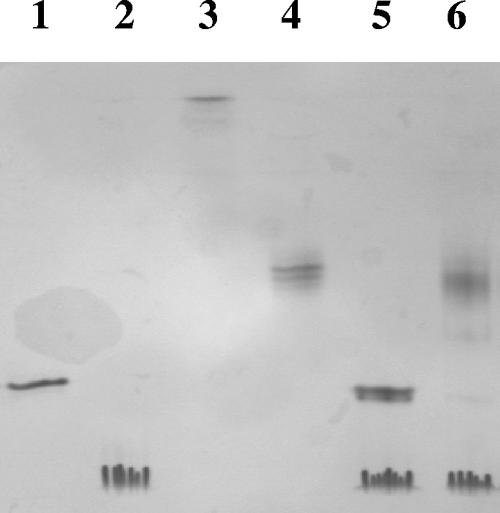
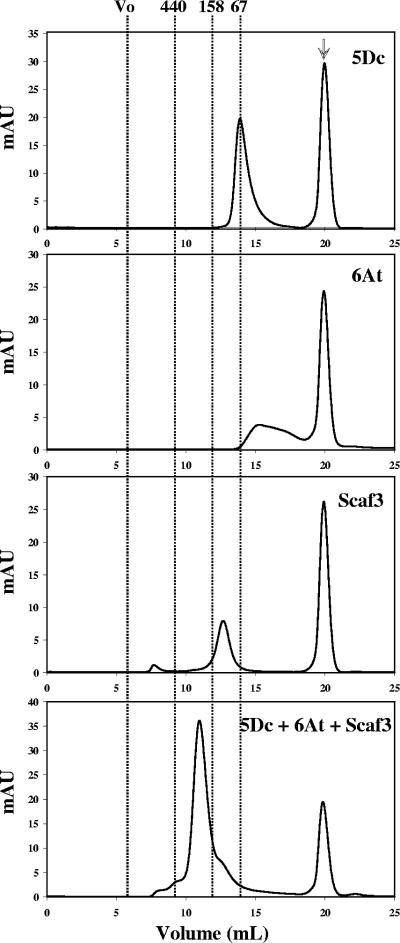
Newly designed 5Dc was mixed stoichiometrically with 6At and Scaf3 or Scaf4 in the presence of CaCl2 to form the corresponding bifunctional minicellulosome. Analyses of the resulting complexes by nondenaturing PAGE, however, revealed that complexation was only partial since both 5Dc and the free binary complex Scaf-6At were observed following interaction of the two hybrid cellulases with both chimeric scaffoldins (analyses of Scaf3-based complexes are shown in Fig. 6). The use of a different order of enzyme incorporation into the scaffoldins did not improve the yield of complexation. This result suggested that either the C. cellulolyticum dockerin of 5Dc was not functional or the interaction occurred but the components of the resulting minicellulosome dissociated during the electrophoresis step, leading to the release of 5Dc. The second hypothesis was confirmed by gel filtration analyses, which indicated that complexation was achieved by mixing stoichiometric amounts of 5Dc, 6At, and either Scaf3 (Fig. 7) or Scaf4 (data not shown), since no trace of free 5Dc was observed. Nevertheless, the partial dissociation of the dockerin grafted to Cel5D induced by nondenaturing PAGE suggests that the interaction with the cohesin partner was somewhat weaker than usually observed for the C. cellulolyticum docking system (14).
FIG. 6.
Nondenaturing PAGE analysis of Scaf3-(6At/5Dc). Lane 1, Scaf3 alone; lane 2, 5Dc alone; lane 3, 6At alone; lane 4, Scaf3 plus 6At; lane 5, Scaf3 plus 5Dc; lane 6, Scaf3 plus 6At plus 5Dc. In each lane, equimolar concentrations (10 μmol/liter) of the indicated proteins were used.
FIG. 7.
Gel filtration analysis of the assembled complex. Injected proteins (100 μl) are indicated on each chromatogram. mAU refers to milliunits of absorbance at 280 nm. Vertical lines indicate the positions of the following molecular mass markers: blue dextran (Vo), >2 MDa; ferritin, 440 kDa; aldolase, 158 kDa; bovine serum albumin, 67 kDa. For each chromatogram, the indicated proteins were at 10 μmol/liter. The peak observed at 20 ml (arrow) in all of the chromatograms corresponds to the maleic acid contained of the sample buffer.
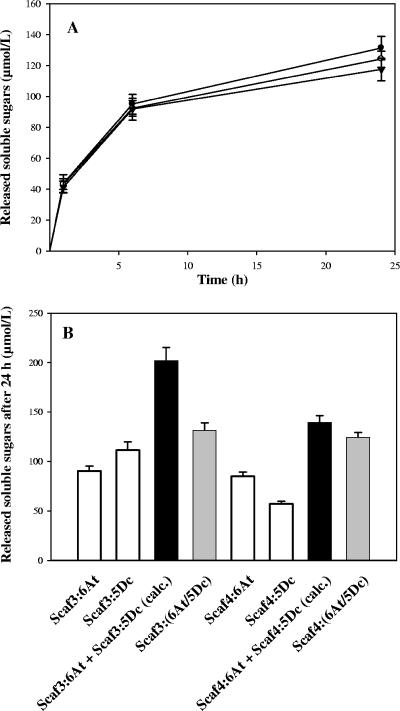
In the free state, the two fungal hybrid enzymes 6At and 5Dc did not display any synergy or competition for the degradation of crystalline cellulose Avicel (data not shown). Likewise, the binding of the fungal enzyme pair to either Scaf3 or Scaf4 had no significant impact (Fig. 8A) on their avicelase activity, which remained in the same range as that of the free-enzyme system. Thus, the fungal GH-5 enzyme in combination with 6At behaved as the bacterial GH-5 endoglucanase. Furthermore, as observed for bacterial 5Ac, binding of 5Dc alone to Scaf3 also improved the avicelase activity of the fungal cellulase (2.4-fold) but this notable CBM effect was erased when 6At was bound jointly on the same chimeric scaffoldin (Fig. 8B). Consequently, the activity of the complex Scaf3-6At/5Dc was 35% lower than the overall activity of the corresponding single-enzyme complexes.
FIG. 8.
Hydrolysis of Avicel by the fungal enzyme pair 6At plus 5Dc in the free states and various complexed states. (A) Kinetics of Avicel hydrolysis by 6At and 5Dc in the free state or bound to Scaf3 or Scaf4. Curves are labeled as follows: free 6At plus 5Dc, ▾; Scaf3-(6At/5Dc), •; Scaf4-(6At/5Dc), ○. The data are the means of four independent experiments (variation within ±5%). (B) Avicelase activities of 6At plus 5Dc bound jointly or individually to Scaf3 and Scaf4. The designated proteins were mixed in stoichiometric amounts, and the status of the protein mixture was verified by gel filtration analysis (data not shown) prior to addition of the substrate. White bars, soluble sugars released by single-enzyme complexes; gray bars, soluble sugars released by Scaf3- and Scaf4-based complexes; black bars, calculated (calc.) sum of soluble sugars released by single-enzyme complexes individually. The data are the means of four independent experiments, and the bars indicate the standard deviations.
Incorporation of 6At into trifunctional chimeric cellulosomes.
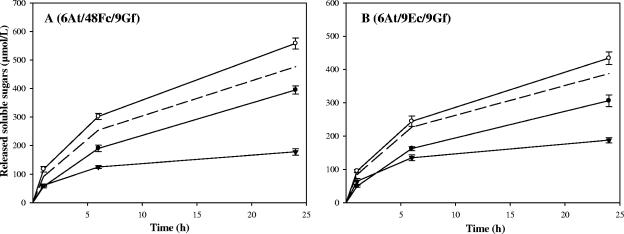
Trifunctional complexes composed of C. cellulolyticum cellulases bound to the chimeric scaffoldin Scaf6 (Fig. 1) were previously assembled. These studies showed that 48F and 9G in complex played a pivotal role in cellulose degradation by chimeric cellulosomes (13). In order to examine whether 6At would participate favorably in such a higher-order chimeric cellulosome complex, the hybrid fungal GH-6 cellulase was also bound to Scaf6, with either the potent enzyme pair 9Gf plus 48Fc or a cellulase pair that included two different modular types of family 9 enzymes, 9Gf plus 9Ec. The kinetics of cellulose degradation by the resulting trifunctional complexes are reported in Fig. 9. The trifunctional complexes Scaf6-6At/48Fc/9Gf (Fig. 9A) and Scaf6-6At/9Ec/9Gf (Fig. 9B) were found to exhibit 3.1- and 2.3-fold enhanced synergistic activity, respectively, compared to the activity of the corresponding free-enzyme systems. Comparison with the kinetics obtained for the bifunctional complexes Scaf6-48Fc/9Gf and Scaf6-9Ec/9Gf also indicates that incorporation of the GH-6 cellulase has a significant impact on the overall activity of the resulting cellulosomes. For the complex Scaf6-6At/48Fc/9Gf, the data obtained suggest that a minor proximity effect occurred between the fungal enzyme and the bacterial cellulases despite the family 3a CBM harbored by Scaf6. The avicelase activity of the trifunctional complex was improved by 18% compared with the calculated activity of Scaf6-48Fc/9Gf plus free 6At (Fig. 9A, dashed line). In the case of Scaf6-6At/9Ec/9Gf, the difference was smaller and may not be significant.
FIG. 9.
Kinetic studies of Avicel hydrolysis by trifunctional complexes versus controls. The designated proteins were mixed in stoichiometric amounts, and the status of the protein mixture was verified by nondenaturing PAGE (data not shown) prior to addition of the substrate. The panels indicate the selected enzymes as follows: panel A, 6At, 9Gf, and 48Fc; and panel B, 6At, 9Gf, and 9Ec. Curves are labeled as follows: free-enzyme systems (▾) composed of 6At, 9Gf, and 48Fc (A) and 6At, 9Gf, and 9Ec (B); bifunctional complexes (•) Scaf6-(48Fc/9Gf) (A) and Scaf6-(9Ec/9Gf) (B); trifunctional complexes (○) Scaf6-(6At/48Fc/9Gf) (A) and Scaf6-(6At/9Ec/9Gf) (B). Dashed line, calculated soluble sugars released by Scaf6-(48Fc/9Gf) plus 6At (A) and Scaf6-(9Ec/9Gf) plus 6At (B). The data are the means of four independent experiments, and the bars indicate the standard deviations.
DISCUSSION
Cohesins and dockerins, the crucial building blocks of bacterial cellulosomes, are usually genuine protein modules that keep their biological properties when they are produced alone (14). This essential property makes it possible to graft these modules to enzymes or proteins which are produced in the free state in vivo. This approach enables the production of engineered forms of the proteins of interest that can be incorporated into cellulosome-like complexes. This approach was originally applied to a free cellulase from C. thermocellum (30), thus conferring on the engineered endoglucanase CelC the ability to bind to the scaffoldin CipA. This strategy has also been successfully applied to a feruloyl esterase from Aspergillus niger (21) and led to the production of an active hemicellulase to which a C. thermocellum dockerin was appended that specifically and strongly interacted with a cohesin module of the corresponding species. More recently, the resident CBM2s of two GH-6 cellulases, an endo- and an exoglucanase, from the thermophilic bacterium Thermobifida fusca have been replaced with dockerins from divergent species (7). The resulting chimeric enzymes displayed CMCase specific activities similar to that of the wild-type enzymes, whereas replacement of CBM2 with a C. thermocellum dockerin improved the activity of the exoglucanase on amorphous cellulose. The grafted dockerin modules were also demonstrated to be functional and to interact with the cognate cohesins (7).
In the present study, a fungal cellulase, Cel6A, was selected for incorporation into chimeric minicellulosomes together with typical bacterial cellulosomal enzymes which belong to the most widespread GH families among prokaryote cellulosomal cellulases. The selected endoglucanase, which is secreted in the free state by N. patriciarum, possesses two characteristics that have yet to be observed among bacterial cellulosomal enzymes, a GH-6 catalytic module and a family 1 CBM, which is characteristic of fungal cellulases.
A C. thermocellum dockerin was therefore added to the fungal cellulase, and the family 1 CBM was maintained in the engineered enzyme. The introduction of a dockerin module had little impact on its activity, since the chimeric cellulase 6At retained approximately 80% of the activity of the parental enzyme 6A on both soluble and crystalline celluloses. The binding of 6At to chimeric scaffoldins, with or without the powerful bacterial CBM3a appended, had essentially no apparent effect on the avicelase activity of the chimeric cellulase.
Incorporation of 6At with C. cellulolyticum cellulases into bifunctional chimeric cellulosomes generated complexes that displayed no stimulation additional to the observed 2.6-fold activity enhancement over that of the free-enzyme systems. Compared to minicellulosomes exclusively composed of bacterial cellulases in which proximity and CBM effects usually occur simultaneously (10), these effects seem to be antagonistic in complexes containing the fungal cellulase. This is, for instance, reflected by the enzyme pair 6At plus 9Gc, which generated the most active complexes on Avicel. No proximity effect seemed to occur between the two enzymes when they were bound to Scaf3, and the enhanced avicelase activity of the minicellulosome was essentially due to the CBM effect induced by the CBM3a of the scaffoldin which mainly applies to 9Gc. Thus, in this complex, the two cellulases appeared to function in an independent manner. The lack of a proximity effect in the Scaf3-based complex may perhaps be explained by the dual binding of 6At to the substrate, mediated by its own CBM1 and by the CBM3a of Scaf3. Such a high level of interaction with the substrate may reduce the mobility of 6At within the minicellulosome, and its capacity to cooperate with the associated cellulase. Indeed, the C. cellulolyticum cellulosomal cellulases characterized to date lack an efficient CBM (10). Moreover, our original report on designer cellulosomes (10) demonstrated that the presence of two CBMs in a single scaffoldin had an inhibitory effect on the cellulose-degrading activity of the resulting minicellulosome. Nevertheless, we chose to maintain CBM1 in the fungal cellulase since this enzyme was selected for its elevated activity on crystalline cellulose and deletion of CBM1 has been shown to reduce the avicelase activity of Cel6A fivefold (9). When 6At and 9Gc were brought together on Scaf4, a strong proximity effect triggered significant synergy between the two enzymes. This proximity effect may complement a secondary CBM effect induced by the family 1 CBM of 6At on 9Gc. The elevated activity of the Scaf4-based hybrid cellulosome also suggests active cooperation (i.e., a proximity effect) between the fungal and bacterial enzymes; the latter complex is significantly more active than the Scaf3-based complex composed of the same enzymes, though the CBM3a harbored by Scaf3 is known to be more efficient than the fungal CBM1 (6) in terms of binding capacity on various crystalline celluloses and enhancement of the activity of the associated cellulase. It will be interesting to determine in the future whether other enzymes that bear a CBM together with a dockerin can replace the scaffoldin-borne CBM, thereby enhancing the activity of designer cellulosomes on crystalline forms of cellulose.
These observations apply to most of the other enzyme pairs tested, for which CBM and proximity effects also appeared to be antagonistic. Both effects were, however, less pronounced than in the case of 6At plus 9Gc, especially the CBM effect. Indeed, the binding of the other GH-9 cellulases and the processive GH-48 enzyme to Scaf3 alone induced inferior levels of enhancement of their avicelase activity than in the case of 9Gc, the activity of which is improved fourfold. Since in Scaf3-based complexes the CBM effect mainly applies to the bacterial cellulase, the reduced activity of the complexes containing the other enzyme pairs bound to Scaf3 is therefore not surprising.
When 6At was bound to Scaf3 or Scaf4 in combination with a bacterial or fungal GH-5 endoglucanase, a very different pattern was obtained. No activity enhancement by complexation onto either chimeric scaffoldin was observed, indicating that no significant proximity or CBM effect occurred. The avicelase activity of both the fungal and bacterial GH-5 cellulases, however, was considerably improved when these enzymes were incorporated alone into Scaf3, thus reflecting a strong CBM effect. Nevertheless, this effect no longer applies when 6At is also present in the complex. One possible explanation could be that the CBM3a of the scaffoldin anchors the minicellulosome on substrate sites for which 6At and the GH-5 cellulases are competitors. In any case, the data indicate that a combination of 6At with a GH-5 endoglucanase is not a suitable enzyme pair to achieve an efficient minicellulosome.
As mentioned above, earlier reports have shown that functional dockerins may be grafted onto noncellulosomal enzymes without impairing their activity (7, 21). To our knowledge, the present report is the first demonstration that incorporation of a noncellulosomal enzyme with a cellulosomal cellulase into a bacterial chimeric cellulosome can generate complexes with improved activity. The complexation of such mixed enzyme pairs, however, generated different effects compared to enzyme pairs composed of cellulosomal cellulases. The CBM effect appeared primarily to apply to the bacterial enzyme, but complexation onto CBM-lacking scaffoldin can trigger significant synergy between the engineered free cellulase and the cellulosomal enzyme(s) included in the complex. In this context, the avicelase activity of the chimeric minicellulosome Scaf4-(6At/9Gc) was superior to that of all Scaf4-based complexes assembled to date with C. cellulolyticum cellulases only. Thus, the technology of chimeric designer cellulosomes (2) is not restricted to enzymes that participate in cellulosome complexes in vivo and may be successfully extended to free enzymes, even those that contain modules (e.g., the family 1 CBM and the family 6 GH) never yet observed in bacterial cellulosomes. For instance, this strategy may perhaps be applied to the fungal cellulase cocktails currently used for biomass saccharification to improve their efficiency. Our results also indicate that free cellulases to which an operational CBM was appended should preferentially be incorporated into chimeric cellulosomes in which the scaffoldin is devoid of CBM3a.
Acknowledgments
We thank Odile Valette for expert technical assistance. We are grateful to Chantal Tardif, Sandrine Pagès, and Raphael Lamed for helpful discussions.
This work was supported by the CNRS, the Conseil Général des Bouches du Rhône, and the Region Provence-Alpes-Côte d'Azur. Additional support was provided by Agence Nationale de la Recherche grant ANR-05-BLAN-0259-01. E.A.B. acknowledges financial support from the Israel Science Foundation (442/05). F.M. holds a fellowship from the French Ministère de l'Enseignement Supérieur et de la Recherche.
Footnotes
Published ahead of print on 27 April 2007.
REFERENCES
- 1.Bayer, E. A., J. P. Belaich, Y. Shoham, and R. Lamed. 2004. The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Annu. Rev. Microbiol. 58:521-554. [DOI] [PubMed] [Google Scholar]
- 2.Bayer, E. A., E. Morag, and R. Lamed. 1994. The cellulosome—a treasure-trove for biotechnology. Trends Biotechnol. 12:379-386. [DOI] [PubMed] [Google Scholar]
- 3.Becker, D., K. S. Johnson, A. Koivula, M. Schulein, and M. L. Sinnott. 2000. Hydrolyses of alpha- and beta-cellobiosyl fluorides by Cel6A (cellobiohydrolase II) of Trichoderma reesei and Humicola insolens. Biochem. J. 345:315-319. [PMC free article] [PubMed] [Google Scholar]
- 4.Belaich, A., G. Parsiegla, L. Gal, C. Villard, R. Haser, and J. P. Belaich. 2002. Cel9M, a new family 9 cellulase of the Clostridium cellulolyticum cellulosome. J. Bacteriol. 184:1378-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger, E., D. Zhang, V. V. Zverlov, and W. H. Schwarz. 2007. Two noncellulosomal cellulases of Clostridium thermocellum, Cel9I and Cel48Y, hydrolyse crystalline cellulose synergistically. FEMS Microbiol. Lett. 268:194-201. [DOI] [PubMed] [Google Scholar]
- 6.Carrard, G., A. Koivula, H. Soderlund, and P. Beguin. 2000. Cellulose-binding domains promote hydrolysis of different sites on crystalline cellulose. Proc. Natl. Acad. Sci. USA 97:10342-10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caspi, J., D. Irwin, R. Lamed, Y. Shoham, H. P. Fierobe, D. Wilson, and E. A. Bayer. 2006. Thermobifida fusca family-6 cellulases as potential designer cellulosome components. Biocatal. Biotransform. 24:3-12. [Google Scholar]
- 8.Damude, H. G., V. Ferro, S. G. Withers, and R. A. Warren. 1996. Substrate specificity of endoglucanase A from Cellulomonas fimi: fundamental differences between endoglucanases and exoglucanases from family 6. Biochem. J. 315:467-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denman, S., G.-P. Xue, and B. Patel. 1996. Characterization of a Neocallimastix patriciarum cellulase cDNA (celA) homologous to Trichoderma reesei cellobiohydrolase II. Appl. Environ. Microbiol. 62:1889-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fierobe, H. P., E. A. Bayer, C. Tardif, M. Czjzek, A. Mechaly, A. Belaich, R. Lamed, Y. Shoham, and J. P. Belaich. 2002. Degradation of cellulose substrates by cellulosome chimeras. Substrate targeting versus proximity of enzyme components. J. Biol. Chem. 277:49621-49630. [DOI] [PubMed] [Google Scholar]
- 11.Fierobe, H. P., C. Gaudin, A. Belaich, M. Loutfi, E. Faure, C. Bagnara, D. Baty, and J. P. Belaich. 1991. Characterization of endoglucanase A from Clostridium cellulolyticum. J. Bacteriol. 173:7956-7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fierobe, H. P., A. Mechaly, C. Tardif, A. Belaich, R. Lamed, Y. Shoham, J. P. Belaich, and E. A. Bayer. 2001. Design and production of active cellulosome chimeras. Selective incorporation of dockerin-containing enzymes into defined functional complexes. J. Biol. Chem. 276:21257-21261. [DOI] [PubMed] [Google Scholar]
- 13.Fierobe, H. P., F. Mingardon, A. Mechaly, A. Belaich, M. T. Rincon, S. Pages, R. Lamed, C. Tardif, J. P. Belaich, and E. A. Bayer. 2005. Action of designer cellulosomes on homogeneous versus complex substrates: controlled incorporation of three distinct enzymes into a defined trifunctional scaffoldin. J. Biol. Chem. 280:16325-16334. [DOI] [PubMed] [Google Scholar]
- 14.Fierobe, H. P., S. Pages, A. Belaich, S. Champ, D. Lexa, and J. P. Belaich. 1999. Cellulosome from Clostridium cellulolyticum: molecular study of the dockerin/cohesin interaction. Biochemistry 38:12822-12832. [DOI] [PubMed] [Google Scholar]
- 15.Fillingham, I. J., P. A. Kroon, G. Williamson, H. J. Gilbert, and G. P. Hazlewood. 1999. A modular cinnamoyl ester hydrolase from the anaerobic fungus Piromyces equi acts synergistically with xylanase and is part of a multiprotein cellulose-binding cellulase-hemicellulase complex. Biochem. J. 343:215-224. [PMC free article] [PubMed] [Google Scholar]
- 16.Gal, L., C. Gaudin, A. Belaich, S. Pages, C. Tardif, and J. P. Belaich. 1997. CelG from Clostridium cellulolyticum: a multidomain endoglucanase acting efficiently on crystalline cellulose. J. Bacteriol. 179:6595-6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaudin, C., A. Belaich, S. Champ, and J. P. Belaich. 2000. CelE, a multidomain cellulase from Clostridium cellulolyticum: a key enzyme in the cellulosome? J. Bacteriol. 182:1910-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harhangi, H. R., A. C. Freelove, W. Ubhayasekera, M. van Dinther, P. J. Steenbakkers, A. Akhmanova, C. van der Drift, M. S. Jetten, S. L. Mowbray, H. J. Gilbert, and H. J. Op den Camp. 2003. Cel6A, a major exoglucanase from the cellulosome of the anaerobic fungi Piromyces sp. E2 and Piromyces equi. Biochim. Biophys. Acta 1628:30-39. [DOI] [PubMed] [Google Scholar]
- 19.Kim, H., K. H. Jung, and M. Y. Pack. 2000. Molecular characterization of xynX, a gene encoding a multidomain xylanase with a thermostabilizing domain from Clostridium thermocellum. Appl. Microbiol. Biotechnol. 54:521-527. [DOI] [PubMed] [Google Scholar]
- 20.Larsson, A. M., T. Bergfors, E. Dultz, D. C. Irwin, A. Roos, H. Driguez, D. B. Wilson, and T. A. Jones. 2005. Crystal structure of Thermobifida fusca endoglucanase Cel6A in complex with substrate and inhibitor: the role of tyrosine Y73 in substrate ring distortion. Biochemistry 44:12915-12922. [DOI] [PubMed] [Google Scholar]
- 21.Levasseur, A., S. Pages, H. P. Fierobe, D. Navarro, P. Punt, J. P. Belaich, M. Asther, and E. Record. 2004. Design and production in Aspergillus niger of a chimeric protein associating a fungal feruloyl esterase and a clostridial dockerin domain. Appl. Environ. Microbiol. 70:6984-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopez-Contreras, A. M., H. Smidt, J. van der Oost, P. A. Claassen, H. Mooibroek, and W. M. de Vos. 2001. Clostridium beijerinckii cells expressing Neocallimastix patriciarum glycoside hydrolases show enhanced lichenan utilization and solvent production. Appl. Environ. Microbiol. 67:5127-5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murray, P. G., C. M. Collins, A. Grassick, and M. G. Tuohy. 2003. Molecular cloning, transcriptional, and expression analysis of the first cellulase gene (cbh2), encoding cellobiohydrolase II, from the moderately thermophilic fungus Talaromyces emersonii and structure prediction of the gene product. Biochem. Biophys. Res. Commun. 301:280-286. [DOI] [PubMed] [Google Scholar]
- 24.Pages, S., A. Belaich, J. P. Belaich, E. Morag, R. Lamed, Y. Shoham, and E. A. Bayer. 1997. Species-specificity of the cohesin-dockerin interaction between Clostridium thermocellum and Clostridium cellulolyticum: prediction of specificity determinants of the dockerin domain. Proteins 29:517-527. [PubMed] [Google Scholar]
- 25.Pages, S., A. Belaich, H. P. Fierobe, C. Tardif, C. Gaudin, and J. P. Belaich. 1999. Sequence analysis of scaffolding protein CipC and ORFXp, a new cohesin-containing protein in Clostridium cellulolyticum: comparison of various cohesin domains and subcellular localization of ORFXp. J. Bacteriol. 181:1801-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park, J. T., and M. J. Johnson. 1949. A submicrodetermination of glucose. J. Biol. Chem. 181:149-151. [PubMed] [Google Scholar]
- 27.Raghothama, S., R. Y. Eberhardt, P. Simpson, D. Wigelsworth, P. White, G. P. Hazlewood, T. Nagy, H. J. Gilbert, and M. P. Williamson. 2001. Characterization of a cellulosome dockerin domain from the anaerobic fungus Piromyces equi. Nat. Struct. Biol. 8:775-778. [DOI] [PubMed] [Google Scholar]
- 28.Reverbel-Leroy, C., A. Belaich, A. Bernadac, C. Gaudin, J. P. Belaich, and C. Tardif. 1996. Molecular study and overexpression of the Clostridium cellulolyticum celF cellulase gene in Escherichia coli. Microbiology 142:1013-1023. [DOI] [PubMed] [Google Scholar]
- 29.Steenbakkers, P. J., X. L. Li, E. A. Ximenes, J. G. Arts, H. Chen, L. G. Ljungdahl, and H. J. Op Den Camp. 2001. Noncatalytic docking domains of cellulosomes of anaerobic fungi. J. Bacteriol. 183:5325-5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tokatlidis, K., P. Dhurjati, and P. Beguin. 1993. Properties conferred on Clostridium thermocellum endoglucanase CelC by grafting the duplicated segment of endoglucanase CelD. Protein Eng. 6:947-952. [DOI] [PubMed] [Google Scholar]
- 31.Xue, G. P., K. S. Gobius, and C. G. Orpin. 1992. A novel polysaccharide hydrolase cDNA (celD) from Neocallimastix patriciarum encoding three multi-functional catalytic domains with high endoglucanase, cellobiohydrolase and xylanase activities. J. Gen. Microbiol. 138:2397-2403. [DOI] [PubMed] [Google Scholar]
- 32.Yaron, S., E. Morag, E. A. Bayer, R. Lamed, and Y. Shoham. 1995. Expression, purification and subunit-binding properties of cohesins 2 and 3 of the Clostridium thermocellum cellulosome. FEBS Lett. 360:121-124. [DOI] [PubMed] [Google Scholar]