Abstract
The identification of optimal genotypes that result in improved production of recombinant metabolites remains an engineering conundrum. In the present work, various strategies to reengineer central metabolism in Escherichia coli were explored for robust synthesis of flavanones, the common precursors of plant flavonoid secondary metabolites. Augmentation of the intracellular malonyl coenzyme A (malonyl-CoA) pool through the coordinated overexpression of four acetyl-CoA carboxylase (ACC) subunits from Photorhabdus luminescens (PlACC) under a constitutive promoter resulted in an increase in flavanone production up to 576%. Exploration of macromolecule complexes to optimize metabolic efficiency demonstrated that auxiliary expression of PlACC with biotin ligase from the same species (BirAPl) further elevated flavanone synthesis up to 1,166%. However, the coexpression of PlACC with Escherichia coli BirA (BirAEc) caused a marked decrease in flavanone production. Activity improvement was reconstituted with the coexpression of PlACC with a chimeric BirA consisting of the N terminus of BirAEc and the C terminus of BirAPl. In another approach, high levels of flavanone synthesis were achieved through the amplification of acetate assimilation pathways combined with the overexpression of ACC. Overall, the metabolic engineering of central metabolic pathways described in the present work increased the production of pinocembrin, naringenin, and eriodictyol in 36 h up to 1,379%, 183%, and 373%, respectively, over production with the strains expressing only the flavonoid pathway, which corresponded to 429 mg/liter, 119 mg/liter, and 52 mg/liter, respectively.
Many plant-derived natural products are important sources for the discovery of new drugs and nutraceuticals (3, 12). However, drug development from natural compounds faces important challenges that discourage the pursuit of such bioactive materials (33). One major limitation is the low concentration of these metabolites in the host organisms, which causes limited supply for rapid research, development, and clinical trials (6). Synthetic or semisynthetic routes are often employed to remedy this problem (30). However, the complicated structure of many natural molecules often results in lengthy chemical procedures that are impractical for large-scale production. Another approach to drug discovery relies on combinatorial chemistry (5). Even though this method can generate large libraries of compounds with common core structures, only a small fraction of bioactive molecules are identified through high-throughput bioassays (33). Biochemical synthesis presents a promising alternative not only for increasing the availability of important natural products but also for diversifying their chemistry. To serve this purpose, engineered microbes are often employed as living biocatalysts (24).
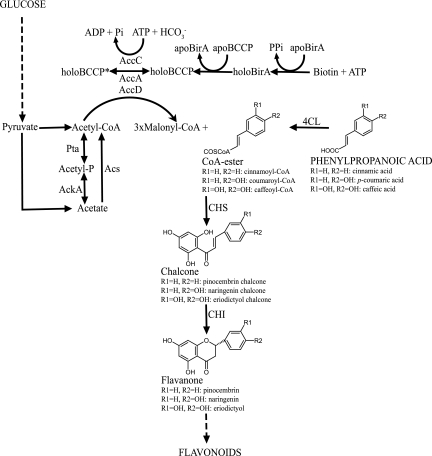
Flavonoids are plant-derived drug candidates and nutraceuticals whose biosynthesis has recently been engineered in recombinant microorganisms (reviewed in references 11 and 24). In plants, flavonoid biosynthesis starts from the conversion of phenylpropanoic acids to the coenzyme A (CoA) esters by the action of 4-coumaroyl-CoA ligase (4CL). Subsequently, three molecules of malonyl-CoA are condensed with one molecule of the CoA ester by chalcone synthase (CHS) to form chalcone. An isomerization reaction catalyzed by chalcone isomerase (CHI) then converts, in a stereospecific manner, chalcone to (2S)-flavanone, which is the first flavonoid molecule that serves as the common precursor of downstream flavonoids (Fig. 1) (44). Engineering flavanone biosynthetic circuits and coexpression with downstream flavonoid enzymes have successfully allowed the synthesis of diverse flavonoid molecules from Escherichia coli (23, 25). It has been demonstrated that, upon introduction of a recombinant plant flavanone biosynthetic circuit, intracellular malonyl-CoA derived from E. coli's native metabolism becomes the limiting precursor molecule (32). Therefore, bacterial recombinant strains can only synthesize relatively low flavonoid amounts, significantly lower than theoretical yields and much lower than alternative approaches employing Saccharomyces cerevisiae (26, 45).
FIG. 1.
Central metabolic pathways connecting malonyl-CoA with the plant flavonoid biosynthesis circuit in E. coli. Dashed arrows represent multiple catalytic steps.
In the present work, rational modifications of the acetate-acetyl-CoA-malonyl-CoA metabolic node were employed in order to improve malonyl-CoA precursor availability in flavanone-producing recombinant E. coli strains. The metabolic engineering approach targeted the multisubunit complex of acetyl-CoA carboxylase (ACC)-biotin ligase (BirA) and enzymes in acetate assimilation pathways in E. coli. The constructed strains were able to produce up to 429 ± 3 mg/liter of flavanone molecules, which is the highest flavonoid production level reported from microbial production platforms. This accounts for an increase of approximately 1,379% over the parental strains.
MATERIALS AND METHODS
Culture media and chemicals.
Luria Broth (LB) and M9 minimal media (1× M9 salts, 1% glucose, 6 nM thiamine, 1 μM MgSO4) were used throughout. Various combinations of ampicillin, kanamycin, chloramphenicol, and streptomycin were added in cultures of plasmid-bearing E. coli. BM4062 complementation medium is described in reference 4. Cinnamic acid, p-coumaric acid, and caffeic acid were purchased from MP Biomedicals Inc. Flavanone standards were purchased from Indofine. Glucose was measured using a OneTouch glucose meter (Johnson & Johnson). Detection of acetate in culture media was performed using an Enzymatische BioAnalytic kit (Roche).
Bacterial strains and plasmids.
E. coli TOP 10F′ and BL21Star (Invitrogen) were used for plasmid propagation and recombinant molecule production, respectively. E. coli K-12 and Photorhabdus luminescens were purchased from ATCC. The E. coli BM4062 mutant strain (4) was obtained from the E. coli Genetic Resource Center, Yale University. Plasmids pCOLADuet-1 and pACYCDuet-1 (Novagen), YEplac195 (ATCC no. 87589), and pTrcHis2 (Invitrogen) were used for cloning and subcloning.
DNA manipulations.
Recombinant DNA techniques were performed according to standard procedures (39). Restriction enzymes and T4 DNA ligase were purchased from New England Biolabs and Promega. Total RNA from E. coli was isolated using an RNeasy Mini kit (QIAGEN) according to the manufacturer's instructions. accA, accBC, accD, P. luminescens birA (birAPl), E. coli birA (birAEc), ackA, pta, and acs cDNA fragments were obtained and amplified by reverse transcription and PCR using SuperScript One-Step RT-PCR with Platinum Taq (Invitrogen). All PCR primers used in this study are described in the supplemental material.
Plasmid constructions.
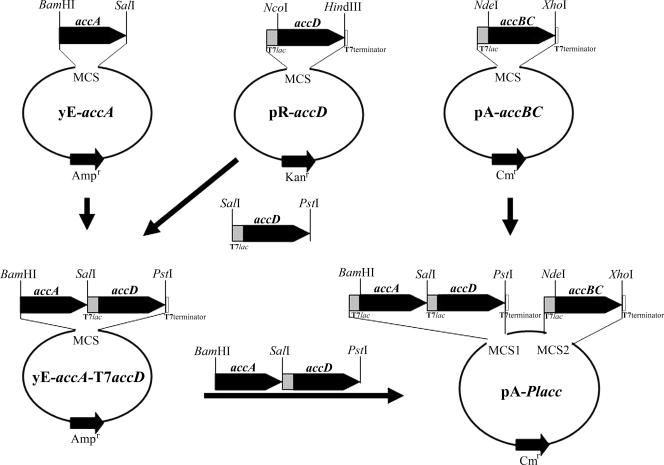
The construction of a plasmid that allowed simultaneous expression in E. coli of the entire four ACC subunits from P. luminescens is illustrated in Fig. 2. Specifically, the PCR product of accA was cloned into YEplac195 between the BamHI and SalI sites, creating plasmid YE-accA. Subsequently, accBC was amplified using forward and reverse primers of accB and accC, respectively, and was cloned into pACYCDuet1 between the NdeI and XhoI sites, resulting in plasmid pA-accBC. accD was first cloned into pRSFDuet1 between the NcoI and HindIII sites, resulting in plasmid pR-accD. Subsequently, a PCR was performed using pR-accD as a template and using a primer pair that allowed the amplification of the T7lac sequence along with the accD structural gene. The PCR product, T7accD was then cloned into YE-accA between the SalI and PstI sites to create YE-accA-T7accD. YE-accA-T7accD was further digested with BamHI and PstI and was inserted into pA-accBC digested with the same enzymes, resulting in plasmid pA-Placc (Fig. 2). In all cases, successful gene cloning was verified by restriction mapping, and the absence of undesired mutations introduced during PCR was verified by direct nucleotide sequencing.
FIG. 2.
Strategic cloning of four PlACC subunits into a low-copy-number plasmid, pACYCDuet-1, for coordinated overexpression. Black bars represent structural genes, gray bars represent T7lac promoter sequence, and white bars represent T7 terminator sequence. MCS signifies a multiple cloning site.
To allow the overexpression of BirA, birAEc was cloned into pCOLADuet-1 between the EcoRI and HindIII sites to create pC-EcBirA. Similarly, birAPl was cloned between the SacI and HindIII sites of pCOLADuet-1 to generate plasmid pC-PlBirA.
For amplification of acetate assimilation pathways, E. coli genes ackA and pta were inserted into pCOLADuet-1 vector in between EcoRI and HindIII and in between BglII and XhoI, respectively, creating plasmid pC-ackA-pta. acs was cloned separately into pCOLADuet-1 in between BamHI and PstI, generating plasmid pC-acs.
Creation of a chimeric biotin ligase.
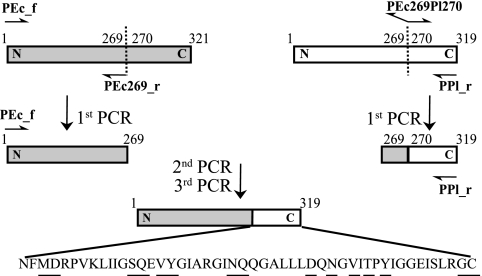
A chimeric gene (birACh) was designed to contain the N terminus of E. coli BirA and the C terminus of BirAPl. In order to determine a functional crossover, the in silico structure-guided recombination method SCHEMA (42) was used to analyze the structure of BirAEc (Protein Data Bank number: 1HXD). Identified by the SCHEMA profiler, a crossover point at residues 269 to 270 of BirAEc allowed the exchange of the C-terminal module with minimum local energy disruption. To initiate the construction of the chimeric construct (BirACh), one primer pair in addition to primers for amplification of birAEc and birAPl was constructed. The reverse primer PEc269 was designed to amplify birAEc up to the 269th codon. Primer PEc269Pl270 contains the reverse complementary sequence of PEc269 followed by a short sequence which begins from the 270th codon of birAPl. The first round of PCR was performed to amplify the N terminus of birAEc and the C terminus of birAPl using primer pair PEc_f (5′-ATGAAGGATAACACCGTGCCACTG-3′) and PEc269_r (5′-ATCCAGCTTTTCCCAGCGCGACAG-3′) and primer pair PEc269Pl270 (5′-CTGTCGCGCTGGGAAAAGCTGGATAATTTTATGGATAGGCCGGTGAAA-3′) and PPl_r (5′-TTAACAACCTCTTAAAGAAATCTCC-3′), respectively, using Expand High Fidelity Taq polymerase (Roche). After isolating the DNA fragments, a second (overlap extension) PCR was performed by using the pool of both truncated DNA fragments from birAEc and birAPl. In this case, no primer was provided in the reaction mixture, and Pfu polymerase (Stratagene) was used to avoid the attachment of adenosine nucleotide at the ends of DNA fragments. The overlap PCR was performed as follows: 1 cycle of 96°C for 1 min 30 s; 25 cycles of 94°C, 65°C, 61°C, 57°C, 53°C, 49°C, 45°C, 41°C, 72°C for 1 min 30 s each; and 1 cycle of 72°C for 7 min. Next, 10 μl of the previous PCR mixture (100 μl) was used as a template for the third PCR to amplify birACh, with PEcbirA_f and PPlbirA_r as primers (Fig. 3).
FIG. 3.
Creation of chimeric BirA (BirACh) by recombination of the N-terminal domain of BirAEc (gray bars) and the C-terminal module of BirAPl (white bars). Underlined amino acids represent residues that are exchanged due to domain swapping.
Heterologous expression and flavonoid production.
Parental E. coli strains E1 and E2, preengineered to produce plant flavanones, were transformed with various plasmids constructed in this study to create different recombinant strains (Table 1). Cell cultures were grown at 37°C unless another specific temperature is indicated and with orbital shaking at 300 rpm. For protein expression and heterologous flavanone production, strains were grown for 18 h in LB. Subsequently, cells were transferred into fresh LB medium at a starting A600 of 0.1 and grown until A600 reached 0.6. At that point, the expression of the recombinant genes was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and incubation continued at 30°C for an additional 3 h. At the end of the incubation period, cells were collected by centrifugation and cultured in fresh M9 minimal medium at A600 of 0.6. At that point, 3 mM phenylpropanoic acid substrates and 1 mM IPTG were added for biotransformation. Incubation continued at 30°C for 36 h prior to analysis of recombinant products.
TABLE 1.
Plasmids and strains used in the present study
| Plasmid or strain | Relevant properties or genetic marker | Source or reference(s) |
|---|---|---|
| Plasmids | ||
| pACYCDuet-1 | P15A(pACYC184), Cmr | Novagen |
| pCOLADuet-1 | ColA, Kmr | Novagen |
| pETDuet-1 | ColE1(pBR322), Ampr | Novagen |
| pCDFDuet-1 | CloDF13, Strr | Novagen |
| pTrcHis2 | pBR322, Ampr | Invitrogen |
| pC-Pc4cl2 | pCDFDuet-1 carrying 4cl-2 from Petroselinum crispum | 25 |
| pE-Phchs-Phchi | pETDuet-1 carrying chs, chi from Petunia hybrida | 23, 25 |
| pE-Phchs-Mschi | pETDuet-1 carrying chs from P. hybrida, chi from Medicago sativa | This study |
| pR-accD | pACYCDuet-1 carrying accD from P. luminescens | |
| yE-accA | YEplac195 carrying accA from P. luminescens | This study |
| yE-accA-T7accD | yE-accA carrying T7lac-accD fusion | This study |
| pA-accBC | pACYCDuet-1 carrying accBC from P. luminescens | This study |
| pA-Placc | pACYCDuet-1 carrying accABCD from P. luminescens | This study |
| pT-PlbirA | pTrcHis2 carrying birA from P. luminescens | This study |
| pT-EcbirA | pTrcHis2 carrying birA from E. coli K-12 | This study |
| pT-ChbirA | pTrcHis2 carrying chimeric birA | This study |
| pC-PlbirA | pCOLADuet-1 carrying birA from P. luminescens | This study |
| pC-EcbirA | pCOLADuet-1 carrying birA from E. coli K-12 | This study |
| pC-ChbirA | pCOLADuet-1 carrying chimeric birA | This study |
| pC-ackA-pta | pCOLADuet-1 carrying ackA, pta from E. coli K-12 | This study |
| pC-acs | pCOLADuet-1 carrying acs from E. coli K-12 | This study |
| Strains | ||
| E. coli BL21Star | F−ompT hsdSB (rB− mB−) gal dcm rne131 (DE3) | Invitrogen |
| E. coli BM4062 | araD139([]) DE(argF-lac)169 LAM TP(bioF-lacZ)501 flhD5301 DE(fruK-yeiR)725(fruA25)relA1 rpsL150(strR) rbsR22 birA85 DE(fimB-fimE)632(::IS1) deoC1 | The E. coli Genetic Resource Center |
| P. luminescens | Type strain | ATCC |
| E. coli K-12 | Type strain | ATCC |
| E1 | BL21Star(pC-Pc4cl2, pE-Phchs-Phchi) | This study |
| E2 | BL21Star(pC-Pc4cl2, pE-Phchs-Mschi) | This study |
| E1A/E2A | E1(pA-Placc)/E2(pA-Placc) | This study |
| E1ABp/E2ABp | E1A(pC-PlbirA)/E2A(pC-PlbirA) | This study |
| E1ABe/E2ABe | E1A(pC-EcbirA)/E2A(pC-EcbirA) | This study |
| E1ABc/E2ABc | E1A(pC-ChbirA)/E2A(pC-ChbirA) | This study |
| E1AAP/E2AAP | E1A(pC-ackA-pta)/E2A(pC-ackA-pta) | This study |
| E1AA/E2AA | E1A(pC-acs)/E2A(pC-acs) | This study |
Analytical methods.
To analyze flavonoid production, small samples of cell culture were collected. E. coli cells were separated through centrifugation, and the culture media were analyzed by high-performance liquid chromatography, using an Agilent 1100 series instrument and a reverse-phase ZORBAX SB-C18 column (4.6 by 150 mm) maintained at 25°C. Flavonoid compounds were separated by elution with an acetonitrile/water gradient at a flow rate of 1.0 ml/min under the following conditions: 10 to 40% (vol/vol) acetonitrile for 10 min, 40 to 60% (vol/vol) acetonitrile for 5 min, and 60 to 10% (vol/vol) acetonitrile for 2 min. The retention times under these conditions for the standard authentic samples were as follows: caffeic acid, 6 min; p-coumaric acid, 7.5 min; eriodictyol 11 min; cinnamic acid, 12.1 min; naringenin, 12.8 min; and pinocembrin, 16.3 min. Recombinant products were detected by monitoring absorbance at 290 nm. Flavanones derived from E. coli were identified by cochromatography and by matching the retention times and UV absorbance profiles with those of authentic compounds as previously described (25, 26, 45). The flavonoid productions from the various recombinant strains were presented as the averages of three independent experiments.
RESULTS
Coordinated overexpression of ACC multisubunit enzyme complex.
Malonyl-CoA is the first committed precursor in fatty acid metabolism in which the ATP-dependent synthesis from acetyl-CoA and bicarbonate is mediated by ACC. In previous reports, it has been suggested that ACC is a rate-limiting enzyme in fatty acid biosynthesis (17). Since malonyl-CoA is also a precursor in flavonoid biosynthesis, our initial strategy was to increase ACC enzyme levels through episomal overexpression of the E. coli enzyme complex. The genes accA, accB, accC, and accD encode the four ACC subunits: carboxyltransferase α, biotin carboxyl carrier protein (BCCP), biotin carboxylase, and carboxyltransferase β, respectively. It is known that the E. coli ACC is feedback inhibited by acyl-acyl carrier proteins (16). Moreover, overproduction of four-subunit E. coli ACC has been shown to affect cell viability (17), and flavonoid production was not improved significantly when the two-subunit ACC from Corynebacterium glutamicum was expressed in E. coli (32).
The effects of the heterologous expression of the entire four ACC subunits from other gram-negative microorganisms in E. coli have not been demonstrated. In this work, we chose to investigate the overexpression of ACC from the gram-negative bacterium P. luminescens, PlACC. PlACC is composed of four subunits, similar to the E. coli ACC. Each PlACC subunit exhibits close homology to that of the E. coli enzyme (87%, 74%, 91%, and 78% identity, respectively, for accA, accB, accC, and accD). In E. coli, accB and accC genes are organized in an operon (27, 29), whereas accA and accD are individually transcribed (28). To avoid growth inhibition due to disproportional expression of ACC (17), the overexpression of the four PlACC subunits was designed according to the transcription mode of native E. coli proteins. Specifically, the expression of each of the P. luminescens ACC carboxyltransferase subunits was regulated individually by the T7lac promoter, whereas the BCCP and biotin carboxylase subunits were expressed as an operon under a single T7lac promoter. Through subcloning, all of the four genes encoding the PlACC proteins were cloned in a single low-copy-number plasmid (pACYCDuet-1, 10 copies) to create plasmid pA-Placc (Fig. 2).
To test the ability of PlACC overexpression to improve flavonoid production, pA-Placc was introduced into two E. coli strains preengineered to allow the biosynthesis of the three common flavonoid precursors, pinocembrin, naringenin, and eriodictyol. E1 strain was constructed by grafting the genes coding for 4CL from Petroselinum crispum (Pc4CL2) and CHS (PhCHS) and CHI (PhCHI) from Petunia hybrida into BL21Star (23). The E2 strain contained the same set of flavonoid genes as E1 with the exception of the gene coding for CHI, which was derived from Medicago sativa (MsCHI) (Table 1). In the recent past, E1 derivatives have been used to produce important downstream flavonoids such as flavones (23) and flavonols (25). Recently, we demonstrated that reconstitution of CHI activity with MsCHI resulted in improvement in flavanone production from E2. The robust activity of MsCHI was identified through comparative biochemical characterization of several CHI enzymes; the more efficient activity is likely due to the differential expression in E. coli and its unique substrate specificity (Y. Yan and M. A. Koffas, unpublished data). Transformation of E1 and E2 with pA-Placc created E1A and E2A, respectively. To assess flavanone biosynthesis, recombinant E. coli strains were cultured in M9 minimal media supplemented with glucose as the sole carbon source and phenylpropanoic acids. Upon IPTG induction, flavonoids secreted in culture media were analyzed using high-performance liquid chromatography after 24 h for optimum flavonoid recombinant production from E. coli (23).
While E. coli ACC overexpression has been shown to result in growth retardation (17), in the present study, upon protein induction of PlACC, no difference in the growth rates of E1A and E2A was observed, compared to rates in the parental strains E1 or E2, respectively. Additionally, colony-forming ability of E1A and E2A was also retained. From monitoring of the flavanone production from the recombinant strains, as shown in Table 2, it is evident that the expression of PlACC increased the production of the three common flavonoid precursors, pinocembrin, naringenin, and eriodictyol. Since the plant biosynthetic pathway in E1A is not as efficient as in E2A (due to the expression of different CHI proteins) (Yan et al., unpublished data), elevation of ACC activity only increased flavanone production modestly. However, in E2A, the recombinant plant pathway did not pose a rate-limiting step and as a result an increase of the intracellular malonyl-CoA pool led to efficient flavanone synthesis. In this case, the synthesis of pinocembrin reached 196 ± 19 mg/liter, a production level that corresponded to a 576% increase over the control E2 production (Table 2).
TABLE 2.
Flavanone production from parental flavanone producer strains (E1 and E2) and engineered strains with PlACC overexpression (E1A and E2A)
| Strain and flavanone | Production (mg/liter) | Growth (A600) | ΔPa |
|---|---|---|---|
| E1 | |||
| Pinocembrin | 1 | 4 | |
| Naringenin | 3 | 4 | |
| Eriodictyol | 0.1 | 5 | |
| E1A | |||
| Pinocembrin | 2 ± 1 | 4 | 100 |
| Naringenin | 4 | 5 | 33 |
| Eriodictyol | 0.1 | 7 | 0 |
| E2 | |||
| Pinocembrin | 29 ± 2 | 2 | |
| Naringenin | 42 ± 1 | 2 | |
| Eriodictyol | 11 ± 1 | 3 | |
| E2A | |||
| Pinocembrin | 196 ± 19 | 3 | 576 |
| Naringenin | 67 | 2 | 60 |
| Eriodictyol | 17 | 4 | 55 |
ΔP, production increase (%) of E1A and E2A relative to E1 and E2, respectively.
Optimization of PlACC activity.
ACC is a biotin-dependent carboxylase that requires covalent linkage of biotin cofactor to one of its four subunits (BCCP) by the action of BirA (43). To test the feasibility of further improving flux through ACC, we aimed at increasing the net rate of ACC biotinylation by overexpressing BirAPl and BirAEc. BM4062 is a birA mutant that is impaired in biotin utilization and can only grow in the presence of high biotin concentrations (4) or after restoration of BirA activity. The functionality of BirAPl and BirAEc was first tested by their ability to complement BM4062. For this purpose, birAPl and birAEc were cloned into pTrcHis2, to create pT-PlbirA and pT-EcbirA, respectively. BM4062 strains were then transformed with these plasmids, and complementation was confirmed by the colony-forming ability at 30°C on minimal salt agar media supplemented with glucose and IPTG, but without biotin (4). Control BM4062, transformed with an empty pTrcHis2 plasmid, did not grow. However, BM4062 transformed with pT-PlbirA and pT-EcbirA formed colonies on biotin-deficient plates containing IPTG, which indicated the complementation by the expression of functional BirAPl and BirAEc, respectively.
To allow the coexpression of PlACC and the recombinant flavonoid pathway, birAPl and birAEc were cloned individually into the multicopy vector pCOLADuet-1 to generate plasmids pC-PlbirA and pC-EcbirA, respectively. In these plasmids, protein expression was regulated by the T7lac promoter. E1A and E2A were then transformed individually with pC-PlbirA or pC-EcbirA to create strains E1ABp, E1ABe, E2ABp, and E2ABe. It has been reported in the past that the intracellular biotin concentration in E. coli is very low (<0.05 nm) (7, 19). Additionally, because the BirA-biotinoyl-5′-AMP complex acts as a repressor of the biotin biosynthetic (bio) operon, the overexpression of BirA severely represses endogenous E. coli biotin biosynthesis (15). For these reasons, biotin availability in these newly generated recombinant strains was postulated to be low. In order to ensure the optimum biotinylation of apo-BCCP (the biotinylated accB subunit of ACC), exogenous biotin was supplemented in the medium at a concentration of 4 μM. Production analysis from the modified recombinant strains, shown in Table 3, demonstrated that flavanone production increased due to BirAPl coexpression. For example, pinocembrin production from E1ABp and E2ABp was enhanced by 100% and 1,166%, respectively, over the control E1 and E2 strains (Table 3). Pinocembrin synthesized from E2ABp increased 87% over E2A, and no significant improvement from E1ABp over E1A was observed. In summary, with this strategy, the maximum production of pinocembrin, naringenin, and eriodictyol reached 367 ± 1 mg/liter, 69 ± 2 mg/liter, and 50 ± 1 mg/liter, respectively. It is important to note however, that the observed flavanone production increase is conditional upon exogenous biotin supplementation. For example, the absence of exogenous biotin in the culture medium resulted in the production of only 230 ± 2 mg/liter of pinocembrin by strain E2ABp, which is 37% lower than the production observed when biotin is present at 4 μM.
TABLE 3.
Flavanone production from engineered strains with PlACC coexpression with BirAPl (E1ABp/E2ABp), BirAEc (E1ABe/E2ABe), or BirACh (E1ABc/E2ABc)a
| Strain and flavanone | BirAPl
|
BirAEc
|
BirACh
|
ΔPp | ΔPe | ΔPc | |||
|---|---|---|---|---|---|---|---|---|---|
| Pp | G | Pe | G | Pc | G | ||||
| E1A | |||||||||
| Pinocembrin | 2 | 3 | 1 | 3 | 2 ± 1 | 3 | 100 | 0 | 100 |
| Naringenin | 5 | 3 | 3 | 3 | 5 | 3 | 67 | 0 | 67 |
| Eriodictyol | 0.2 | 4 | 0.1 | 4 | 0.1 | 4 | 100 | 0 | 0 |
| E2A | |||||||||
| Pinocembrin | 367 ± 1 | 3 | 226 ± 17 | 3 | 355 ± 2 | 3 | 1,166 | 679 | 1,124 |
| Naringenin | 69 ± 2 | 3 | 61 ± 3 | 3 | 69 ± 2 | 3 | 64 | 45 | 64 |
| Eriodictyol | 50 ± 1 | 4 | 29 ± 1 | 3 | 30 | 3 | 355 | 164 | 173 |
Pp, Pe, Pc, production (mg/liter) from E1ABp/E2ABp, E1ABe/E2ABe, and E1ABc/E2ABc, respectively; G, growth level (A600); ΔPp, production increase (%) of E1ABp and E2ABp relative to E1 and E2, respectively; ΔPe, production increase (%) of E1ABe and E2ABe relative to E1 and E2, respectively; ΔPc, production increase (%) of E1ABc and E2ABc relative to E1 and E2, respectively.
Interestingly enough, flavanone synthesis from E1ABe did not improve. For example, pinocembrin production from E2ABe only improved by 679% over E1 and E2, respectively (Table 3). The lower production improvement from E. coli strains coexpressing PlACC with BirAEc compared to those coexpressing PlACC with BirAPl suggested that the level of enhancement of the PlACC activity depended on the biotinylating protein. In the past, computational analysis based on structural determination postulated that the small C-terminal domain of BirA mediated the interactions with BCCP (43). To provide further evidence that the differential PlACC activity was due to the interaction with BirA, a chimeric BirA (BirACh) consisting of the N terminus of BirAEc and the C terminus of BirAPl was created. The design of BirACh through domain swapping was aided by the SCHEMA profiler, which identifies fragments of proteins that can be exchanged without compromising the integrity of protein folding (42). BirAEc is composed of 321 residues, and the SCHEMA analysis at the C terminus identified that a recombination point at position 270 resulted in minimum energy disruption. Based on this prediction, BirACh was created by recombining the first 269 residues of BirAEc with the last 50 residues of BirAPl (319 amino acids) (Fig. 3). The functionality of BirACh was tested by the ability to complement BM4062 after cloning into pTrcHis2 plasmid. BM4062 expressing the C-terminal truncation of BirAEc did not grow on selective media without biotin. However, biotin auxotrophy was recovered by BM4062 expressing BirACh, which demonstrated the functionality of the chimera. To investigate the activity of BirACh with PlACC, BirACh was cloned into pCOLADuet-1 vector, creating pC-ChbirA. This construct was then introduced into E1A and E2A to create E1ABc and E2ABc, respectively (Table 1). These strains were cultured and induced as previously described to achieve flavanone production. As shown in Table 3, coexpression of PlACC with BirACh restored production improvements. For example, pinocembrin production from E1ABc and E2ABc increased 100% and 1,124%, respectively, over E1 and E2, and corresponded to 100% and 57% increases over E1ABe and E2ABe, respectively. These results demonstrated that when the C terminus of BirAEc was replaced with the BirAPl module, the reduction of flavanone synthesis from the coexpression of BirAEc and PlACC could be rescued. It can also be suggested that the interaction of ACC with BirA is important for ACC activity, with the C-terminal module of BirA playing a key role in this protein coupling.
Increasing acetyl-CoA availability.
Acetyl-CoA is an intermediate in glycolysis and is involved in myriad metabolic functions. With glucose as a carbon source, acetyl-CoA metabolism in E. coli under aerobic conditions also leads to acetate formation (31). Acetate is a toxic by-product and has been shown to inhibit the growth rate of E. coli (31, 36, 38). In E. coli, acetate is naturally recycled to form acetyl-CoA through two distinct pathways. The first pathway interconverts acetate and acetyl-CoA through two enzymes, AckA and Pta (20, 34). The second pathway uptakes acetate through the action of Acs.
In order to achieve further flavonoid production improvement while simultaneously reducing acetate accumulation in the fermentation broth, we increased the expression of the two acetate assimilation pathways in E. coli separately. For this purpose, ackA and pta were cloned into pCOLADuet-1 to create pC-ackA-pta. Similarly, acs was also cloned into pCOLADuet-1 vector to create pC-acs. The overexpression of the acetate assimilation enzymes was individually regulated by the T7lac promoter. Introduction of pC-ackA-pta and pC-acs separately into E1 and E2 strains expressing PlACC created strains E1AAP, E1AA, E2AAP, and E2AA, respectively (Table 1).
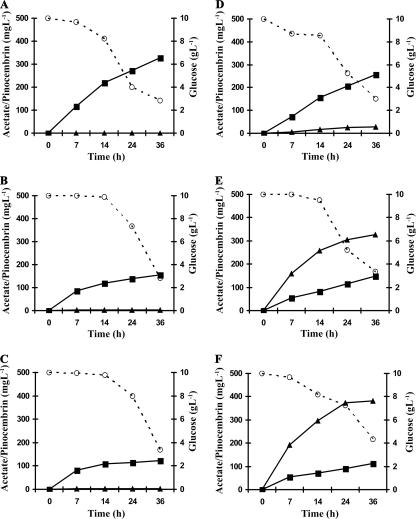
To monitor the availability of extracellular acetate, the recombinant strains were cultured in minimal salt medium supplemented with glucose over the course of 36 h of protein induction and cinnamic acid biotransformation. As shown in Fig. 4, the amount of secreted acetate from all of the recombinant strains increased as a function of time. However, after a 14-h period, acetate availability was significantly lower in cultures that overexpressed the acetate assimilation pathways than in the control strains E1 or E2. From these results, it also appeared that the overexpression of Acs resulted in more efficient acetate assimilation than in the double AckA Pta overexpression. Moreover, in general, acetate production was lower from E2 derivatives (Fig. 4D to F) than from E1 strains (Fig. 4A to C). Glucose uptake by E2 derivatives (Fig. 4D to F) was also lower than with E1 derivatives (Fig. 4A to C).
FIG. 4.
Dynamics of acetate, glucose, and pinocembrin in minimal media upon culturing of recombinant E. coli strains. (A) E1; (B) E1AAP; (C) E1AA; (D) E2; (E) E2AAP; (F) E2AA. Glucose, circles; acetate, squares; and pinocembrin, triangles.
Next, the level of flavanone production through improvement of acetate assimilation was monitored. As shown in Table 4 and Fig. 4, flavanone synthesis from recombinant E. coli overexpressing acetate assimilation enzymes was greater than that of the control strains harboring only the plant pathway or together with ACC. For example, the synthesis of pinocembrin from E2AA improved by 1,221% and 95% relative to E2 and E2A, respectively. The use of acetate as a single carbon source resulted in marked toxicity and severely inhibited cell growth (results not shown). However, incorporation of 2 g/liter acetate into glucose-containing media did not negatively affect cell growth and further increased flavanone productions (Table 5). In general, together with ACC, the overexpression of Acs enhanced flavanone synthesis more prominently than the double AckA Pta overexpression. The highest production levels of pinocembrin, naringenin, and eriodictyol from E1AA were 4 mg/liter, 8 mg/liter, and 3 mg/liter, respectively. From E2AA, peak production of pinocembrin, naringenin, and eriodictyol reached 429 ± 3 mg/liter, 119 ± 0 mg/liter, and 52 ± 1 mg/liter, respectively, which corresponded to 1,379%, 183%, and 373% increases over control E2 production from glucose. Correspondingly, the levels of pinocembrin, naringenin, and eriodictyol synthesis from E2AA were 119%, 78%, and 205%, respectively, higher than that from E2A (Table 2).
TABLE 4.
Flavanone production from engineered strains with PlACC coexpression with AckA and Pta (E1AAP/E2AAP) or Acs (E1AA/E2AA)a
| Strain and flavanone | AckA Pta
|
Acs
|
ΔPap | ΔPaa | ||
|---|---|---|---|---|---|---|
| Pap | G | Paa | G | |||
| E1A | ||||||
| Pinocembrin | 2 | 3 | 3 | 3 | 100 | 200 |
| Naringenin | 7 | 5 | 8 ± 1 | 6 | 133 | 167 |
| Eriodictyol | 0.1 | 5 | 0.2 | 4 | 0 | 100 |
| E2A | ||||||
| Pinocembrin | 328 ± 23 | 3 | 383 ± 18 | 3 | 1,031 | 1,221 |
| Naringenin | 85 ± 2 | 3 | 87 ± 3 | 3 | 102 | 107 |
| Eriodictyol | 44 ± 2 | 3 | 51 ± 1 | 3 | 300 | 364 |
Pap, Paa, production (mg/liter) from E1AAP/E2AAP and E1AA/E2AA, respectively; G, growth level (A600); ΔPap, production increase (%) of E1AAP and E2AAP relative to E1 and E2, respectively; ΔPaa, production increase (%) of E1AA and E2AA relative to E1 and E2, respectively.
TABLE 5.
Flavanone production from engineered strains with PlACC coexpression with AckA and Pta (E1AAP/E2AAP) or Acs (E1AA/E2AA)a
| Strain and flavanone | AckA Pta
|
Acs
|
ΔPap | ΔPaa | ||
|---|---|---|---|---|---|---|
| Pap | G | Paa | G | |||
| E1A | ||||||
| Pinocembrin | 3 | 3 | 4 | 3 | 200 | 300 |
| Naringenin | 8 ± 1 | 4 | 8 | 4 | 167 | 167 |
| Eriodictyol | 2 | 3 | 3 | 3 | 1,900 | 2,900 |
| E2A | ||||||
| Pinocembrin | 405 ± 11 | 3 | 429 ± 3 | 3 | 1,297 | 1,379 |
| Naringenin | 104 | 3 | 119 | 3 | 148 | 183 |
| Eriodictyol | 45 ± 4 | 3 | 52 ± 1 | 3 | 309 | 373 |
Pap, Paa, production (mg/liter) from E1AAP/E2AAP and E1AA/E2AA, respectively; G, growth level (A600); ΔPap, production increase (%) of E1AAP and E2AAP relative to E1 and E2, respectively; ΔPaa, production increase (%) of E1AA and E2AA relative to E1 and E2, respectively. Cultures were grown in minimal media supplemented with 2 g/liter acetate in addition to glucose.
DISCUSSION
Microbes have been engineered for a diverse array of biotechnological applications (2, 9, 13, 18, 22, 24, 37). Within this context, metabolic engineering of E. coli has allowed the biosynthesis of high-value plant flavonoids (reviewed in reference 24). However, low productivity remains a bottleneck for large-scale applications and low-cost production. We hypothesized that the low titer of flavonoid production from recombinant E. coli is partially due to the low intracellular concentration of malonyl-CoA (41). In this work, we presented a number of metabolic engineering strategies directed at increasing carbon flux toward malonyl-CoA.
In the first strategy, we engineered the heterologous expression of ACC from a gram-negative bacterium, P. luminescens. Surprisingly, this expression did not result in growth inhibition, unlike the overexpression of the native E. coli ACC protein complex (17). A recent study reported that the expression of the two-subunit ACC from the gram-positive bacterium C. glutamicum resulted in pinocembrin and naringenin synthesis of up to approximately 1 mg/liter and 0.7 mg/liter, respectively (32), from a flavanone-producing E. coli. In order to elevate production levels, the authors first cultivated the recombinant E. coli in a large volume of rich media. In the next step, biomass was harvested and resuspended in minimal media to a final concentration of 50 g/liter. Through this approach, pinocembrin and naringenin production reached approximately 60 mg/liter in 36 h (32). In the present work, the coordinated overexpression of the four-subunit enzyme assembly from the gram-negative bacterium P. luminescens resulted in flavanone production of up to 196 ± 19 mg/liter, 67 ± 0 mg/liter, and 17 ± 0 mg/liter for pinocembrin, naringenin, and eriodictyol, respectively, without the need of biomass concentration.
The lack of significant amounts of endogenous BirA could also be a limiting factor in the synthesis of active holo-PlACC. To ensure optimum biotinylation of holo-PlACC, BirAEc and BirAPl were overproduced through a high-copy-number episomal expression (20 to 40 copies). This resulted in a further increase of the flavanone production from the recombinant strains. For example, pinocembrin synthesis increased by 87% from E2ABp over E2A. To our surprise, flavanone production from recombinant strains coexpressing PlACC and BirAEc did not improve considerably (Table 3). In general, even though biotin ligases demonstrate relatively low protein sequence homology (for example, BirAPl and BirAEc have a 67% homology at the protein level), BirA proteins are interchangeable between organisms, as all of them have been shown to complement E. coli birA mutants (8). However, various studies in the past indicated that the recognition and specificity in protein biotinylation were mediated through the assembly of a protein complex structure (10, 35). In fact, the ACC-BirA complex has been isolated from a thermostable bacterium but not from enterobacteria due to metastability (14). We aimed at validating that the differential coupling between PlACC with BirA affected ACC activity, and hence flavanone production. For this purpose, a chimeric BirA consisting of the N terminus of BirAEc and the C terminus of BirAPl was created (Fig. 3). By coexpressing PlACC with BirACh, the reduction of flavanone synthesis due to BirAEc coexpression was rescued. For example, pinocembrin production was increased by 81% compared to the single PlACC expression in E2, which corresponded to a 57% increase over the coexpression of PlACC and BirAEc in E2 (Table 3). These results could provide evidence that the interaction of ACC with BirA can affect the activity of the former enzyme and that the C-terminal unit of BirA plays a key role in supporting the protein complex. Overall, auxiliary overproduction of ACC with BirA further increased flavanone productions (Table 3), although exogenous biotin was required for optimum flavanone production.
An alternative strategy to increase flux toward malonyl-CoA through elevation of acetyl-CoA was also explored. Specifically, we chose to increase flavonoid productivity while simultaneously reducing acetate toxicity by amplifying the two E. coli acetate assimilation pathways. The results indicated that when PlACC was coexpressed with AckA and Pta or with Acs, flavanone generation increased up to 1,221% over the control strain with glucose as the sole carbon source. Improvement of flavanone synthesis correlated to pronounced acetate utilization in these strains (Fig. 4). In general, higher acetate accumulation was observed from strains expressing the AckA-Pta pathway than those expressing Acs, which is likely due to the reversibility of the AckA-Pta pathway (21). Nevertheless, since acetate still accumulated upon Acs overproduction, it was suggested that the activity of acetate uptake enzymes could still be improved. From these engineered strains, exogenous supplementation of the inexpensive acetate resulted in the synthesis of pinocembrin, naringenin, and eriodictyol up to 429 ± 3 mg/liter, 119 ± 0 mg/liter, and 52 ± 1 mg/liter, respectively, in 36 h. To the best of our knowledge, these are the highest flavanone production levels from any recombinant system that has been reported to date.
The presented metabolic engineering strategies of increasing flavanone precursor metabolites will open the way for high-level production of other downstream flavonoids, as well as unnatural analogues. In perspective, exploration of various solutions is especially important since cellular complexities complicate the identification of a singular approach that can be used for strain improvements (1, 2, 40). The various approaches presented in this work can provide comprehensive assessments of optimum “atom economy,” which are especially necessary for large-scale microbial production of flavonoids.
Supplementary Material
Acknowledgments
This work was partially supported by a grant from the U.S. National Science Foundation (BES-0331404) to M.A.G.K. and the New York State Professional Development Award and the Mark Diamond Research Grant (SUM-05-14) to E.L.
The assistance of Sei-Fei Tan and Christopher Renzi is gratefully acknowledged.
Footnotes
Published ahead of print on 27 April 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alper, H., Y. S. Jin, J. F. Moxley, and G. Stephanopoulos. 2005. Identifying gene targets for the metabolic engineering of lycopene biosynthesis in Escherichia coli. Metab. Eng. 7:155-164. [DOI] [PubMed] [Google Scholar]
- 2.Alper, H., K. Miyaoku, and G. Stephanopoulos. 2005. Construction of lycopene-overproducing E. coli strains by combining systematic and combinatorial gene knockout targets. Nat. Biotechnol. 23:612-616. [DOI] [PubMed] [Google Scholar]
- 3.Balunas, M. J., and A. D. Kinghorn. 2005. Drug discovery from medicinal plants. Life Sci. 78:431-441. [DOI] [PubMed] [Google Scholar]
- 4.Barker, D. F., and A. M. Campbell. 1980. Use of bio-lac fusion strains to study regulation of biotin biosynthesis in Escherichia coli. J. Bacteriol. 143:789-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burke, M. D., E. M. Berger, and S. L. Schreiber. 2004. A synthesis strategy yielding skeletally diverse small molecules combinatorially. J. Am. Chem. Soc. 126:14095-14104. [DOI] [PubMed] [Google Scholar]
- 6.Butler, M. S. 2004. The role of natural product chemistry in drug discovery. J. Nat. Prod. 67:2141-2153. [DOI] [PubMed] [Google Scholar]
- 7.Campbell, A., A. Del Campillo-Campbell, and R. Chang. 1972. A mutant of Escherichia coli that requires high concentrations of biotin. Proc. Natl. Acad. Sci. USA 69:676-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campeau, E., and R. A. Gravel. 2001. Expression in Escherichia coli of N- and C-terminally deleted human holocarboxylase synthetase. Influence of the N-terminus on biotinylation and identification of a minimum functional protein. J. Biol. Chem. 276:12310-12316. [DOI] [PubMed] [Google Scholar]
- 9.Chang, M. C., and J. D. Keasling. 2006. Production of isoprenoid pharmaceuticals by engineered microbes. Nat. Chem. Biol. 2:674-681. [DOI] [PubMed] [Google Scholar]
- 10.Chapman-Smith, A., T. W. Morris, J. C. Wallace, and J. E. Cronan, Jr. 1999. Molecular recognition in a post-translational modification of exceptional specificity. Mutants of the biotinylated domain of acetyl-CoA carboxylase defective in recognition by biotin protein ligase. J. Biol. Chem. 274:1449-1457. [DOI] [PubMed] [Google Scholar]
- 11.Chemler, J. A., Y. Yan, and M. A. Koffas. 2006. Biosynthesis of isoprenoids, polyunsaturated fatty acids and flavonoids in Saccharomyces cerevisiae. Microb. Cell Fact. 5:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chin, Y. W., M. J. Balunas, H. B. Chai, and A. D. Kinghorn. 2006. Drug discovery from natural sources. AAPS J. 8:E239-E253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cirino, P. C., J. W. Chin, and L. O. Ingram. 2006. Engineering Escherichia coli for xylitol production from glucose-xylose mixtures. Biotechnol. Bioeng. 95:1167-1176. [DOI] [PubMed] [Google Scholar]
- 14.Clarke, D. J., J. Coulson, R. Baillie, and D. J. Campopiano. 2003. Biotinylation in the hyperthermophile Aquifex aeolicus. Eur. J. Biochem. 270:1277-1287. [DOI] [PubMed] [Google Scholar]
- 15.Cronan, J. E., Jr. 1988. Expression of the biotin biosynthetic operon of Escherichia coli is regulated by the rate of protein biotination. J. Biol. Chem. 263:10332-10336. [PubMed] [Google Scholar]
- 16.Davis, M. S., and J. E. Cronan, Jr. 2001. Inhibition of Escherichia coli acetyl coenzyme A carboxylase by acyl-acyl carrier protein. J. Bacteriol. 183:1499-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davis, M. S., J. Solbiati, and J. E. Cronan, Jr. 2000. Overproduction of acetyl-CoA carboxylase activity increases the rate of fatty acid biosynthesis in Escherichia coli. J. Biol. Chem. 275:28593-28598. [DOI] [PubMed] [Google Scholar]
- 18.Demain, A. L. 2006. From natural products discovery to commercialization: a success story. J. Ind. Microbiol. Biotechnol. 33:486-495. [DOI] [PubMed] [Google Scholar]
- 19.Eisenburg, M. A., B. Mee, O. Prakash, and M. R. Eisenburg. 1975. Properties of α-dehydrobiotin-resistant mutants of Escherichia coli K-12. J. Bacteriol. 122:66-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.el-Mansi, E. M., and W. H. Holms. 1989. Control of carbon flux to acetate excretion during growth of Escherichia coli in batch and continuous cultures. J. Gen. Microbiol. 135:2875-2883. [DOI] [PubMed] [Google Scholar]
- 21.Kirkpatrick, C., L. M. Maurer, N. E. Oyelakin, Y. N. Yoncheva, R. Maurer, and J. L. Slonczewski. 2001. Acetate and formate stress: opposite responses in the proteome of Escherichia coli. J. Bacteriol. 183:6466-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee, W., T. K. Wood, and W. Chen. 2006. Engineering TCE-degrading rhizobacteria for heavy metal accumulation and enhanced TCE degradation. Biotechnol. Bioeng. 95:399-403. [DOI] [PubMed] [Google Scholar]
- 23.Leonard, E., J. Chemler, K. H. Lim, and M. A. Koffas. 2006. Expression of a soluble flavone synthase allows the biosynthesis of phytoestrogen derivatives in Escherichia coli. Appl. Microbiol. Biotechnol. 70:85-91. [DOI] [PubMed] [Google Scholar]
- 24.Leonard, E., Z. L. Fowler, and M. A. G. Koffas. 2007. Metabolic engineering, p. 301-359. In M. Al-Rubeai and M. Fusseneger (ed.), Systems biology, vol. 5. Springer, London, United Kingdom. [Google Scholar]
- 25.Leonard, E., Y. Yan, and M. A. Koffas. 2006. Functional expression of a P450 flavonoid hydroxylase for the biosynthesis of plant-specific hydroxylated flavonols in Escherichia coli. Metab. Eng. 8:172-181. [DOI] [PubMed] [Google Scholar]
- 26.Leonard, E., Y. Yan, K. H. Lim, and M. A. Koffas. 2005. Investigation of two distinct flavone synthases for plant-specific flavone biosynthesis in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 71:8241-8248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, S. J., and J. E. Cronan, Jr. 1992. The gene encoding the biotin carboxylase subunit of Escherichia coli acetyl-CoA carboxylase. J. Biol. Chem. 267:855-863. [PubMed] [Google Scholar]
- 28.Li, S. J., and J. E. Cronan, Jr. 1992. The genes encoding the two carboxyltransferase subunits of Escherichia coli acetyl-CoA carboxylase. J. Biol. Chem. 267:16841-16847. [PubMed] [Google Scholar]
- 29.Li, S. J., and J. E. Cronan, Jr. 1993. Growth rate regulation of Escherichia coli acetyl coenzyme A carboxylase, which catalyzes the first committed step of lipid biosynthesis. J. Bacteriol. 175:332-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lombardino, J. G., and J. A. Lowe III. 2004. The role of the medicinal chemist in drug discovery—then and now. Nat. Rev. Drug Discov. 3:853-862. [DOI] [PubMed] [Google Scholar]
- 31.Luli, G. W., and W. R. Strohl. 1990. Comparison of growth, acetate production, and acetate inhibition of Escherichia coli strains in batch and fed-batch fermentations. Appl. Environ. Microbiol. 56:1004-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyahisa, I., M. Kaneko, N. Funa, H. Kawasaki, H. Kojima, Y. Ohnishi, and S. Horinouchi. 2005. Efficient production of (2S)-flavanones by Escherichia coli containing an artificial biosynthetic gene cluster. Appl. Microbiol. Biotechnol. 68:498-504. [DOI] [PubMed] [Google Scholar]
- 33.Paterson, I., and E. A. Anderson. 2005. Chemistry. The renaissance of natural products as drug candidates. Science 310:451-453. [DOI] [PubMed] [Google Scholar]
- 34.Pruss, B. M., and A. J. Wolfe. 1994. Regulation of acetyl phosphate synthesis and degradation, and the control of flagellar expression in Escherichia coli. Mol. Microbiol. 12:973-984. [DOI] [PubMed] [Google Scholar]
- 35.Reche, P. A., M. J. Howard, R. W. Broadhurst, and R. N. Perham. 2000. Heteronuclear NMR studies of the specificity of the post-translational modification of biotinyl domains by biotinyl protein ligase. FEBS Lett. 479:93-98. [DOI] [PubMed] [Google Scholar]
- 36.Roe, A. J., D. McLaggan, I. Davidson, C. O'Byrne, and I. R. Booth. 1998. Perturbation of anion balance during inhibition of growth of Escherichia coli by weak acids. J. Bacteriol. 180:767-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rui, L., L. Cao, W. Chen, K. F. Reardon, and T. K. Wood. 2004. Active site engineering of the epoxide hydrolase from Agrobacterium radiobacter AD1 to enhance aerobic mineralization of cis-1,2-dichloroethylene in cells expressing an evolved toluene ortho-monooxygenase. J. Biol. Chem. 279:46810-46817. [DOI] [PubMed] [Google Scholar]
- 38.Russell, J. B., and F. Diez-Gonzalez. 1998. The effects of fermentation acids on bacterial growth. Adv. Microb. Physiol. 39:205-234. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 40.Stephanopoulos, G., and A. J. Sinskey. 1993. Metabolic engineering—methodologies and future prospects. Trends Biotechnol. 11:392-396. [DOI] [PubMed] [Google Scholar]
- 41.Takamura, Y., and G. Nomura. 1988. Changes in the intracellular concentration of acetyl-CoA and malonyl-CoA in relation to the carbon and energy metabolism of Escherichia coli K12. J. Gen. Microbiol. 134:2249-2253. [DOI] [PubMed] [Google Scholar]
- 42.Voigt, C. A., C. Martinez, Z. G. Wang, S. L. Mayo, and F. H. Arnold. 2002. Protein building blocks preserved by recombination. Nat. Struct. Biol. 9:553-558. [DOI] [PubMed] [Google Scholar]
- 43.Weaver, L. H., K. Kwon, D. Beckett, and B. W. Matthews. 2001. Competing protein:protein interactions are proposed to control the biological switch of the E coli biotin repressor. Protein Sci. 10:2618-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winkel-Shirley, B. 2001. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126:485-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yan, Y. J., A. Kohli, and M. A. G. Koffas. 2005. Biosynthesis of natural flavanones in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 71:5610-5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.