Abstract
To be able to function as a probiotic, bacteria have to survive the passage through the gastrointestinal tract. We have examined survival and gene expression of Lactobacillus reuteri ATCC 55730 after a sudden shift in environmental acidity to a pH close to the conditions in the human stomach. More than 80% of the L. reuteri cells survived at pH 2.7 for 1 h. A genomewide expression analysis experiment using microarrays displayed 72 differentially expressed genes at this pH. The early response to severe acid shock in L. reuteri differed from long-term acid adaptation to milder acid stress studied in other lactic acid bacteria. The genes induced included the following: clpL, genes putatively involved in alterations of the cell membrane and the cell wall; genes encoding transcriptional regulators; phage genes; and genes of unknown function. Two genes, clpL, encoding an ATPase with chaperone activity, and lr1516, encoding a putative esterase, were selected for mutation analyses. The mutants were significantly more sensitive to acid than the wild type was. Thus, these genes could contribute to the survival of L. reuteri in the gastrointestinal tract.
Lactobacillus reuteri is a commensal lactic acid bacterium that commonly inhabits the gastrointestinal tract of humans as well as of animals. No pathogenic properties have been linked to this species; instead strains of L. reuteri have been suggested to possess health-promoting, i.e., probiotic, properties (52, 60). To be able to function as a probiotic, bacteria have to survive the passage through the gastrointestinal tract. When entering the stomach, the environment changes rapidly and the bacteria are suddenly exposed to stress factors, such as various enzymes and an extremely acidic pH. Each day, approximately 2.5 liters of gastric juice, containing hydrochloric acid, is produced in the human stomach. Consequently, the fasting gastric pH is approximately 1.5, and the feeding pH is between 3.0 and 5.0 (14).
Several bacterial species are unable to survive the harsh conditions in the stomach, while others can survive the passage by using different defense mechanisms. These mechanisms often involve changes in gene expression and phenotype. A number of strategies to adapt to acidic environments have been reported in gram-positive bacteria; these strategies include pumps removing protons from the cytoplasm, such as the F1Fo ATPase proton pump or the glutamate decarboxylase system; production of general stress proteins and chaperones to repair and stabilize proteins and DNA; altered metabolism and cell envelope composition; and alkalization of the external environment, for example, with urease or through the arginine deiminase (ADI) pathway (reviewed in references 14, 17, and 57).
The aim of this study was to examine the response of L. reuteri ATCC 55730 after a shift in environmental acidity to a pH close to the conditions in the human stomach. Survival of L. reuteri after a transfer from pH 5.1 to pH 2.7 was monitored, and the early changes in gene expression were measured with oligonucleotide DNA microarrays. In this analysis, approximately 70 genes were differentially expressed in the acidic pH. The induced genes included the stress response gene clpL and genes encoding putative cell envelope-altering proteins.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used are listed in Table 1. Lactobacillus reuteri ATCC 55730 (earlier named SD2112), a strain isolated from mother's milk was grown in de Man-Rogosa-Sharpe (MRS) broth (Oxoid) at 37°C (unless stated otherwise) in plastic tubes, in which anaerobic conditions were obtained within 1 hour as indicated by an anaerobic indicator (BD). For acid shock experiments, the pH of the MRS broth was adjusted with hydrochloric acid (HCl). When solid growth medium was utilized, L. reuteri was grown on MRS agar (Oxoid) and incubated at 37°C (unless stated otherwise) under anaerobic conditions obtained with anaerobic system envelopes (GasPak; BD). When required, antibiotics were added (erythromycin, 5 μg ml−1 [Sigma]; chloramphenicol, 7.5 μg ml−1 [Sigma]). Escherichia coli was grown at 37°C in Luria-Bertani broth (LB) (51). When required, antibiotics were added (erythromycin, 400 μg ml−1 [Sigma]; kanamycin, 40 μg ml−1 [Sigma]).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain, plasmid, or primer | Description or primer sequencea | Reference or source |
|---|---|---|
| Strains | ||
| E. coli EC1000 | Strain harboring a copy of the pWV01 repA gene in the chromosome; Kanr | Used by the kind permission of J. Kok, University of Groningen (29a) |
| L. reuteri ATCC 55730 (earlier named SD2112 or MM53) | Isolated from mother's milk | Used by the kind permission of Biogaia AB, Sweden |
| L. reuteri ATCC 55730 clpL mutant | Variant with a mutated clpL | This study |
| L. reuteri ATCC 55730 lr1516 mutant | Variant with a mutated lr1516 | This study |
| L. reuteri ATCC 55730(pORI28) | Variant harboring both pVE6007 and pORI28 | This study |
| Vectors and constructions | ||
| pVE6007 | CmrrepA-positive temperature-sensitive derivative of pWV01 | Used by the kind permission of E. Maguin, INRA (34a) |
| pORI28 | EmrrepA-negative derivative of pWV01 | Used by the kind permission of J. Kok, University of Groningen (29a) |
| pORI28-lr1864 | pORI28 + 738-bp insert from lr1864 (clpL) | This study |
| pORI28-lr1516 | pORI28 + 558-bp insert from lr1516 | This study |
| Primers (purpose) | ||
| pORI28-b (detection of pORI28 + insert) | 5′-TTGTTGTTTTTATGATTACAAAGTGA-3′ | This study |
| pORI28-e (detection of pORI28 + insert) | 5′-TTGGTTGATAATGAACTGTGCTG-3′ | This study |
| pVE6007-p (detection of pVE6007) | 5′-GTTTTCCCAGTCACGACGTT-3′ | This study |
| pVE6007-s (detection of pVE6007) | 5′-GGCCGCTCTAGAACTAGTGGA-3′ | This study |
| lr1864-f (generation of gene fragment) | 5′-TGACTGGATCCTAATCAATTAATGGGTGGCATGA-3′ (starts at position 54) | This study |
| lr1864-r (generation of gene fragment) | 5′-TGACTGAATTCGCCTTGCTTGGAGCATTAAC-3′ (starts at position 791 [rev.]) | This study |
| lr1864-cf (detection of insertion) | 5′-ATGGCTCAAAACCCAATGAA-3′ (starts at position 1) | This study |
| lr1864-cr (detection of insertion) | 5′-AGCAGCTTGGAGAACGTCAT-3′ (starts at position 879 [rev.]) | This study |
| lr1516-f (generation of gene fragment) | 5′-TGACTGGATCCTAATGATCATCAACATACGATTAAGGAA-3′ (starts at position 84) | This study |
| lr1516-r (generation of gene fragment) | 5′-TGACTGAATTCTCCTCCACATACATTTTCGTA-3′ (starts at position 410 [rev.]) | This study |
| lr1516-cf (detection of insertion) | 5′-TGGTTAAAGAGGGAGTTGTTCC-3′ (starts at position 41) | This study |
| lr1516-cr (detection of insertion) | 5′-CATCCGCATATTTGATTTGG-3′ (starts at position 442 [rev.]) | This study |
| 469F3 (RT-PCR) | 5′-GGTGGATTGGAGAATTACTCTTTATTTG-3′ (starts at position 184) | This study |
| 469R3 (RT-PCR) | 5′-CCTACTTCTCCTTGTTTACTCCATTCA-3′ (starts at position 314 [rev.]) | This study |
| RTs0501F (RT-PCR) | 5′-ACCACGGCGAACGTTTGA-3′ (starts at position 132) | This study |
| RTs0501R (RT-PCR) | 5′-GTTGAAACACCCCTGATTTTTC-3′ (starts at position 184 [rev.]) | This study |
| RTs0537F (RT-PCR) | 5′-AGCAGCGATTACTGGTGCAGTT-3′ (starts at position 33) | This study |
| RTs0537R (RT-PCR) | 5′-ACCTGCTTGCACCGTGTAAATACT-3′ (starts at position 120 [rev.]) | This study |
| RT819F (RT-PCR) | 5′-TATTGCTGCTGGGCCTGATC-3′ (starts at position 411) | This study |
| RT819R (RT-PCR) | 5′-AAGCGTCAAGGTTATTTTCAACTTGT-3′ (starts at position 520) [rev.]) | This study |
| RTs0858F (RT-PCR) | 5′-CAGCATCATACCTTAACGGTGACTACT-3′ (starts at position 476) | This study |
| RTs0858R (RT-PCR) | 5′-TTTTGCCATGAACCGTAACGA-3′ (starts at position 566) [rev.]) | This study |
| RTs1797F (RT-PCR) | 5′-CCCTTGAGCTCGATCGGTTA-3′ (starts at position 212) | This study |
| RTs1797R (RT-PCR) | 5′-CCAAATACACCAGCATCATTTTT-3′ (starts at position 296) [rev.]) | This study |
| 1864F (RT-PCR) | 5′-AATGGGTGGCATGAATGGTTT-3′ (starts at position 60) | This study |
| 1864R (RT-PCR) | 5′-TGGCAATTGACCAGTTCTACGATA-3′ (starts at position 168 [rev.]) | This study |
| RTs1879F (RT-PCR) | 5′-CTGAAAAGAAACTTGATTGGTTCCA-3′ (starts at position 155) | This study |
| RTs1879R (RT-PCR) | 5′-TGCGATTACAACTTCATCCTTTGA-3′ (starts at position 231 [rev.]) | This study |
| RTs2076F (RT-PCR) | 5′-GCAGCGGCAGATGATTTGTT-3′ (starts at position 244) | This study |
| RTs2076R (RT-PCR) | 5′-ACGCCGGCAATTGTTGAA-3′ (starts at position 308 [rev.]) | This study |
Underlined nucleotides in the primer sequences indicate inserted restriction enzyme cleaving sites (BamHI and EcoRI), and nucleotides underlined twice indicate inserted stop codons. The positions where the sequences start are indicated in parentheses. rev., reverse.
Acid shock experiments.
For acid shock experiments, MRS at pH 5.8 was inoculated with an overnight culture of L. reuteri to an optical density at 600 nm (OD600) of 0.005, and the cells were grown at 37°C to an OD600 of 1.0, i.e., late exponential phase. At this OD600, the pH of the culture was 5.1. To lower the pH, the cells were diluted 1:10 in prewarmed MRS at pH 2.3, which yielded a final pH of 2.7 (referred to hereafter as acid-treated cells). For a control, cells were diluted 1:10 in prewarmed MRS at pH 5.1 (referred to hereafter as treated control cells). For a reference for the diluted samples, untreated cells were collected at time zero (referred to hereafter as untreated cells). For expression analyses, transcription was stopped after 5 and 15 min at 37°C by addition of 2/3 volume of ice-cold methanol. The cells were then harvested by centrifugation at 4,000 × g for 10 min, the supernatant was decanted, and the pellets were stored at −70°C and later utilized for isolation of RNA. To monitor survival of L. reuteri, untreated cells (time zero) and cells diluted in MRS at pH 2.3 or MRS at pH 5.1 were plated on MRS agar after 5, 15, 30, and 60 min (n = 3). The plates were incubated under anaerobic conditions for 48 h.
Isolation of RNA.
The cells from the acid shock experiments were suspended in STE (6.7% sucrose, 50 mM Tris [pH 8.0], 1 mM EDTA), harvested by centrifugation, and resuspended in STE. After transfer to tubes with Lysing Matrix B (Qbiogene), the cells were disrupted in a Fast prep instrument (Qbiogene) at the speed setting of 6.0 for 40 seconds. The tubes were centrifuged for 5 min at 16,000 × g at 2°C, and the supernatant was used for isolation of RNA with QIAGEN RNeasy kit. DNA was removed on the columns with QIAGEN RNase-free DNase.
Construction of an L. reuteri microarray.
Long oligonucleotides (60-mers) were designed and constructed for 1,864 open reading frames from a draft genome sequence of L. reuteri ATCC 55730 (5) and 15 open reading frames encoding known extracellular proteins from L. reuteri DSM 20016 (58) using OligoArray 1.0 software. Six control 60-mer oligonucleotides were also included. These controls are identical to DNA sequences from E. coli genes (yacF, ybaS, yciC, yfiF, ygjU, and yjcG) and have no sequence similarity to genes in the L. reuteri genome. Once the oligonucleotide was synthesized, oligonucleotide concentrations were normalized to a concentration of 25 μM and spotted onto Corning UltraGAPS-II slides using an OmniGrid robot (GeneMachines). Each gene was represented once on the microarray. All six of the control spots were represented eight times on the array, once in each subgrid. Oligonucleotide design and synthesis and array construction were performed at the Research Technology Support Facility at Michigan State University, East Lansing.
Oligonucleotide microarray experiment.
The design of the microarray experiment is presented in Fig. 1. The experiment was performed as direct comparisons between untreated, acid-treated, and treated control cells. Four biological replicates were performed with dye swaps for the 5-min samples and three biological replicates with dye swaps for the 15-min samples. For synthesis of cDNA, 5 to 10 μg of RNA in a volume of 12.8 μl was mixed with 5 μl (0.5 μg μl−1) random hexamers (QIAGEN). After incubation at 70°C for 10 min, 1.5 μl Superscript III (200 U μl−1) (Invitrogen), 0.5 μl RnaseOut (40 U μl−1) (Invitrogen), and 1.2 μl nucleotides (12.5 mM dATP, 12.5 mM dCTP, 12.5 mM dGTP, 7.5 mM dTTP [Invitrogen], 5 mM aminoallyl dUTP [Sigma]) were added. The reverse transcription reaction was performed at 25°C for 5 min, 50°C for 60 min, and 70°C for 15 min. To remove RNA, 15 μl of 0.1 M NaOH was added, and the samples were incubated at 70°C for 10 min. After neutralization by the addition of 15 μl of 0.1 M HCl, the cDNA was purified with the QIAGEN PCR mini kit. Next the cDNA was labeled with Cy3 or Cy5 (CyDye postlabeling reactive dye pack; Amersham) for 1 h in the dark, and the labeled cDNA for each comparison was combined and purified again by using the QIAGEN kit. To the combined sample, 1 μg salmon testes DNA (Sigma) and 1 μg yeast tRNA (Sigma) were added. After incubation at 100°C, formamide buffer (50% formamide, 10× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% sodium dodecyl sulfate [SDS]) was added to the sample, and the samples were applied to a microarray slide, which had been prehybridized in prehybridization buffer (10 g liter−1 bovine serum albumin, 5× SSC, 0.1% SDS) for 45 min, washed in water, and dried by centrifugation at 150 × g. The slide was hybridized at 42°C overnight in the dark. The slides were washed first in 2× SSC plus 2.5% SDS and 0.2× SSC plus 2.5% SDS and then repeatedly in 0.2× SSC. Finally, the microarray slides were scanned with a GenePix 4000B scanner (Amersham Pharmacia Biotech).
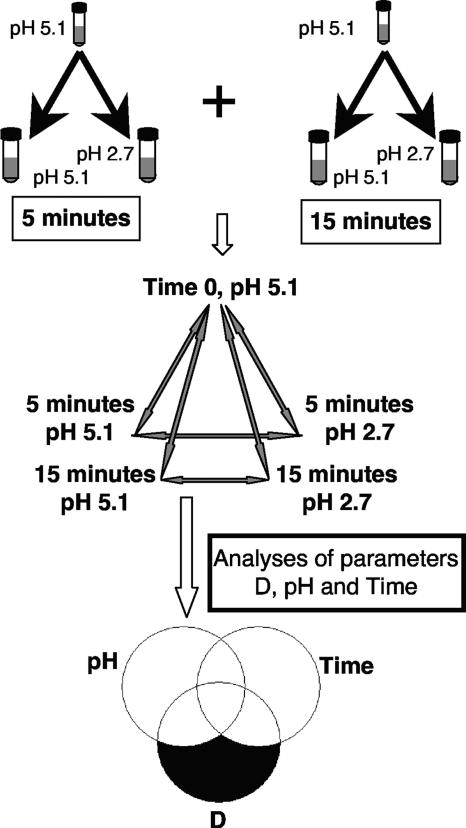
FIG. 1.
Experimental design for the gene expression studies. Cells of L. reuteri ATCC 55730 were diluted into MRS broth at pH 2.7 or MRS broth at pH 5.1 and incubated for 5 or 15 min. Direct microarray comparisons were performed for both time points between untreated cells at time zero, acid-treated cells at pH 2.7, and the treated control cells at pH 5.1. Four biological replicates were performed for 5-min incubation, and three biological replicates were performed for 15-min incubation. The data were analyzed with an empirical Bayes approach with respect to the three parameters pH, time, and dilution (D). Genes primarily affected by dilution were excluded from further studies.
Microarray and bioinformatic analyses.
GenePix Pro 4.0.12 software was utilized for image analysis of the microarrays. The raw data of spots were stored and analyzed using BASE (50), a database system for analyses of microarray data, which had been modified at the Linneaus Centre of Bioinformatics, Uppsala University, Sweden. Statistical analysis was performed in the analysis environment of Linneaus Centre of Bioinformatics Data Warehouse (http://www.lcb.uu.se./lcbdw.php) using LIMMA package in R (http://www.r-project.org) from Bioconductor (24). First, systematic variation was removed by print-tip lowess normalization (63) of the data sets. Genes represented by spots of low quality on more than 15% of the arrays were excluded from further analysis. Using LIMMA, a linear model was fit to the data in order to estimate the effects of pH, time, and dilution (Fig. 1). The genes were then ranked by a parametric empirical Bayes approach (33) to find the genes most likely to be differentially expressed. The cutoff values for the posterior log odds ratio (the B score) were set to 20 for pH, −2.5 for time, and 10 for dilution, since these values isolated distinct subgroups of genes, and for M (i.e., mean log2 ratio) for pH and dilution, respectively, to a ‖M‖ of >0.75. In addition, genes that changed less than twofold after 5 or 15 min in the expression ratio between pH 2.7 or pH 5.1 and the untreated cells at time zero were excluded from further analyses. Of the control spots, one gene (yacF) indicated expression on all slides and was not regarded in the analyses. The remaining control spots gave signals of low intensity and were used for estimation of background signals. Genes corresponding to spots of low intensity on the majority of the slides were excluded from the analyses. The function of previously uncharacterized proteins, encoded by the selected genes, were predicted with COG (Clusters of Orthologous Groups of proteins) (55, 56) and pfam (4) identified using BlastP (2) at the National Centre for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov/BLAST/). Signal peptides were searched with SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/) (7, 40, 41) for gram-positive bacteria, and transmembrane helices were searched with TMHMM (http://www.cbs.dtu.dk/services/TMHMM-2.0/) (28).
Real-time RT-PCR experiment.
To validate microarray data, the expression of eight genes was analyzed with real-time reverse transcription-PCR (RT-PCR). Genes with various gene expression ratios 15 min after acid shock or genes in the treated control were selected: lr0501 (EF534265; Spo0J-like protein), lr0537 (Apf-like protein), lr0858 (Apf-like protein), hpk3 (lr0819, encoding a histidine sensor kinase), lr1797 (putative phosphatidylglycerophosphatase), lr1864 (ClpL, ATPase with chaperone activity), lr1879 (PduD, dehydratase medium subunit) and lr2076 (transcriptional regulator). As a reference gene, lr0469 (DQ074824; SecY, protein translocase subunit), a gene exhibiting high signals on the microarrays but not displaying any changes in expression was used. Primers, which are presented in Table 1, were designed with Primer Express Software v2.0 (Applied Biosystems). After an extra DNase treatment, cDNA was constructed from 5 ng RNA and RNA removed with NaOH as described above. The cDNA was stored at −20°C. Real-time RT-PCR mixtures contained 12.5 μl Power SYBR green PCR master mix (Applied Biosystems), 4 μl diluted cDNA, 80 μM of each primer, and distilled H2O to a final volume of 25 μl. The PCR was performed at 50°C for 2 min, 95°C for 10 min, followed by 40 cycles of 95°C for 0.15 min and 60°C for 1 min in an ABI Prism 7000 sequence detection system (Applied Biosystems). Three biological replicates were analyzed using three technical replicates for each sample. For relative quantification, a calibration curve with serial dilutions (0.0626, 0.25, and 1 ng) of L. reuteri ATCC 55730 chromosomal DNA for each primer pair was generated. The results were evaluated by the method of Fredlund et al. (22). Briefly, the efficiency of cDNA amplification was calculated by the method of Wilhelm and Pingoud (61). The changes in expression between acid-treated cells or the treated control and untreated cells (time zero) were calculated by comparing the relative gene expression of the target gene to the reference gene by the method of Pfaffl (45).
Construction and analyses of a clpL mutant and an esterase mutant.
Two genes from the expression analyses were selected for construction of disruption mutants: clpL, which is similar to ATPases involved in stress response, and a putative esterase, strongly up-regulated at pH 2.7. Plasmids and primers used are listed in Table 1. The mutants were constructed principally by the method described by Walter et al. (59). The genes were inactivated by site-specific integration of plasmid pORI28 into the L. reuteri ATCC 55730 chromosome by using the temperature-sensitive plasmid pVE6007 as a helper plasmid. Internal fragments of the genes were amplified by PCR (using the insertion primers in Table 1) and cloned into pORI28 (using the EcoRI and BamHI sites) using E. coli EC1000 as the host. The helper plasmid pVE6007 was electrotransformed into L. reuteri ATCC 55730 by the protocol of Ahrné et al. (1). The phenotypic expression and incubation were performed at 35°C. In the next step, pORI28 with the insert was transformed using the same method. Bacteria carrying both plasmids were grown overnight at 35°C in the presence of erythromycin and chloramphenicol. Fifty microliters of this culture was inoculated into 10 ml of prewarmed (44°C) MRS broth without antibiotics. After incubation for 8 h at 44°C, bacteria were plated on prewarmed MRS agar plates containing erythromycin and incubated at 44°C overnight. Clones lacking pVE6007 and possessing an integrated pORI28 were isolated by replica plating on MRS plates containing either erythromycin or chloramphenicol. The integration of the pORI28 plasmids into target genes was checked by PCR using primers flanking the target region (detection primers in Table 1) and primers flanking the multicloning site of pORI28 (Table 1).
In order to test the survival at low pH, the L. reuteri ATCC 55730 clpL mutant, esterase mutant, and wild type were grown overnight at 37°C in MRS. The bacteria were diluted to an OD600 of 0.1 in 10 ml prewarmed MRS and incubated at 37°C until an OD600 of 1.0 was reached. After addition of 10 μl culture to 10 ml synthetic stomach juice (8.3 g liter−1 proteose peptone [Oxoid], 3.5 g liter−1 glucose, 2.05 g liter−1 NaCl, 0.6 g liter−1 KH2PO4, 0.11 g liter−1 CaCl2, 0.37 g liter−1 KCl, adjusted to pH 2.0 with HCl, a modification from the synthetic stomach juice of Cotter et al. [13] lacking enzymes and bile), the tubes were incubated at 37°C, and samples were removed after 20 and 50 min. Samples were also taken before addition to the synthetic stomach juice. The samples were diluted in phosphate-buffered saline and spread on MRS plates, which were incubated anaerobically for 24 h at 37°C. The experiment was repeated on three occasions, and duplicate samples were analyzed each time. The differences between the values for the mutant and wild-type strains were tested statistically by Student's t test.
RESULTS
Survival of L. reuteri after acid shock.
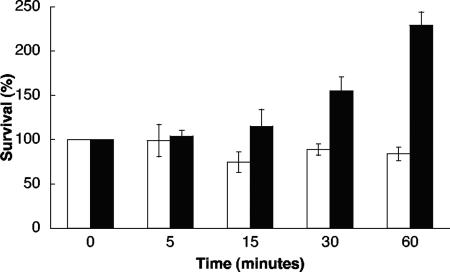
The survival of Lactobacillus reuteri ATCC 55730 after a sudden acid shock is presented in Fig. 2. More than 80% of the cells treated with acid survived at pH 2.7 for 1 h. After 2 h, approximately 60% of the cells were viable (data not shown). In the control, the cells started to grow within 30 min, and after 1 h, the number of cells had more than doubled (Fig. 2).
FIG. 2.
Survival of L. reuteri ATCC 55730 at pH 2.7 (white bars) and at pH 5.1 (black bars). The percentage of cells surviving compared to the number of cells at the start of the experiment (n = 3) is shown on the y axis. Error bars represent standard deviations.
Gene expression analyses.
Since dilution likely would affect the gene expression as well, direct comparisons between untreated cells, treated control cells (diluted into MRS at pH 5.1), and acid-treated cells (diluted into MRS at pH 2.7) were performed. The effect of dilution was estimated using a linear model, and consequently, only genes affected by dilution could be separated. Using the parametric empirical Bayes approach (33), which can account for multiple variables, three parameters, pH, time, and dilution, were considered (Fig. 1). The B score cutoff resulted in 343, 248, and 291 selected genes for the pH, time, and dilution parameters, respectively. Genes affected only by the dilution parameter were not further analyzed. To reduce the number of genes and to isolate strongly affected genes, a cutoff value for M was set at a ‖M‖ of >0.75 for pH. From the statistical ranking of the pH and time parameters, genes with at least a twofold difference in expression after 5 or 15 min compared to the untreated cells were selected. This resulted in 72 differentially expressed genes at pH 2.7 (Table 2). Of these, 34 genes were induced and 38 were repressed.
TABLE 2.
Lactobacillus reuteri genes induced or repressed during acid shock at pH 2.7
| Gene and function | Accession no.a | Description of encoded proteinb | Expression ratioc
|
Mean M pHd | |
|---|---|---|---|---|---|
| 5 min | 15 min | ||||
| Genes up-regulated at pH 2.7 | |||||
| Stress response | |||||
| lr1864 | DQ219976 | ClpL ATPase with chaperone activity | N | ++ | 1.1 |
| lr1797 | DQ219975 | Phosphatidylglycerophosphatase A and related proteins | N | ++ | 1.0 |
| Regulation and signal transduction | |||||
| lr1474 | DQ219969 | Rrf2 transcriptional regulator | ++ | +++ | 1.3 |
| lr1468 | DQ219968 | Transcriptional regulator | N | +++ | 0.83 |
| lr1933 | DQ219977 | Transcriptional regulator | N | ++ | 0.91 |
| lr1993 | DQ219980 | Transcriptional regulator | N | +++ | Te |
| lr2076 | DQ219989 | Transcriptional regulator | N | ++++ | T |
| rr7 (lr1804) | DQ219942 | Two-component signal transduction response regulator | N | ++ | 0.78 |
| Transport and metabolism | |||||
| lr1028 | DQ219961 | 6-Phosphogluconolactonase/glucosamine-6-phosphate isomerase/deaminase | N | ++ | 1.3 |
| lr0811 | DQ219956 | Deoxyxylulose-5-phosphate synthase | N | ++ | 0.87 |
| Translation and posttranslational modifications | |||||
| lr0807 | DQ219955 | Cysteinyl-tRNA synthetase | N | ++ | 0.90 |
| lr0597 | DQ219952 | Thioredoxin domain-containing protein | N | ++ | T |
| Cell envelope biogenesis | |||||
| lr2064 | DQ219988 | Endopeptidases/phage-associated protein | N | ++ | 0.83 |
| lr1516 | DQ219970 | Putative esterase | ++ | ++++ | 3.2 |
| Phage-associated genes | |||||
| lr1463 | DQ074903 | Phage terminase-like protein, large subunit | ++ | +++ | 1.4 |
| lr2045 | DQ219981 | Phage-associated protein | N | ++ | 1.1 |
| lr2047 | DQ219982 | Phage-associated protein | N | ++ | 1.1 |
| lr2051 | DQ219983 | Phage terminase | N | ++ | 0.82 |
| lr2054 | DQ219984 | Phage head maturation protease | ++ | ++ | 1.4 |
| lr2055 | DQ219985 | Phage φC31 gp36 major capsid-like protein | N | ++ | 1.1 |
| lr2057 | DQ219986 | Phage protein | + | ++ | 1.1 |
| lr2058 | DQ219987 | Phage protein | N | ++ | 0.93 |
| Unknown function | |||||
| lr0922 | DQ074860 | Extracellular hydrolase | N | ++ | 1.1 |
| lr1158 | DQ074883 | Extracellular hydrolase | N | ++ | 0.77 |
| lr1794 | DQ219974 | Flavoprotein | N | ++ | 1.1 |
| lr1937 | DQ219979 | Conserved intracellular protein of unknown function | N | ++ | 1.1 |
| lr1139 | AY970988 | Conserved intracellular protein of unknown function | N | ++ | 1.5 |
| lr2103 | DQ219991 | Conserved intracellular protein of unknown function | + | +++ | 1.2 |
| lr0997 | DQ219958 | Conserved membrane protein of unknown function | + | ++++ | 2.1 |
| lr1191 | DQ219999 | Conserved membrane protein of unknown function | N | ++ | 1.3 |
| lr1515 | DQ074905 | Unconserved extracellular protein of unknown function | ++ | ++++ | 2.5 |
| lr2117 | DQ220006 | Unconserved intracellular protein of unknown function | ++ | +++ | 1.8 |
| lr2090 | DQ219990 | Conserved membrane protein of unknown function | ++ | ++ | 1.6 |
| lr1934 | DQ219978 | Conserved intracellular protein of unknown function | N | ++ | T |
| Genes down-regulated at pH 2.7 | |||||
| Stress response | |||||
| lr0722 | DQ219953 | Cold shock protein | − | −− | −1.6 |
| Regulation and signal transduction | |||||
| lr0021 | DQ220000 | Guanosine polyphosphate pyrophosphohydrolases/ synthetases | −−− | −−− | −2.4 |
| lr0190 | DQ219995 | Transcriptional regulator | N | −−− | T |
| lr1578 | DQ219971 | Transcriptional regulator | − | −− | −0.82 |
| Transport and metabolism | |||||
| lr0382 | AY971000 | Branched-chain amino acid transport protein | −− | −− | −1.6 |
| lr0153 | DQ219993 | Arabinose efflux permease | N | −− | T |
| lr0115 | DQ219946 | Deoxynucleoside kinase | − | −− | −2.5 |
| lr1001 | DQ219959 | ADP-ribose pyrophosphatase | − | −− | −0.89 |
| lr1297 | DQ219963 | Thymidine kinase | − | −− | −1.3 |
| lr1401 | DQ219964 | Xanthine/uracil permeases | −− | −−− | −1.7 |
| lr1433 | DQ219965 | Cytidylate kinase | −− | −− | −1.6 |
| lr0320 | DQ219998 | Transport protein | −− | −−− | −2.0 |
| lr0562 | DQ220002 | MntH, Mn2+ and Fe2+ transporter of the NRAMPf family | −− | −− | −1.3 |
| Energy production and conversion | |||||
| lr0229 | DQ219949 | Flavodoxin | −− | −− | −1.4 |
| DNA replication, recombination, and repair | |||||
| lr0119 | DQ219948 | DNA polymerase III, delta subunit | N | −−− | −1.6 |
| lr1014 | DQ220003 | Site-specific recombinase XerD | N | −− | −1.3 |
| lr1240 | DQ219962 | Recombinational DNA repair ATPase | N | −− | −1.6 |
| Translation and posttranslational modification | |||||
| lr0118 | DQ219947 | Ribosomal protein S20 | N | −−− | −1.3 |
| lr0277 | DQ219997 | Ribosomal protein L10 | −− | −−− | −1.8 |
| lr0862 | DQ219957 | Asp-tRNAAsn/Glu-tRNAGln amidotransferase C subunit | −− | −− | −1.3 |
| lr1432 | DQ220005 | Ribosomal protein S1 | −− | −−−− | −1.8 |
| lr1690 | DQ219973 | Ribosomal protein L31 | N | −− | −1.4 |
| lr0251 | DQ219950 | RNase P-protein component | N | −− | T |
| lr1002 | DQ219960 | Peroxiredoxin | −− | −− | −0.95 |
| lr1465 | DQ219967 | Protease subunit of ATP-dependent Clp proteases | −− | −−− | −1.5 |
| Cell division | |||||
| lr1411 | DQ220004 | Actin-like ATPase involved in cell morphogenesis | −− | N | −1.1 |
| lr0145 | DQ219992 | Cell division protein | N | −− | −0.89 |
| Cell envelope biogenesis | |||||
| lr0537 | DQ074830 | Apf-like protein | −−−− | −−− | −4.3 |
| lr0858 | DQ074851 | Apf-like protein | −− | −− | −2.2 |
| Intracellular trafficking and secretion | |||||
| lr1103 | AY970986 | Protein translocase subunit YajC | N | −− | −0.87 |
| Unknown function | |||||
| lr0546 | DQ220001 | Recombination protein | −− | N | −1.1 |
| lr1422 | DQ074901 | Conserved extracellular protein of unknown function | − | −− | −1.4 |
| lr0318 | DQ219951 | Conserved intracellular protein of unknown function | −− | −−− | −2.0 |
| lr0733 | DQ219954 | Conserved intracellular protein of unknown function | N | −− | −1.5 |
| lr1628 | DQ219972 | Conserved intracellular protein, MarZ, of unknown function | −− | − | −1.0 |
| lr1434 | DQ074902 | Unconserved extracellular protein of unknown function | −− | −−− | −1.7 |
| lr2118 | DQ220007 | Unconserved intracellular protein of unknown function | −− | −− | −1.3 |
| lr0195 | DQ219996 | 5-Formyltetrahydrofolate cycloligase or methenyl-tetrahydrofolate synthetase | −− | −− | −1.4 |
The GenBank accession numbers are given.
Based on COG classes or high similarity to characterized proteins.
Ratio of expression for cells after 5 or 15 min at pH 2.7 and cells at time zero. Comparisons between pH 5.1 and time zero or between pH 2.7 and pH 5.1 are not displayed. The symbols show the degree of up-regulation (+) or down-regulation (−) of the genes as follows: +, 1.75 to 2-fold up-regulation; ++, 2-fold to 3-fold up-regulation; +++, 3-fold to 5-fold up-regulation; ++++, more than 5-fold up-regulation of the gene; −, 1.75 to 2-fold down-regulation; −−, 2-fold to 3-fold down-regulation; −−−, 3-fold to 5-fold down-regulation; −−−−, more than 5-fold down-regulation. N, the expression is up- or down-regulated but the changes were less than 1.75-fold.
Mean M (log2) for parameter pH.
The gene is selected because it is likely to be changed by time (parameter T).
NRAMP, natural resistance-associated macrophage protein.
In addition, 85 of the genes affected by the pH or time parameter, changed expression more than twofold in the treated control (i.e., after dilution in MRS at pH 5.1) compared to the untreated cells, i.e., they were also affected by dilution (see Table S1 in the supplemental material). A selection of these genes is presented in Table 3. Six of these genes were identical to the genes in Table 2. For example, a gene encoding a putative esterase, lr1516, was induced at pH 2.7 but repressed in the treated control. However, the majority of the genes in Table 3 were not affected or only displayed minor changes at pH 2.7 compared to untreated cells. Although affected by both the pH and dilution parameters, for this group of genes, the effect of the lowered pH counteracted the effect of dilution. These genes were consequently higher or alternatively, lower, expressed at pH 2.7 in comparison to the treated control (Table 3). This can be considered an indirect effect of the lowered pH on these genes.
TABLE 3.
Expression profiles of selected Lactobacillus reuteri genes differently regulated at pH 2.7 and in the treated control
| Gene | Accession no.a | Description of encoded proteinb | Expression ratioc
|
|||||
|---|---|---|---|---|---|---|---|---|
| 5 min at the indicated pH
|
15 min at the indicated pH
|
|||||||
| pH 2.7/T0 | pH 5.1/T0 | pH 2.7/pH 5.1 | pH 2.7/T0 | pH 5.1/T0 | pH 2.7/pH 5.1 | |||
| lr0115 | DQ219946 | Deoxynucleoside kinase | − | +++ | −−− | −− | ++ | −−−− |
| lr1516 | DQ219970 | Putative esterase | ++ | −−− | ++ | ++++ | −− | ++++ |
| lr0537 | DQ074830 | Apf-like protein | −−−− | ++++ | −−−− | −−− | ++ | −−−− |
| lr0858 | DQ074851 | Apf-like protein | −− | ++ | −−− | −− | N | −− |
| lr0318 | DQ219951 | Conserved intracellular protein of unknown function | −− | ++ | −−− | −−− | N | −−− |
| lr0733 | DQ219954 | Conserved intracellular protein of unknown function | N | ++ | −− | −− | N | −− |
| hpk3 (lr0819) | DQ219931 | Histidine sensor kinase | N | −− | +++ | N | −−− | ++++ |
| lr0628 | DQ233673 | LuxS, autoinducer-production protein | N | −− | ++ | N | −−−− | +++ |
| lr1518 | DQ233704 | ArgR, arginine repressor | N | −−− | +++ | N | −−− | ++ |
| lr1020 | DQ233707 | Ornithine carbamoyltransferase | N | −− | ++ | N | −−− | ++ |
| lr1517 | DQ233695 | ArcA, arginine deiminase | N | −−−− | ++++ | N | −−−− | ++ |
| lr1731 | DQ233708 | ArgE, acetylornithine deacetylase | N | −− | ++ | N | −− | +++ |
| lr1870 | DQ233714 | PduO | N | N | N | N | −−− | ++ |
| lr1871 | DQ233715 | PduN | N | N | + | N | −− | +++ |
| lr1872 | DQ233716 | PduM | N | N | ++ | N | −−− | ++++ |
| lr1873 | DQ233717 | PduL | N | − | + | N | −−− | +++ |
| lr1874 | DQ233718 | PduJ | N | N | N | N | −− | ++ |
| lr1875 | DQ233719 | PduK | N | −− | ++ | N | −−−− | ++++ |
| lr1876 | DQ233720 | PduH | N | − | ++ | N | −− | +++ |
| lr1877 | DQ233721 | PduG | N | −− | ++ | N | −−− | +++ |
| lr1879 | DQ233723 | PduD, dehydratase medium subunit | N | −− | ++ | N | −−−− | ++++ |
| lr1880 | DQ233724 | PduC, dehydratase large subunit | N | −− | ++ | N | −−− | ++++ |
| lr1881 | DQ233725 | PduB | N | −− | ++ | N | −−− | N |
The GenBank accession numbers are given.
Based on COG classes or high similarity to characterized proteins.
Ratio of expression for cells after 5 or 15 min at pH 2.7 or 5.1 and cells at time zero (T0). The symbols show the degree of up-regulation (+) or down-regulation (−) of the genes as follows: +, 1.75 to 2-fold up-regulation; ++, 2-fold to 3-fold up-regulation; +++, 3-fold to 5-fold up-regulation; ++++, more than 5-fold up-regulation of the gene; −, 1.75 to 2-fold down-regulation; −−, 2-fold to 3-fold down-regulation; −−−, 3-fold to 5-fold down-regulation; −−−−, more than 5-fold down-regulation. N, the expression is up- or down-regulated but the changes were less than 1.75-fold.
Expression profiles.
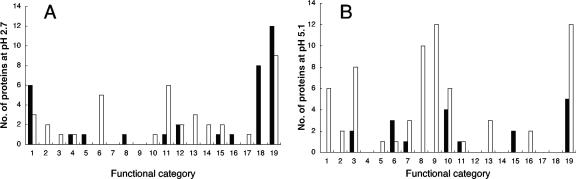
The predicted cellular functions of the genes in L. reuteri ATCC 55730 that directly or indirectly changed expression due to the acidic pH are presented in Fig. 3. Of the genes induced at pH 2.7, several were encoding regulatory, phage-associated, or metabolic proteins. The gene clpL (lr1864), which encodes an ATPase with putative chaperone activity, was also up-regulated at pH 2.7. Further, two genes belonged to COG (55, 56) classes related to lipid metabolism. These were encoding a putative deoxyxylulose-5-phosphate synthase (lr0811) and a putative phosphatidylglycerophosphatase (lr1797). The induced genes also included genes potentially involved in cell envelope biogenesis. The protein encoded by lr2064 was a putative endopeptidase, similar (35% identity) to enterolysin A from Enterococcus faecalis. Enterolysin A is a bacteriocin with cell wall-degrading ability (42). In addition, lr1516, which was strongly induced (more than sixfold) at pH 2.7, encoded a putative esterase belonging to a class of penicillin-binding proteins (beta-lactamase family class C) according to COG. The functions of several genes induced at the low pH were unknown, although for some of them, a general function was predicted. The gene lr0997 encoded a conserved membrane protein that is widespread in bacteria. This gene also exhibited similarities with the gdmH/epiH genes found in species such as Staphylococcus gallinarum and Staphylococcus epidermidis.
FIG. 3.
Predicted functions of the proteins encoded by genes induced or repressed at pH 2.7 (A) and in the treated control (B), based on COG or pfam or high similarity to characterized proteins. The number of proteins is shown on the y axis in both panels. Up-regulated genes (black bars) and down-regulated genes (white bars) are shown. Functional categories: 1, regulation and signal transduction; 2, transport; 3, amino acid transport and metabolism; 4, carbohydrate transport and metabolism; 5, coenzyme metabolism/lipid metabolism; 6, nucleotide transport and metabolism; 7, inorganic ion transport and metabolism; 8, lipid metabolism; 9, secondary metabolite biosynthesis, transport, catabolism/energy production, and conversion; 10, energy production and conversion; 11, translation, ribosomal structure, and biogenesis; 12, posttranslational modifications, protein turnover, and chaperones; 13, DNA replication, recombination, and repair; 14, cell division and chromosome partitioning; 15, cell envelope biogenesis; 16, defense mechanisms; 17, intracellular trafficking and secretion; 18, phage proteins; 19, proteins of unknown function.
In contrast to the genes induced at pH 2.7, the majority of the genes repressed at the same pH could be assigned a function. Interestingly, one of the down-regulated genes, lr0021, was similar to relA. In gram-positive bacteria, RelA is involved in both synthesis and hydrolysis of ppGpp, a signal molecule known to be involved in the stringent response and induction of tolerance against different types of stress (37, 48). Furthermore, genes involved in transport and metabolism, energy conversion and production; cell division proteins; genes involved in replication, regulation, translation, ribosome biogenesis or posttranslational modification, and the translocational subunit YajC were down-regulated (Fig. 3).
The genes changing expression at pH 2.7 clearly belonged to other classes than those differentially expressed in the treated control (Fig. 3). In the treated control, the majority of the induced genes were involved in transport and metabolism or energy production. Of the repressed genes, several were involved in transport, lipid metabolism, biosynthesis of secondary metabolites, and catabolic pathways. Among these genes, genes in the pdu operon, genes encoding components in arginine deiminase pathway, and luxS were found (Table 3).
Validation of microarray data with real-time RT-PCR.
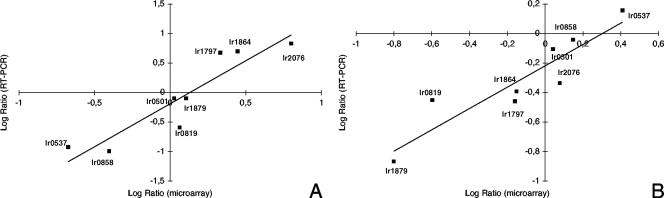
In order to confirm the results obtained with microarray hybridization, the expression of eight genes was measured with real-time RT-PCR. The relative expression ratios after 15 min between acid-treated cells or the treated control cells and untreated cells (time zero) were calculated using lr0469 (SecY) as an internal reference. This gene displayed high signals on the microarrays, although the expression did not change. In Fig. 4, the gene expression ratios of the eight genes, determined by microarray hybridizations and real-time RT-PCR, are compared. The results from the two methods displayed good correlation with Pearson correlation coefficients of r = 0.92 and r = 0.915 for the acid-treated and treated control cells, respectively.
FIG. 4.
Comparison of gene expression measurements by microarray and real-time RT-PCR. Changes in gene expression in L. reuteri ATCC 55730 after 15 min are presented as ratios (log transformed in base 10) between acid-treated cells (A) or treated control cells (B) and untreated cells. The correlation coefficients (r) were 0.92 and 0.915 for the acid-treated and treated control, respectively.
Acid tolerance of the clpL and esterase mutants.
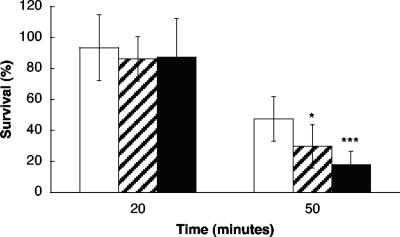
Because of the resemblance to ATPases involved in general stress response and the strong inducement at pH 2.7, clpL and a putative esterase, lr1516, respectively, were selected for functional analyses. Inactivation of the genes did not notably affect the growth of L. reuteri in MRS, although the growth rate of the esterase mutant in exponential phase was slightly lower than that of the wild type, i.e., the doubling time increased by approximately 9%. When studied under the microscope, no differences in morphology were detected between the wild type and the mutants (data not shown). Further, the survival of the mutants at low pH was tested and compared to the wild type. There were no clear differences between the wild type and the mutants when diluted in MRS to a final pH of 2.7 (data not shown). The survival was also examined in synthetic gastric juice (modified from that of Cotter et al. [13]; lacking enzymes and bile) at pH 2.0 (Fig. 5). After 20 minutes of incubation, no notable difference between the mutants and the wild type were found. However, after 50 min, the number of surviving cells of the clpL mutant and the esterase mutant were lower than that of the wild type. Approximately 47% of the wild type, 30% of the clpL mutant, and 18% of the esterase mutant cells were viable after the incubation. The differences between the wild type and mutants were statistically significant (P < 0.05 for the clpL mutant and P < 0.001 for the esterase mutant).
FIG. 5.
Survival of L. reuteri ATCC 55730 wild type (white bars), clpL mutant (shaded bars), and esterase mutant (black bars) in synthetic gastric juice at pH 2.0. The data are presented as a percentage of the number of cells at the start of the experiment (n = 3). Asterisks indicate level of statistical significance (*, P < 0.05; ***, P < 0.001) in comparison to the wild type in Student's t test. Error bars represent standard deviations.
DISCUSSION
Response of L. reuteri ATCC 55730 to acid shock.
The cell surface of bacteria is a shield against environmental stresses, such as an acidic environment. To increase the acid tolerance, bacteria can change the composition of the cell membrane or cell wall in order to decrease the permeability for protons. In Streptococcus mutans, the levels of monosaturated fatty acids and longer-chain fatty acids increase during growth at pH 5 in comparison to growth at pH 7 (47). Inhibition of membrane fatty acid alterations in this species also results in decreased tolerance to acid (21). Other oral bacteria, for example, Lactobacillus casei, shift the fatty acid composition of the membrane in response to low pH as well (20). In L. reuteri, lr1797 encoding a putative phosphatidylglycerophosphatase was induced at pH 2.7. Phosphatidylglycerophosphatase is a key enzyme in the synthesis of phosphatidylglycerol and cardiolipin, which are major acidic phospholipids of bacterial membranes (18). The up-regulation of lr1797 is thus likely part of a membrane adaptation to acidic conditions. In L. reuteri strain CRL 1098, growth with bile salts induces changes in the fraction of glycolipids and phospholipids and in the fatty acid composition of the cell membrane. The presence of bile also reduces the survival under acid stress and freezing (54). In addition, a putative esterase gene, lr1516, belonging to the COG beta-lactamase family of penicillin-binding proteins was strongly induced in L. reuteri under acidic conditions. Penicillin-binding proteins are usually involved in peptidoglycan synthesis (3). In S. mutans, the dltC gene, which is involved in synthesis and esterification of d-alanyl-lipoteichoic acid, has been studied (8). The dltC mutant is highly sensitive to acid treatment and more permeable to protons than the wild type. Furthermore, one of the genes encoding regulatory proteins induced after the acid treatment in L. reuteri was rr7 (lr1804), producing the response regulator of a two-component signal transduction system. This gene is part of an operon homologous to the yycFG operon in Bacillus subtilis (19), Streptococcus pneumoniae, Staphylococcus aureus (11), and Lactobacillus sakei (39). In B. subtilis, this system regulates genes involved in cell wall metabolism, such as components of teichoic acid biosynthesis (27). In addition, the response regulator of this system in S. pneumoniae is involved in fatty acid biosynthesis and the fatty acid composition in the cell membrane (38). Up-regulation of lr1797 and lr1516 and of the genes lr2064 and lr0811 putatively involved in cell wall biogenesis and lipid metabolism, respectively, indicates that cell envelope alterations are important for the response to acid shock in L. reuteri as well as in other bacteria.
In addition, other cell surface-associated genes were up-regulated after acid shock. The gene lr0997, encoding a conserved membrane protein, had similarities with the gdmH/epiH genes found in Staphylococcus gallinarum and Staphylococcus epidermidis for example. In S. gallinarum, GdmH has been reported to be involved in translocation of the lantibiotic as an accessory factor to the ATP-binding cassette transporter GdmT (44) and in immunity to gallidermin (26). Although lacking in several other lantibiotic gene clusters, gdmH-like genes are present in various bacterial species, implying that these genes play a role in several microbial processes (26). However, to our knowledge, no connection to acid tolerance of this gene family has been described.
Other well-known mechanisms to adapt to high acidity include inducement of general stress responses. In L. reuteri ATCC 55730, the gene clpL (lr1864), which encodes an ATPase with putative chaperone activity, was up-regulated at pH 2.7. Many members of the Clp protein family are chaperones that in association with ClpP are also involved in degradation of damaged proteins (23). Several studies have demonstrated up-regulation of the expression of clp genes in response to stress. Northern blot analysis and RT-PCR of clpP and clpL in Oenococcus oeni revealed that the both genes are induced by heat stress and in the presence of 10% ethanol (6). In Lactobacillus rhamnosus, the clpL1 and clpL2 genes were induced by heat stress (53). Further, the ClpL protein in S. mutans was produced in larger amounts when cells were grown at pH 5.0 than cells grown at pH 7.0 as detected by two-dimensional gels (31).
Activation of phage-associated genes may be due to activation of the integrated phage as an escape mechanism. Eight of the induced phage-associated genes were located in a cluster on the L. reuteri chromosome. Interestingly, one of these genes was lr2064, which may be involved in cell wall degradation. In pathogenic bacteria, phage genes are sometimes associated with genetic islands harboring virulence and colonization factors. Examples of such islands are genes encoding cholera toxin in Vibrio cholerae (36) and strain-specific genes of the enterohemorrhagic E. coli O157:H7 (25). Horizontal gene transfer is an important mechanism for bacteria to evolve and adapt to new environments. The induction of phage-associated genes at low pH could indicate that L. reuteri has obtained genes involved in acid tolerance from phages.
The majority of the genes down-regulated at pH 2.7 in L. reuteri ATCC 55730 were involved in basic cellular processes, such as replication, cell division, and translation (Fig. 3). Several species of lactic acid bacteria are able to survive under acidic conditions (57). Survival experiments (Fig. 2) showed that L. reuteri survived but did not proliferate at pH 2.7. This probably explains why genes involved in cell division or replication were down-regulated under acidic conditions. Down-regulation of genes encoding ribosomal proteins in response to acidic pH is also observed in the gram-negative bacterium Shewanella oneidensis (30). In a study on Lactobacillus plantarum, Pieterse et al. (46) demonstrate that growth rate has a large impact on gene expression under lactic acid stress.
Previous studies on acid stress in lactic acid bacteria have reported induction of heat shock proteins. However, the focus of these studies is on long-term acid stress rather than acid shock. In Lactobacillus acidophilus, the heat shock proteins DnaK, DnaJ, GrpE, GroES, and GroEL are produced as a response to acid adaptation (34). Further, the production of GroES, GroEL, and DnaK increases in Lactobacillus delbrueckii subsp. bulgaricus grown at pH 4.75 compared to cells grown at pH 6 (32). However, in Lactobacillus sanfranciscensis, only GrpE increases in acid-tolerant mutants and acid-adapted cells, while the amounts of DnaJ, DnaK, and GroES do not change (16), and in S. oneidensis (30), the expression of dnaK, dnaJ, groES-groEL, and grpE is repressed under acidic conditions. L. reuteri ATCC 55730 possesses the heat shock proteins, GroES, GroEL, DnaK, DnaJ, and GrpE (data not shown), but none of the corresponding genes were up- or down-regulated in the early response to acid shock in this study. Also, the FoF1 ATPase operon is present in L. reuteri (data not shown), but the expression of the genes in this operon did not change significantly. This system is known to mediate the extrusion of protons from the cytoplasm, and other studies on lactobacilli have demonstrated that it is involved in survival at low pH (12, 29). The genes encoding the FoF1 ATPase were up-regulated in L. plantarum when exposed to bile (10), but not under lactic acid stress (46). In addition, microarray analyses of the acid tolerance response in S. pneumoniae display an increase of less than twofold of the FoF1 ATPase operon 200 min after acid shock (35).
Indirect effects of the lowered pH.
Survival data were considered when designing the experimental setup for expression analysis (Fig. 1). Lowering the pH by direct addition of HCl to a final pH of 2 notably decreased the survival of the cells in comparison to resuspending the cells in MRS at pH 2 (data not shown). Therefore, and in order to mimic the entrance of bacteria into the gastrointestinal tract, the cells were diluted into MRS at a lower pH in the experiments. This experimental design resembles the environmental changes bacteria may experience after oral intake, both regarding the sudden shift in pH and time and the effects of dilution (Fig. 1). L. reuteri produces substantial amounts of lactate and acetate when fermenting food products (43). When the L. reuteri bacteria grow, in MRS broth, the bacteria mainly produce lactate (data not shown), but MRS also contains acetate. After the decrease in pH, most of the lactate produced by the bacteria and the acetate from the MRS broth will convert to the more antimicrobial undissociated forms of lactic acid and acetic acid, respectively. This was likely affecting the bacteria in the experiment. The detected response to the lowered pH was thus dependent on the acidity but probably also the formation of undissociated organic acids. However, the contribution of the latter could be regarded an indirect effect of the low pH resembling intake of fermented food products. Properties of the food matrix, such as content of organic acids and buffering capacity, are important to consider when discussing the response of probiotic bacteria to the acidic pH connected to ingestion.
Interestingly, the effect of dilution was not the same at pH 2.7 as in the treated control. Instead, on many genes, the lowered pH had an indirect effect, which counteracted the effect of dilution (Table 3). Most notably, several genes were repressed in the treated control although unchanged at pH 2.7. For example, these genes were involved in transport, lipid metabolism, biosynthesis of secondary metabolites, and catabolic pathways (Fig. 3). In addition, some genes in this group were also known to be important for low-pH tolerance. Four genes encoding the arginine deiminase pathway components, arginine deiminase (lr1517), arginine repressor (lr1518), acetylornithine deacetylase (lr1731), and ornithine carbamoyltransferase (lr1020), were identified. This pathway produces ammonia and contributes to alkalization of the environment. The system has been shown to be regulated by the combined effect of arginine availability, energy depletion, catabolite repression, and oxygenation rather than by low pH (reviewed in reference 57). In the L. reuteri strain CRL 1098, the ADI system probably has a role in the acid tolerance response. Adaptation at low pH in the presence of arginine in the exponential phase and energy depletion in the stationary phase seem to activate the pathway (49). The arginine/ornithine antiporter gene, arcD, connected to the ADI pathway was also induced in another L. reuteri strain, LTH5531, during sourdough fermentation (15). Furthermore, the repressed genes in the treated control contained several genes likely to be changed by decreased cell density. An example of this is the quorum-sensing gene luxS (62). The reason why these genes were not down-regulated in the acid-treated cells, as in the treated control, remains to be revealed.
The experimental design in this study provides the ability to estimate how three parameters (pH, time, and dilution) affect gene expression. When entering the gastrointestinal tract, bacteria experience a complete change in environment. The regulation of genes under these conditions will most likely be of an intricate nature. Studying complex interactions leads the focus to the experimental design and the use of relevant controls. If the untreated cells had been excluded from this analysis, the outcome would have been thoroughly different.
Role of ClpL and the putative esterase under acid shock.
As the first report on microarray analyses of Lactobacillus reuteri, this study describes the response to acid shock after a severe reduction of pH. This response differed from the responses to milder long-term acid stress studied in other species. However, clpL, earlier identified to be involved in stress tolerance, was up-regulated at low pH. Several genes putatively involved in cell envelope alterations, such as a putative esterase (lr1516) and a putative phosphatidylglycerophosphatase (lr1797), were also up-regulated after the shift in pH. Disruption of clpL or lr1516 significantly increased the sensitivity to acid shock (Fig. 5). Hence, although not essential for surviving at low pH, these genes clearly were of importance for the response of L. reuteri to acid shock. The roles of clp genes have not been studied in detail in L. reuteri, but as a member of this family (23), ClpL is likely to function as a chaperone. In Lactobacillus plantarum, in vivo experiments using an in vivo expression technology system demonstrated that clpC (lp_1019) is induced in the gastrointestinal tracts of mice (9). The corresponding gene to lr1516 in L. plantarum (lp_3312) was also induced in this experiment (9). As mentioned above, lr1516 possibly is involved in changing the cell wall and thus increasing the tolerance of the cells towards acid. Taken together, this implies that the L. reuteri clpL and lr1516 have roles in managing life in the gastrointestinal tract. The influence of lr1516 on cell wall biogenesis is currently unknown. Therefore, detailed studies of the enzymatic function, molecular mechanisms, and cellular role of this gene are required. Further studies of the composition of the cell membrane and cell wall of L. reuteri and the genes altering and regulating these properties would bring the adaptation to acidic environments to light.
Supplementary Material
Acknowledgments
This study was supported by the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (50.0288/00 and 20.9/2001-2432) and BioGaia AB, Stockholm, Sweden. R. A. Britton is supported by grants from the Rackham Foundation and the Center for Microbial Pathogenesis at Michigan State University.
We are very grateful to Anette Hagberg, Department for Plant Biology and Forest Genetics, SLU, Uppsala, Sweden, and Hanna Göransson, WCN expression array platform, Rudbeck Laboratory, Uppsala, Sweden, for support with the analyses of the microarray data and for critically reading the manuscript. We also thank the Linneaus Centre of Bioinformatics, Uppsala University, Sweden, for the opportunity to use BASE and the LCB data warehouse. We also thank Kristi Whitehead for assistance in the development of the DNA microarrays.
Footnotes
Published ahead of print on 20 April 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ahrné, S., G. Molin, and L. Axelsson. 1992. Transformation of Lactobacillus reuteri with electroporation: studies on the erythromycin resistance plasmid pLUL631. Curr. Microbiol. 24:199-205. [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Arbeloa, A., J. E. Hugonnet, A. C. Sentilhes, N. Josseaume, L. Dubost, C. Monsempes, D. Blanot, J. P. Brouard, and M. Arthur. 2004. Synthesis of mosaic peptidoglycan cross-bridges by hybrid peptidoglycan assembly pathways in gram-positive bacteria. J. Biol. Chem. 279:41546-41556. [DOI] [PubMed] [Google Scholar]
- 4.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Båth, K., S. Roos, T. Wall, and H. Jonsson. 2005. The cell surface of Lactobacillus reuteri ATCC 55730 highlighted by identification of 126 extracellular proteins from the genome sequence. FEMS Microbiol. Lett. 253:75-82. [DOI] [PubMed] [Google Scholar]
- 6.Beltramo, C., C. Grandvalet, F. Pierre, and J. Guzzo. 2004. Evidence for multiple levels of regulation of Oenococcus oeni clpP-clpL locus expression in response to stress. J. Bacteriol. 186:2200-2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 8.Boyd, D. A., D. G. Cvitkovitch, A. S. Bleiweis, M. Y. Kiriukhin, D. V. Debabov, F. C. Neuhaus, and I. R. Hamilton. 2000. Defects in d-alanyl-lipoteichoic acid synthesis in Streptococcus mutans results in acid sensitivity. J. Bacteriol. 182:6055-6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bron, P. A., C. Grangette, A. Mercenier, W. M. de Vos, and M. Kleerebezem. 2004. Identification of Lactobacillus plantarum genes that are induced in the gastrointestinal tract of mice. J. Bacteriol. 186:5721-5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bron, P. A., D. Molenaar, W. M. de Vos, and M. Kleerebezem. 2006. DNA micro-array-based identification of bile-responsive genes in Lactobacillus plantarum. J. Appl. Microbiol. 100:728-738. [DOI] [PubMed] [Google Scholar]
- 11.Clausen, V. A., W. Bae, J. Throup, M. K. Burnham, M. Rosenberg, and N. G. Wallis. 2003. Biochemical characterization of the first essential two-component signal transduction system from Staphylococcus aureus and Streptococcus pneumoniae. J. Mol. Microbiol. Biotechnol. 5:252-260. [DOI] [PubMed] [Google Scholar]
- 12.Corcoran, B. M., C. Stanton, G. F. Fitzgerald, and R. P. Ross. 2005. Survival of probiotic lactobacilli in acidic environments is enhanced in the presence of metabolizable sugars. Appl. Environ. Microbiol. 71:3060-3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cotter, P. D., C. G. Gahan, and C. Hill. 2001. A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol. Microbiol. 40:465-475. [DOI] [PubMed] [Google Scholar]
- 14.Cotter, P. D., and C. Hill. 2003. Surviving the acid test: responses of gram-positive bacteria to low pH. Microbiol. Mol. Biol. Rev. 67:429-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dal Bello, F., J. Walter, S. Roos, H. Jonsson, and C. Hertel. 2005. Inducible gene expression in Lactobacillus reuteri LTH5531 during type II sourdough fermentation. Appl. Environ. Microbiol. 71:5873-5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Angelis, M., L. Bini, V. Pallini, P. S. Cocconcelli, and M. Gobbetti. 2001. The acid-stress response in Lactobacillus sanfranciscensis CB1. Microbiology 147:1863-1873. [DOI] [PubMed] [Google Scholar]
- 17.De Angelis, M., and M. Gobbetti. 2004. Environmental stress responses in Lactobacillus: a review. Proteomics 4:106-122. [DOI] [PubMed] [Google Scholar]
- 18.Dowhan, W. 1997. Molecular basis for membrane phospholipid diversity: why are there so many lipids? Annu. Rev. Biochem. 66:199-232. [DOI] [PubMed] [Google Scholar]
- 19.Fabret, C., and J. A. Hoch. 1998. A two-component signal transduction system essential for growth of Bacillus subtilis: implications for anti-infective therapy. J. Bacteriol. 180:6375-6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fozo, E. M., J. K. Kajfasz, and R. G. Quivey, Jr. 2004. Low pH-induced membrane fatty acid alterations in oral bacteria. FEMS Microbiol. Lett. 238:291-295. [DOI] [PubMed] [Google Scholar]
- 21.Fozo, E. M., and R. G. Quivey, Jr. 2004. Shifts in the membrane fatty acid profile of Streptococcus mutans enhance survival in acidic environments. Appl. Environ. Microbiol. 70:929-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fredlund, E., C. Beerlage, P. Melin, J. Schnurer, and V. Passoth. 2006. Oxygen and carbon source-regulated expression of PDC and ADH genes in the respiratory yeast Pichia anomala. Yeast 23:1137-1149. [DOI] [PubMed] [Google Scholar]
- 23.Frees, D., K. Savijoki, P. Varmanen, and H. Ingmer. 2007. Clp ATPases and ClpP proteolytic complexes regulate vital biological processes in low GC, Gram-positive bacteria. Mol. Microbiol. 63:1285-1295. [DOI] [PubMed] [Google Scholar]
- 24.Gentleman, R. C., V. J. Carey, D. M. Bates, B. Bolstad, M. Dettling, S. Dudoit, B. Ellis, L. Gautier, Y. Ge, J. Gentry, K. Hornik, T. Hothorn, W. Huber, S. Iacus, R. Irizarry, F. Leisch, C. Li, M. Maechler, A. J. Rossini, G. Sawitzki, C. Smith, G. Smyth, L. Tierney, J. Y. Yang, and J. Zhang. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 26.Hille, M., S. Kies, F. Gotz, and A. Peschel. 2001. Dual role of GdmH in producer immunity and secretion of the staphylococcal lantibiotics gallidermin and epidermin. Appl. Environ. Microbiol. 67:1380-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howell, A., S. Dubrac, K. K. Andersen, D. Noone, J. Fert, T. Msadek, and K. Devine. 2003. Genes controlled by the essential YycG/YycF two-component system of Bacillus subtilis revealed through a novel hybrid regulator approach. Mol. Microbiol. 49:1639-1655. [DOI] [PubMed] [Google Scholar]
- 28.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 29.Kullen, M. J., and T. R. Klaenhammer. 1999. Identification of the pH-inducible, proton-translocating F1F0-ATPase (atpBEFHAGDC) operon of Lactobacillus acidophilus by differential display: gene structure, cloning and characterization. Mol. Microbiol. 33:1152-1161. [DOI] [PubMed] [Google Scholar]
- 29a.Law, J., G. Buist, A. Haandrikman, J. KoK, G. Venema, and K. Leenhouts. 1995. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J. Bacteriol. 177:7011-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leaphart, A. B., D. K. Thompson, K. Huang, E. Alm, X. F. Wan, A. Arkin, S. D. Brown, L. Wu, T. Yan, X. Liu, G. S. Wickham, and J. Zhou. 2006. Transcriptome profiling of Shewanella oneidensis gene expression following exposure to acidic and alkaline pH. J. Bacteriol. 188:1633-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Len, A. C., D. W. Harty, and N. A. Jacques. 2004. Stress-responsive proteins are upregulated in Streptococcus mutans during acid tolerance. Microbiology 150:1339-1351. [DOI] [PubMed] [Google Scholar]
- 32.Lim, E. M., S. D. Ehrlich, and E. Maguin. 2000. Identification of stress-inducible proteins in Lactobacillus delbrueckii subsp. bulgaricus. Electrophoresis 21:2557-2561. [DOI] [PubMed] [Google Scholar]
- 33.Lönnstedt, I., and T. Speed. 2002. Replicated microarray data. Stat. Sin. 12:31-46. [Google Scholar]
- 34.Lorca, G. L., G. Font de Valdez, and A. Ljungh. 2002. Characterization of the protein-synthesis dependent adaptive acid tolerance response in Lactobacillus acidophilus. J. Mol. Microbiol. Biotechnol. 4:525-532. [PubMed] [Google Scholar]
- 34a.Maguin, E., P. Duwat, T. Hege, D. Ehrlich, and A. Gruss. 1992. New thermosensitive plasmid for gram-positive bacteria. J. Bacteriol. 174:5633-5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin-Galiano, A. J., K. Overweg, M. J. Ferrandiz, M. Reuter, J. M. Wells, and A. G. de la Campa. 2005. Transcriptional analysis of the acid tolerance response in Streptococcus pneumoniae. Microbiology 151:3935-3946. [DOI] [PubMed] [Google Scholar]
- 36.McLeod, S. M., H. H. Kimsey, B. M. Davis, and M. K. Waldor. 2005. CTXphi and Vibrio cholerae: exploring a newly recognized type of phage-host cell relationship. Mol. Microbiol. 57:347-356. [DOI] [PubMed] [Google Scholar]
- 37.Mechold, U., and H. Malke. 1997. Characterization of the stringent and relaxed responses of Streptococcus equisimilis. J. Bacteriol. 179:2658-2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mohedano, M. L., K. Overweg, A. de la Fuente, M. Reuter, S. Altabe, F. Mulholland, D. de Mendoza, P. Lopez, and J. M. Wells. 2005. Evidence that the essential response regulator YycF in Streptococcus pneumoniae modulates expression of fatty acid biosynthesis genes and alters membrane composition. J. Bacteriol. 187:2357-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morel-Deville, F., F. Fauvel, and P. Morel. 1998. Two-component signal-transducing systems involved in stress responses and vancomycin susceptibility in Lactobacillus sakei. Microbiology 144:2873-2883. [DOI] [PubMed] [Google Scholar]
- 40.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 41.Nielsen, H., and A. Krogh. 1998. Prediction of signal peptides and signal anchors by a hidden Markov model. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6:122-130. [PubMed] [Google Scholar]
- 42.Nilsen, T., I. F. Nes, and H. Holo. 2003. Enterolysin A, a cell wall-degrading bacteriocin from Enterococcus faecalis LMG 2333. Appl. Environ. Microbiol. 69:2975-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Østlie, H. M., J. Treimo, and J. A. Narvhus. 2005. Effect of temperature on growth and metabolism of probiotic bacteria in milk. Int. Dairy J. 15:989-997. [Google Scholar]
- 44.Peschel, A., N. Schnell, M. Hille, K. D. Entian, and F. Gotz. 1997. Secretion of the lantibiotics epidermin and gallidermin: sequence analysis of the genes gdmT and gdmH, their influence on epidermin production and their regulation by EpiQ. Mol. Gen. Genet. 254:312-318. [DOI] [PubMed] [Google Scholar]
- 45.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pieterse, B., R. J. Leer, F. H. Schuren, and M. J. van der Werf. 2005. Unravelling the multiple effects of lactic acid stress on Lactobacillus plantarum by transcription profiling. Microbiology 151:3881-3894. [DOI] [PubMed] [Google Scholar]
- 47.Quivey, R. G., Jr., R. Faustoferri, K. Monahan, and R. Marquis. 2000. Shifts in membrane fatty acid profiles associated with acid adaptation of Streptococcus mutans. FEMS Microbiol. Lett. 189:89-92. [DOI] [PubMed] [Google Scholar]
- 48.Rallu, F., A. Gruss, S. D. Ehrlich, and E. Maguin. 2000. Acid- and multistress-resistant mutants of Lactococcus lactis: identification of intracellular stress signals. Mol. Microbiol. 35:517-528. [DOI] [PubMed] [Google Scholar]
- 49.Rollan, G., G. L. Lorca, and G. Font de Valdez. 2003. Arginine catabolism and acid tolerance response in Lactobacillus reuteri isolated from sourdough. Food Microbiol. 20:313-319. [Google Scholar]
- 50.Saal, L. H., C. Troein, J. Vallon-Christersson, S. Gruvberger, A. Borg, and C. Peterson. 2002. BioArray Software Environment (BASE): a platform for comprehensive management and analysis of microarray data. Genome Biol. 3:software0003.1-0003.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 52.Shornikova, A. V., I. A. Casas, E. Isolauri, H. Mykkanen, and T. Vesikari. 1997. Lactobacillus reuteri as a therapeutic agent in acute diarrhea in young children. J. Pediatr. Gastroenterol. Nutr. 24:399-404. [DOI] [PubMed] [Google Scholar]
- 53.Suokko, A., K. Savijoki, E. Malinen, A. Palva, and P. Varmanen. 2005. Characterization of a mobile clpL gene from Lactobacillus rhamnosus. Appl. Environ. Microbiol. 71:2061-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taranto, M. P., M. L. Fernandez Murga, G. Lorca, and G. F. de Valdez. 2003. Bile salts and cholesterol induce changes in the lipid cell membrane of Lactobacillus reuteri. J. Appl. Microbiol. 95:86-91. [DOI] [PubMed] [Google Scholar]
- 55.Tatusov, R. L., N. D. Fedorova, J. D. Jackson, A. R. Jacobs, B. Kiryutin, E. V. Koonin, D. M. Krylov, R. Mazumder, S. L. Mekhedov, A. N. Nikolskaya, B. S. Rao, S. Smirnov, A. V. Sverdlov, S. Vasudevan, Y. I. Wolf, J. J. Yin, and D. A. Natale. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tatusov, R. L., E. V. Koonin, and D. J. Lipman. 1997. A genomic perspective on protein families. Science 278:631-637. [DOI] [PubMed] [Google Scholar]
- 57.van de Guchte, M., P. Serror, C. Chervaux, T. Smokvina, S. D. Ehrlich, and E. Maguin. 2002. Stress responses in lactic acid bacteria. Antonie Leeuwenhoek 82:187-216. [PubMed] [Google Scholar]
- 58.Wall, T., S. Roos, K. Jacobsson, A. Rosander, and H. Jonsson. 2003. Phage display reveals 52 novel extracellular and transmembrane proteins from Lactobacillus reuteri DSM 20016(T). Microbiology 149:3493-3505. [DOI] [PubMed] [Google Scholar]
- 59.Walter, J., P. Chagnaud, G. W. Tannock, D. M. Loach, F. Dal Bello, H. F. Jenkinson, W. P. Hammes, and C. Hertel. 2005. A high-molecular-mass surface protein (Lsp) and methionine sulfoxide reductase B (MsrB) contribute to the ecological performance of Lactobacillus reuteri in the murine gut. Appl. Environ. Microbiol. 71:979-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weizman, Z., G. Asli, and A. Alsheikh. 2005. Effect of a probiotic infant formula on infections in child care centers: comparison of two probiotic agents. Pediatrics 115:5-9. [DOI] [PubMed] [Google Scholar]
- 61.Wilhelm, J., and A. Pingoud. 2003. Real-time polymerase chain reaction. Chembiochem 4:1120-1128. [DOI] [PubMed] [Google Scholar]
- 62.Xavier, K. B., and B. L. Bassler. 2003. LuxS quorum sensing: more than just a numbers game. Curr. Opin. Microbiol. 6:191-197. [DOI] [PubMed] [Google Scholar]
- 63.Yang, Y. H., S. Dudoit, P. Luu, D. M. Lin, V. Peng, J. Ngai, and T. P. Speed. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.