Abstract
Here we used a multigene sequencing approach for the identification and molecular typing of environmental mycobacteria of the fast-growing subgroup. Strains were isolated from hemodialysis water and clinical samples. Eleven type strains of related species of the genus were also included in this study. To gain further insight into the diversity of the environmental mycobacteria, we analyzed several housekeeping genes (16S rRNA, ITS1, gyrB, hsp65, recA, rpoB, and sodA). No individual phylogenetic tree allowed good discrimination of all of the species studied. However, a concatenated and a consensus analysis, combining the genes, allowed better discrimination of each strain to the species level, and the increase in sequence size also led to greater tree robustness. This approach is useful not only for the discrimination and identification of environmental mycobacteria but also for their molecular typing and studies of population genetics. Our results demonstrate high genetic diversity among the isolates obtained, which are probably new species of the genus.
In recent years, there has been a marked increase in the number of mycobacterial species for which valid descriptions are available (41, 42). The genus Mycobacterium currently has more than 125 species, as denoted in J. P. Euzéby's List of Prokaryotic Names with Standing in Nomenclature (http://www.bacterio.cict.fr). This genus includes organisms that cause serious diseases in humans and animals. A first classification system on the basis of pigmentation and growth rate was introduced. Slow-growing mycobacteria, defined as those that showed growth in >7 days, were divided into the following categories: group I, photochromogens; group II, scotochromogens; group III, nonphotochromogens. Rapid-growing mycobacteria (RGM), defined as those showing growth in <7 days, were designated group IV. This classification is consistent with the genotypic taxonomy defined for the genus. Most pathogenic mycobacteria are slow growers. Other Mycobacterium species, some common in water, are included in the RGM group and occasionally infect humans. When isolated from samples taken from patients, these organisms are referred to as “atypical,” “nontuberculous mycobacteria,” or simply “environmental mycobacteria” (EM). EM colonize a wide range of surfaces, and because of their resistance to disinfectants they can survive in hospitals and cause nosocomial outbreaks (45). EM have been increasingly implicated in human diseases, including nosocomial infections, pulmonary and disseminated diseases in immunocompromised patients, and a variety of other diseases (5, 49). EM are ubiquitous (8, 13) and are found in many ecosystems, such as dust or soil (20, 26). However, in most of these instances, water is proposed as a possible common vehicle of transmission. These bacteria have been isolated from treated drinking water (9, 25, 35), hospital and clinic water systems (44), water supplies in hemodialysis centers (6, 16), and various other sites, including dental units (34), ice machines (15, 24), swimming pool and hot tub water (12, 19, 27), metalworking fluid (28, 36), and aerosols generated during the dismantling of moisture-damaged buildings (43). This group of mycobacteria is heterogeneous in terms of epidemiology, clinical disease spectrum, and drug susceptibility and is now considered a group of emerging waterborne pathogens.
In the clinical laboratory, it is still difficult to identify and differentiate closely related species of mycobacteria by phenotypic and biochemical tests, particularly for some very common species. Phenotypic methods are relatively slow, require expertise, and often use nonstandardized reagents. The reference molecular method for identification is determination of the sequences of the 16S rRNA genes, but there are instances in which the sequences of these genes have been found to be very similar, if not identical, between species of the genus, thereby making it necessary to find alternative specific sequences. Other DNA sequences or genes present in all mycobacteria have been described for the differentiation of species, such as internal transcribed sequences between the 16S and 23S rRNA genes (ITS1) (32); dnaJ (39); hsp65, which encodes the 65-kDa heat shock protein (31, 38); gyrB, which encodes the β subunit of DNA gyrase (10); recA, which has a crucial function in homologous DNA recombination; DNA damage repair, and induction of the SOS response (4); rpoB, which encodes the β subunit of RNA polymerase (1); the gene for the 32-kDa protein (37); secA1, which codes for the essential protein SecA1, a key component of the major pathway of protein secretion across the cytoplasmic membrane (47); and sodA, which encodes the superoxide dismutase (48). However, information gained from a single gene may be unreliable in indicating genetic relatedness among isolates, and none of these genes alone can, at present, differentiate all Mycobacterium species. The combined use of sequences of several genes opens up the possibility of increasing discriminatory power for identification and typing purposes, as proposed by Adékambi and Drancourt (2) and Devulder et al. (11).
In a previous study, 60 isolates in R2A medium from hemodialysis water were grouped into 16 patterns of restriction fragment length polymorphism (RFLP) of the 16S rRNA gene after digestion with TaqI and HaeIII (16). Twenty isolates (one, two, or three of each RFLP group) were selected for further analysis by a multigene sequencing approach, together with seven mycobacteria isolated from clinical specimens and previously classified phenotypically as atypical mycobacteria. We sequenced the fragments of seven genes, the 16S rRNA gene, ITS1, gyrB, hsp65, recA, rpoB, and sodA (making a total of 2,994 to 3,174 nucleotides), and analyzed them for the 27 strains and for 11 related type strains. Data on other type strains were retrieved from publicly available databases. Here we examined genetic diversity and phylogenetic results by comparing the phylogenetic trees resulting from these data. We demonstrate how the concatenation or consensus analysis of several genes allows increased discriminatory power and provides a more robust phylogenetic tree. The usefulness of the sequenced genes for population genetic studies and molecular typing is also discussed.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
A total of 60 EM were isolated in a previous study from a hemodialysis water distribution system at three sampling points, before and after a hyperchlorination step. Sixteen RFLP patterns in the 16S rRNA gene were detected. A minimum of one representative strain of each RFLP pattern was chosen for further analysis. The strains used were as follows: six strains from 17 isolates at point 1, five from 20 isolates at point 2, and one from 5 isolates at point 3 before hyperchlorination and two strains selected from 5 isolates at point 1 and four from 11 isolates at point 3 after hyperchlorination (16). Seven additional isolates obtained from clinical samples at the Hospital Son Dureta (Palma de Mallorca, Spain) were also included, together with the most closely related 11 type strains of the genus, i.e., M. abscessus CCUG20993T (CIP104536T), M. aubagnense CCUG50186T (U8T = CIP108543T), M. bolletii CCUG50184T (BDT = CIP108541T), M. chelonae CCUG47445T (CIP104535T), M. fortuitum CCUG27973T (CIP104534T), M. immunogenum CCUG47286T (CIP106684T), M. massiliense CCUG48898T (CIP108297T), M. mucogenicum CCUG47451T (ATCC 49650T = CIP105223T), M. peregrinum CCUG27976T (CIP105382T), M. phocaicum CCUG50185T (N4T = CIP108542T), and M. septicum CCUG43574T (CIP106642T = ATCC 700731T). All of the strains used in this study are shown in Table 1.
TABLE 1.
Bacterial strains used in this study
| Straina | Source | Geographic origin | Yr of isolation |
|---|---|---|---|
| MG1 | Hemodialysis water | Mallorca, Spain | 2003 |
| MG2 | Hemodialysis water | Mallorca, Spain | 2002 |
| MG3 | Hemodialysis water | Mallorca, Spain | 2003 |
| MG4 | Hemodialysis water | Mallorca, Spain | 2002 |
| MG5 | Hemodialysis water | Mallorca, Spain | 2003 |
| MG6 | Hemodialysis water | Mallorca, Spain | 2003 |
| MG7 | Hemodialysis water | Mallorca, Spain | 2003 |
| MG8 | Hemodialysis water | Mallorca, Spain | 2003 |
| MG9 | Hemodialysis water | Mallorca, Spain | 2003 |
| MG10 | Hemodialysis water | Mallorca, Spain | 2004 |
| MG11 | Hemodialysis water | Mallorca, Spain | 2003 |
| MG12 | Hemodialysis water | Mallorca, Spain | 2003 |
| MG13 | Hemodialysis water | Mallorca, Spain | 2003 |
| MG14 | Hemodialysis water | Mallorca, Spain | 2003 |
| MG15 | Hemodialysis water | Mallorca, Spain | 2003 |
| MG16 | Hemodialysis water | Mallorca, Spain | 2003 |
| MG17 | Hemodialysis water | Mallorca, Spain | 2003 |
| MG18 | Hemodialysis water | Mallorca, Spain | 2003 |
| MG19 | Hemodialysis water | Mallorca, Spain | 2004 |
| MG20 | Hemodialysis water | Mallorca, Spain | 2003 |
| MHSD2 | Human sputum | Mallorca, Spain | 2004 |
| MHSD3 | Human tissue biopsy sample | Mallorca, Spain | 2004 |
| MHSD4 | Human sputum | Mallorca, Spain | 2003 |
| MHSD5 | Human sputum | Mallorca, Spain | 2003 |
| MHSD6 | Unknown | Mallorca, Spain | 2002 |
| MHSD11 | Human sputum | Mallorca, Spain | 2000 |
| MHSD12 | Human sputum | Mallorca, Spain | 2000 |
| M. abscessus CCUG20993T | Unknown | Unknown | 1987 |
| M. alvei CIP103464T | Water (Llobregat River) | Barcelona, Spain | 1981 |
| M. aubagnense CCUG50186T | Human bronchial aspirate | Aubagne, France | 2004 |
| M. bolletii CCUG50184T | Human bronchial lavage fluid | Marseille, France | 2004 |
| M. chelonae CCUG47445T | Tortoise tubercle | Unknown | 1923 |
| M. conceptionense CCUG50187T | Human bone tissue biopsy sample | Reunion Island | 2002 |
| M. farcinogenes NCTC10955T | Bovine lymph node | Unknown | 1973 |
| M. fortuitum CCUG27973T | Human cold abscess | Rio de Janeiro, Brazil | 1937 |
| M. goodii ATCC 700504T | Human blood | Tromsö, Norway | 2005 |
| M. houstonense ATCC 49403T | Face wound | Houston, TX | 2004 |
| M. immunogenum CCUG47286T | Metalworking fluid | Missouri | 1990 |
| M. mageritense CIP104973T | Human sputum | Madrid, Spain | 1987 |
| M. massiliense CCUG48898T | Human sputum and bronchial alveolar lavage fluid | Marseille, France | 2005 |
| M. mucogenicum CCUG47451T | Thyroglossal duct cyst | Midland, TX | 1995 |
| M. neworleansense ATCC 49404T | Scalp wound | New Orleans, LA | 2004 |
| M. peregrinum CCUG27976T | Human bronchial aspiration | Mexico | 1991 |
| M. phocaicum CCUG50185T | Human bronchial aspirate | Marseille, France | 2004 |
| M. porcinum CIP105392T | Swine lymph node | Japan | 1996 |
| M. senegalense CIP104941T | Bovine lymph node | Senegal | 1973 |
| M. septicum CCUG43574T | Catheter tip | Melbourne, Australia | 1999 |
| M. smegmatis ATCC 19420T | Industrial plant | Beltsville, MD | 1950 |
| M. wolinskyi ATCC 700010T | Human postsurgical abscess | Switzerland | 1999 |
A superscript capital T indicates a type strain.
Strains were cultured in R2A medium (30) containing 0.5 g yeast extract, 0.5 g acid hydrolysate of casein, 0.5 g glucose, 0.5 g starch, 0.3 g K2HPO4, 0.3 g sodium pyruvate, 0.25 g pancreatic digest of casein, 0.25 g peptic digest of animal tissue, and 0.0492 g MgSO4 · 7H2O per liter and supplemented with 1.5% (wt/vol) agar for the plates. The bacteria were incubated for 3 to 4 days at 30°C.
DNA extraction.
Bacterial genomic DNA for PCR amplifications was obtained by lysis with sodium dodecyl sulfate at 1%, proteinase K (150 mg/ml), lysozyme (3 mg/ml), and mutanolysin (1.7 mg/ml). DNA was purified by the cetyltrimethylammonium bromide method, followed by two successive extractions with phenol-chloroform-isoamyl alcohol (25:24:1) and a final extraction with chloroform-isoamyl alcohol (24:1). Total DNA was precipitated with absolute ethanol and sodium acetate at 3 M, pH 5.2. The pellet was washed with 70% ethanol and resuspended in a small volume of milliQ water (46).
PCR amplification and DNA sequencing.
The 16S rRNA, ITS1, gyrB, hsp65, recA, rpoB, and sodA genes of the 38 strains were amplified and sequenced with the primers indicated in Table 2, by following previously described procedures. PCR amplification was performed with a DNA thermocycler (Eppendorf). Individual reaction mixtures contained 5 μl of a PCR buffer (Amersham Pharmacia Biotech, Inc.), 8 μl of each of the nucleoside triphosphates (Roche Diagnostics GmbH) at a concentration of 100 μM, and 2.5 μl of each of the primers at a concentration of 10 μM. For sequencing reactions, the same primers and conditions were used. The amplified products were purified with montage PCR centrifugal filter devices (Millipore) and sequenced with an automatic AB3730 DNA analyzer (Applied Biosystems).
TABLE 2.
Primers used in this study
| Gene and primer | Sequence (5′ → 3′)a | Reference(s) |
|---|---|---|
| 16S rRNA | ||
| 16F27 | AGAGTTTGATCMTGGCTCAG | 23 |
| 16R1492 | TACGGYTACCTTGTTACGACTT | |
| ITS1 | ||
| sp1 | ACCTCCTTTCTAAGGAGCACC | 32 |
| sp2 | GATGCTCGCAACCACTATCCA | |
| gyrB | ||
| gyrBA | GAGTTGGTGCGGCGTAAGAGC | 10 |
| gyrBE | CGGCCATCCARCACGATCTTG | |
| hsp65 | ||
| TB11 | ACCAACGATGGTGTGTCCAT | 38 |
| TB12 | CTTGTCGAACCGCATACCCT | |
| recA | ||
| recF3 | GGCAARGGYTCGGTSATGC | 4 |
| recF4 | TCGACCAACTACTTCTARCG | |
| recR2 | TTGATCTTCTTCTCGATCTC | |
| rpoB | ||
| MycoF | GGCAAGGTCACCCCGAAGGG | 1 |
| MycoR | AGCGGCTGCTGGGTGATCATC | |
| MycoseqF | GAAGGGTGAGACCGAGCTGAC | |
| MycoseqR | GCTGGGTGATCATCGAGTACGG | |
| sodA | ||
| Z205F | ACGTTCACCACAGCAAGCACCA | 11, 48 |
| Z212R | TCGGCCCAGTTCACGACGTT | |
| GSOD2R | TCGGCCAGTTCACGACGTTCCA |
M = A or C; N = A, C, G, or T; R = A or G; S = C or G; Y = C or T.
Sequence analysis.
Gene sequences were aligned with the closest relatives retrieved from the BLAST nucleotide sequence database. The alignment was done by a hierarchical method for multiple alignments implemented in the CLUSTAL X computer program (40). Automatically aligned sequences were checked manually. Evolutionary distances derived from sequence pair dissimilarities (Jukes-Cantor correction [22]) were calculated with the DNADIST program included in the Phylogenetic Inference Package (PHYLIP version 3.5c) (14). Phylogenetic trees were constructed by the neighbor-joining distance method. Bootstrap analysis (1,000 replicates) was done with the PHYLIP package. Bootstrap values higher than 500 are indicated in the corresponding trees. Topologies of the trees were visualized with the TreeView program (29). In addition to the trees for each gene, a concatenated analysis (11) and a consensus analysis of the sequenced gene fragments were performed (7).
Allele diversity, nucleotide diversity, and statistical analysis.
Allele and nucleotide diversities were calculated with the DnaSP package, version 3.51 (Faculty of Biology, University of Barcelona; http://www.ub.es/DnaSP) (33). For identification purposes, distinct allele sequences were assigned arbitrary allele numbers for each locus. For each isolate, the combination of alleles obtained at each locus defined its allelic profile. Each allelic profile constitutes a sequence type (ST). Clustering of STs was performed with the Sequence Type Analysis and Recombinational Tests (START) program (21). The matrix of pairwise distances between the allelic profiles was converted to NEXUS files, and the split decomposition was analyzed with the SplitsTree program (www.splitstree.org) (18).
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study (see Table S1 in the supplemental material) have been deposited in the EMBL database under the following accession numbers: 16S rRNA gene, AM421246 to AM421252; ITS1, AM421253 to AM421293; gyrB, AM421294 to AM421331; hsp65, AM421332 to AM421355; recA, AM421356 to AM421378 and AM501932; rpoB, AM421379 to AM421404; sodA, AM421405 to AM421429.
RESULTS
Phylogenetic analysis.
A total of 3,187 nucleotides, corresponding to seven genes, were determined for most of the strains studied (see Table S1 in the supplemental material), i.e., the 16S rRNA gene (715 to 1,400 bp), ITS1 (140 to 193 bp), gyrB (218 bp), hsp65 (307 bp), recA (664 bp), rpoB (643 to 658 bp), and sodA (325 bp). Interspecies sequence similarities for the type strains varied from 95.52 to 100% (16S rRNA gene), 85.91 to 96.67% (gyrB), 84.09 to 100% (hsp65), 85.05 to 100% (recA), 82.93 to 100% (rpoB), and 86.18 to 100% (sodA).
Two bands of ITS1 were detected in three clinical isolates (MHSD5, MHSD6, and MHSD11) and three type strains (M. fortuitum, M. mucogenicum, and M. peregrinum). In these three isolates and in the M. peregrinum type strain, the double bands were separated by agarose gel electrophoresis, eluted, and sequenced. In the other two type strains, both bands were separated and eluted but only one could be sequenced. The primers and conditions used did not allow the sequencing of the ITS1 region in one clinical isolate (MHSD12). tRNA coding sequences were not found in any of the ITS1 genes studied.
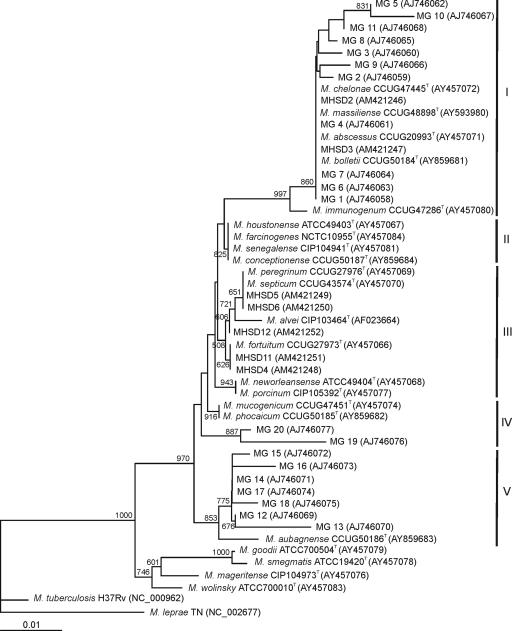
As a first step, individual trees were analyzed separately. The strains studied were considered as RGM and fell within the RGM phylogenetic branch, as defined by Devulder et al. (11) when considering the 16S rRNA gene sequences. Five clearly differentiated groups supported by high bootstrap values were delineated in the 16S phylogenetic tree (Fig. 1), i.e., group I (including the M. chelonae-like species M. abscessus, M. bolletii, M. chelonae, M. immunogenum, and M. massiliense), group II (M. conceptionense, M. farcinogenes, M. houstonense, and M. senegalense), group III (M. alvei, M. fortuitum, M. neworleansense, M. peregrinum, M. porcinum, and M. septicum), group IV (M. mucogenicum and M. phocaicum), and group V (M. aubagnense). Hemodialysis water isolates belonged to group I (11 strains), group IV (2 strains), and group V (7 strains). Clinical isolates belonged to group I (two strains) and group III (five strains). Intragroup phylogenetic distances were very low and did not allow the identification of the new strains to the species level.
FIG. 1.
Phylogenetic tree based on the 16S rRNA gene sequences of the environmental and clinical Mycobacterium strains studied. The bar indicates sequence divergence. Bootstrap values of 500 or more (from 1,000 replicates) are indicated at the nodes. Sequence accession numbers are in parentheses. Roman numerals indicate phylogenetic groupings.
The phylogenetic tree of the ITS1 region was clearly split into two clusters, maintaining the same groupings as defined in the 16S tree, excepting the M. mucogenicum and M. aubagnense type strains, which clustered with group I. Phylogenetic distances between strains were much higher, thereby providing better discrimination. In the case of double bands, they were not identical in sequence and were located in the same phylogenetic branch, but in some cases close to two distinct type strains; one band of MHSD11 clustered with M. fortuitum, and the other clustered with M. porcinum (see Fig. S1a in the supplemental material).
Similarity values between type strains were lower and were more discriminative for the gyrB gene than for the 16S rRNA gene (see Fig. S1b in the supplemental material). Clinical isolates in group III were affiliated with the same phylogenetic branch as in the 16S rRNA gene. MHSD5 and MHSD6 were more closely related to M. peregrinum than to M. septicum in the gyrB tree, as well as in the other gene trees studied. MHSD4 and MHSD11 were grouped with the M. fortuitum type strain in all of the genes studied, except sodA and rpoB. Environmental strains of groups I and V were grouped in the same branch as M. chelonae. Strains MG20 and MG19 of group IV clustered separately with M. aubagnense.
In the hsp65 analysis (see Fig. S1c in the supplemental material), group I isolates were split into two branches, one with the M. chelonae and M. immunogenum type strains and the other with M. abscessus, M. bolletii, and M. massiliense, with similarities lower than 90%. One strain of group I (MG8) was affiliated with the group V strains.
In the recA analysis (see Fig. S1d in the supplemental material), all strains maintained the same groupings, except MG14 of group V, which clustered with the group I strains. The recA gene, together with hsp65, also allowed better discrimination of group I strains than did the 16S rRNA gene.
In the rpoB analysis (see Fig. S1e in the supplemental material), the same groupings as in the 16S rRNA gene analysis were maintained, with only one exception: MG5 of group I clustered with the strains of group V. The strains of groups IV and V were not well differentiated and clustered in the same phylogenetic branch.
The group V strains were in the same phylogenetic branch in the sodA analysis (see Fig. S1f in the supplemental material). Strains MHSD5 and MHSD6 were closely related to M. peregrinum. However, the other new strains, environmental and clinical, were grouped with M. fortuitum, a member of group III. The type strains of group I, as well as MHSD2, were affiliated in the same branch.
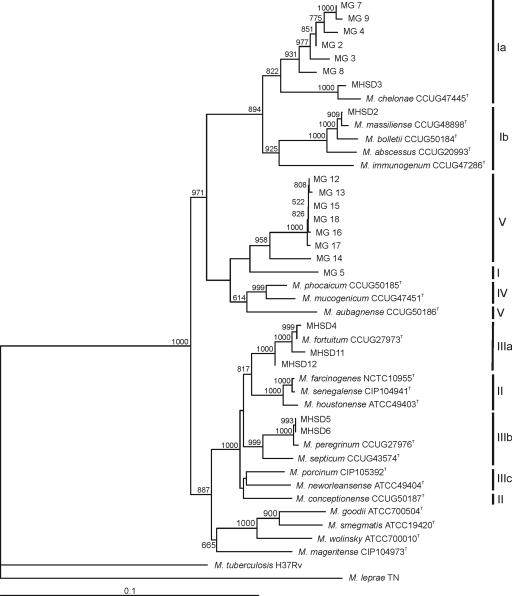
The congruence of individual trees, despite individual cases like MHSD12, allowed the concatenation of the sequences studied in order to construct a more robust phylogeny of these environmental and clinical RGM. The concatenated analysis considered five genes (approximately 2,731 bp); the ITS1 and gyrB data sets were incomplete and were not included. Figure 2 shows the concatenated tree. Six strains (MG6, MG10, MG20, MG11, MG1, and MG19) were not included in the global analysis because we were unable to sequence some genes (see Table S1 in the supplemental material). The robustness of the concatenated tree was demonstrated by the high bootstrap values at all branches. All of the strains clustered in the same groups as in the 16S rRNA gene analysis, except MG5 (from group I to group V) and M. aubagnense (from group V to group IV). Moreover, groups I and III were clearly split into two or three subgroups, i.e., Ia, Ib, IIIa, IIIb, and IIIc. M. conceptionense from group II clustered with the group IIIc strains. The grouping of environmental strains of group V was maintained.
FIG. 2.
Hypothetical multilocus concatenated tree showing the molecular evolutionary relationships of the gyrB, hsp65, recA, rpoB, sodA, and 16S rRNA genes of the environmental and clinical Mycobacterium strains studied. The bar indicates sequence divergence. Bootstrap values of 500 or more (from 1,000 replicates) are indicated at the nodes. Roman numerals indicate phylogenetic groupings in the 16S rRNA gene analysis.
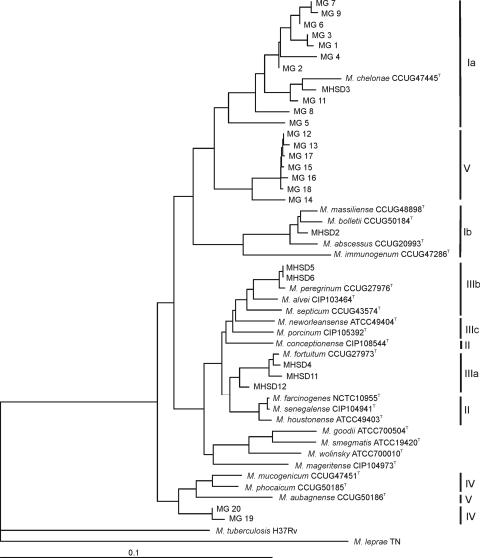
Another type of analysis, allowing the inclusion of all of the genes and most of the strains, is the construction of a consensus or agreement tree (7), as indicated in Fig. 3. MG10 was not included in the analysis because only three genes were sequenced. The topology of the resulting tree was the same as in the concatenated analysis. MG19 and MG20, which were not in the concatenated tree, clustered in the consensus tree with the type strains of groups IV (M. mucogenicum and M. phocaicum) and V (M. aubagnense).
FIG. 3.
Hypothetical multilocus consensus tree showing the molecular evolutionary relationships of the gyrB, hsp65, recA, rpoB, sodA, and 16S rRNA genes of the environmental and clinical Mycobacterium strains studied. The bar indicates sequence divergence. Roman numerals indicate phylogenetic groupings in the 16S rRNA gene analysis.
Multilocus sequence typing.
The sequenced genes were analyzed to determine their usefulness for typing purposes and for genetic population analysis. Type strains of group II (M. conceptionense, M. farcinogenes, M. houstonense, and M. senegalense), some of group III (M. neworleansense and M. porcinum), and type strains not closely related to our EM and clinical isolates (M. goodii, M. leprae, M. mageritense, M. smegmatis, M. tuberculosis, and M. wolinskyi) were excluded from further analysis.
The number of polymorphic sites in the seven loci studied varied from 53 (gyrB) to 191 (ITS1) in the 38 strains (Table 3). The inferred nucleotide and genetic diversity observed is adequate for genetic population analysis. The number of alleles per locus varied accordingly, from 17 (hsp65) to 28 (sodA), and the allelic profile of each strain is shown in Table 4. No two isolates had identical allelic profiles, except MHSD5 and MHSD6, which were identical in the six genes studied. These two strains were isolated from sputum specimens from two patients.
TABLE 3.
Genetic diversity of the selected loci among the Mycobacterium strains analyzed in this study
| No. of strains | Locus | Fragment length (bp)a | No. of alleles | Avg genetic diversity ± SD | No. of polymorphic sites | No. of nucleotide substitutions/nucleotide site | Avg nucleotide diversity ± SD |
|---|---|---|---|---|---|---|---|
| 37 | gyrB | 218 (216) | 25 | 0.928 ± 0.036 | 53 | 24.53 | 0.071 ± 0.005 |
| 36 | hsp65 | 307 | 17 | 0.925 ± 0.026 | 62 | 20.19 | 0.076 ± 0.003 |
| 35 | recA | 664 (656) | 23 | 0.953 ± 0.022 | 168 | 25.61 | 0.093 ± 0.002 |
| 38 | rpoB | 662 (640) | 26 | 0.954 ± 0.021 | 156 | 24.37 | 0.086 ± 0.002 |
| 36 | sodA | 325 | 28 | 0.970 ± 0.019 | 69 | 21.23 | 0.061 ± 0.004 |
| 38 | 16S rRNA | 716 (715) | 22 | 0.920 ± 0.034 | 66 | 9.23 | 0.018 ± 0.001 |
| 40 | ITS1b | 218 | 21 | 0.9474 | 191 | 87.61 |
Values in parentheses are the total numbers of sites, excluding gaps or missing data.
ITS1 genetic diversity was calculated for all of the sequences, including the cases of double bands.
TABLE 4.
STs at the seven loci examined in the 26 strains of this study and 12 closely related type strains
| Straina | Group | Allele at locus:
|
ST | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 16S rRNA | ITS1 | gyrB | hsp65 | recA | rpoB | sodA | |||
| MG4 | I | 7 | 10 | 7 | 5 | 3 | 5 | 8 | 1 |
| MG2 | I | 6 | 11 | 1 | 9 | 1 | 1 | 7 | 2 |
| MG19 | IV | 14 | 8 | 22 | 10 | 15 | 3 | ||
| MHSD11 | III | 12 | 5 | 18 | 14 | 22 | 21 | 1 | 4 |
| MHSD12 | III | 11 | 10 | 14 | 23 | 22 | 15 | 5 | |
| MHSD2 | I | 7 | 18 | 13 | 15 | 9 | 8 | 21 | 6 |
| MHSD3 | I | 7 | 13 | 6 | 6 | 4 | 3 | 10 | 7 |
| MHSD4 | III | 12 | 6 | 17 | 14 | 21 | 20 | 11 | 8 |
| MHSD5 | III | 9 | 1 | 9 | 11 | 18 | 23 | 13 | 9 |
| MHSD6 | III | 9 | 1 | 9 | 11 | 18 | 23 | 13 | 9 |
| MG5 | I | 1 | 14 | 5 | 8 | 6 | 12 | 1 | 10 |
| MG15 | V | 16 | 7 | 2 | 1 | 13 | 12 | 24 | 11 |
| MG16 | V | 17 | 7 | 4 | 1 | 13 | 13 | 25 | 12 |
| MG18 | V | 19 | 7 | 3 | 1 | 13 | 12 | 24 | 13 |
| MG14 | V | 18 | 7 | 7 | 1 | 3 | 14 | 24 | 14 |
| MG12 | V | 20 | 7 | 1 | 1 | 13 | 12 | 24 | 15 |
| MG13 | V | 21 | 7 | 1 | 1 | 13 | 12 | 24 | 16 |
| MG20 | IV | 15 | 8 | 21 | 10 | 15 | 4 | 17 | |
| MG6 | I | 7 | 11 | 1 | 1 | 1 | 5 | 18 | |
| MG17 | V | 18 | 7 | 1 | 1 | 14 | 12 | 24 | 19 |
| MG3 | I | 4 | 11 | 1 | 15 | 1 | 1 | 1 | 20 |
| MG7 | I | 7 | 11 | 1 | 4 | 2 | 1 | 2 | 21 |
| MG9 | I | 5 | 11 | 1 | 4 | 1 | 2 | 3 | 22 |
| MG1 | I | 7 | 11 | 1 | 15 | 1 | 1 | 23 | |
| MG8 | I | 3 | 11 | 1 | 1 | 1 | 4 | 12 | 24 |
| MG11 | I | 2 | 14 | 6 | 7 | 6 | 6 | 25 | |
| M. abscessus | I | 7 | 21 | 14 | 17 | 11 | 9 | 18 | 26 |
| M. alvei | III | 10 | 13 | 25 | 16 | 27 | |||
| M. aubagnense | V | 22 | 16 | 23 | 2 | 15 | 16 | 28 | 28 |
| M. bolletii | I | 7 | 20 | 15 | 16 | 10 | 10 | 20 | 29 |
| M. chelonae | I | 7 | 12 | 8 | 6 | 5 | 1 | 23 | 30 |
| M. fortuitum | III | 12 | 4 | 19 | 14 | 21 | 19 | 9 | 31 |
| M. immunogenum | I | 8 | 15 | 20 | 7 | 12 | 11 | 22 | 32 |
| M. massiliense | I | 7 | 19 | 16 | 15 | 8 | 7 | 19 | 33 |
| M. mucogenicum | IV | 13 | 17 | 25 | 3 | 17 | 17 | 27 | 34 |
| M. peregrinum | III | 9 | 2 | 11 | 11 | 19 | 24 | 14 | 35 |
| M. phocaicum | IV | 13 | 9 | 24 | 3 | 16 | 18 | 26 | 36 |
| M. septicum | III | 9 | 3 | 12 | 12 | 20 | 26 | 17 | 37 |
Type strains: M. abscessus CCUG20993T, M. alvei CIP103463T, M. aubagnense CCUG50186T, M. bolletii CCUG50184T, M. chelonae CCUG47445T, M. fortuitum CCUG27973T, M. immunogenum CCUG47286T, M. massiliense CCUG48898T, M. mucogenicum CCUG47451T, M. peregrinum CCUG27976T, M. phocaicum CCUG50185T, and M. septicum CCUG43574T.
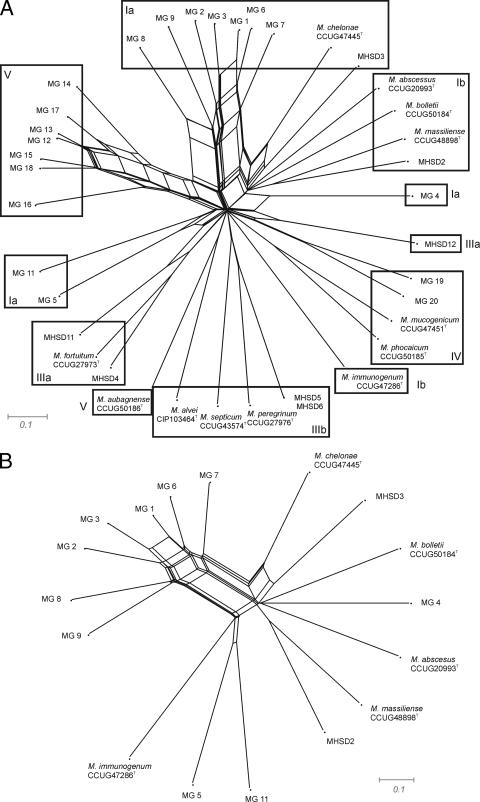
A splits tree was constructed to determine possible lateral gene transfers in the population of mycobacteria studied. Strains of groups Ia and V, defined in the consensus and concatenated trees, might have two or three identical genes (Fig. 4). Groups IIIb and IV represent two different clonal populations. The only alleles that might be shared by two type strains were the 16S rRNA gene and hsp65 (M. mucogenicum and M. phocaicum), but no recombination was detected between type strains. M. chelonae shared only the 16S rRNA gene allele with EM isolates of group Ia and the hsp65 allele with clinical isolate MHSD3 of the same phylogenetic group, Ia. MG5 also had the 16S rRNA gene allele in common with MHSD11 of group IIIa. Group V isolates remained clustered, showing a range of possible recombinations in the gyrB, hsp65, recA, rpoB, and sodA alleles.
FIG. 4.
Splits tree showing the distribution of all of the environmental and clinical isolates studied, as well as the closely related type strains (A). (B) Detail of the group I network.
DISCUSSION
In most studies of EM in water distribution systems, isolates are not identified to the species level. The difficulty in isolate identification has been widely reported, and in most cases isolates are ascribed to taxa with no standing in the nomenclature, such as “M. chelonae-like organisms,” or as members of the “M. fortuitum complex.” In the present work, we demonstrate that 16S rRNA gene sequence analysis alone is a valuable tool for the characterization of EM but is not sufficient to identify most of the environmental isolates. 16S rRNA gene partial sequence analysis shows that several species have identical sequences, even considering the complete sequence, i.e., M. abscessus-M. bolletii-M. chelonae-M. massiliense, M. septicum-M. peregrinum, and most of the species of the M. fortuitum complex. Therefore, we integrated additional housekeeping genes into the analysis (gyrB, hsp65, recA, rpoB, and sodA) which are relatively variable and present in all mycobacteria species. ITS1 is a good region for strain discrimination, but the presence of double bands in some cases does not allow a consensus analysis with the other genes.
rpoB has been proposed as an additional method to 16S rRNA gene-based identification of mycobacteria (3). This method has been used to support the delineation of new species; however, in some circumstances it does not show enough discriminatory power.
The hsp65 gene is also more variable than the 16S rRNA gene. Species from group I were better discriminated from each other. For example, the M. abscessus, M. bolleti, and M. massiliense type strains differed by nearly 30 nucleotides from M. chelonae, whereas their partial 16S rRNA genes were identical. As described by Ringuet et al. (31), nucleotide differences are found along the length of the amplified hsp65 fragment but are frequent in two regions, probably hypervariable regions of the hsp65 gene.
Nucleotide sequences of gyrB were clearly more variable than the 16S rRNA gene, with 53 polymorphic sites of 218 nucleotides (24.53%). This difference was observed not only between type strains of group I (M. abscessus-M. bolletii-M. chelonae) and the M. fortuitum group (M. fortuitum-M. peregrinum) but also between the species within each group. These differences were not as evident in our environmental isolates (except MG19 and MG20), which clustered in the same branch close to M. chelonae, independently of their initial affiliation in the 16S rRNA gene sequence.
Analysis of the recA gene revealed the presence of a large number of nucleotide substitutions among the species tested. Unlike the 16S rRNA gene or hsp65, in which variability is confined to certain areas of the gene, the variability occurred throughout the recA gene.
The analysis of the sodA gene also showed that it is a variable gene with 69 polymorphic sites out of 325 (21.23%). This analysis allowed good discrimination for group V strains but not for members of groups I and IV.
The discriminatory power of the concatenated and consensus trees was higher than that observed with only the 16S rRNA gene. The absence of discriminatory power of some genes related to certain species is compensated for by that of others. Members of group V clustered in all of the genes analyzed, and one gene might be enough for their identification. However, for the characterization of members of group I, additional genes were considered. Therefore, the use of several genes allowed the discrimination of each strain to the species level; furthermore, the increase in sequence size led to a considerable increase in tree robustness, thereby increasing bootstrap values in nodes.
Taking a distance of 1% as a discriminative species value in the consensus tree, we assigned our clinical isolates to the species M. fortuitum (MHSD4 and MHSD11) and M. peregrinum (MHSD5 and MHSD6). However, the hemodialysis water isolates, with a minimal distance of 4% from the closest type strain, cannot be ascribed to any known species and on the basis of phylogenetic distances could be considered representatives of at least four species. However, the definitive proposal of a new species requires a detailed phenotypic identification, which has not yet been done.
The 20 EM studied are representative of the 60 isolates obtained in a previous study. All showed distinct allele combinations; that is, each strain is representative of a different ST, indicating high genetic diversity of the isolates. The same situation can be devised for five of the clinical isolates, and only two (MHSD5 and MHSD6) had identical allelic profiles. Possible recombination events were detected only between isolates of group Ia or between isolates of group IV. The possibility of genetic exchange in each group, but not with strains of a distinct group, supports the hypothesis that these two groups represent genetically closely related strains of new species that have not been described to date.
On the basis of our results, we conclude that the diversity of EM in hemodialysis water is very high and that these EM have not been described previously for this type of oligotrophic environment. The detection of these new types of EM could be attributed to the procedure used for their isolation. In contrast to the medium usually used for mycobacteria, such as Middlebrook or Lowenstein-Jensen medium, R2A medium is nonselective. Moreover, the samples were not “decontaminated” with NaOH, a procedure which is commonly used when isolating mycobacteria. This decontamination step might result in a loss of sensitive mycobacteria and a bias in their recovery (17).
We demonstrate the high discriminatory power of the multigene approach used in this study, thereby confirming its usefulness not only for species discrimination and identification in EM but also for their molecular typing. This typing scheme is required when testing water as a source of waterborne pathogens in epidemiological studies.
Supplementary Material
Acknowledgments
This work was supported in part by CICYT (Spain) grant REN2002-04035-CO3-01 and by the I Plà Balear de Recerca I Desenvolupament Tecnològic de les Illes Balears. M. Gomila was the recipient of a predoctoral fellowship from the Spanish Ministerio de Educación y Ciencia.
We are grateful to CCUG for providing type strains of Mycobacterium. We thank J. Gascó (Hospital Son Llàtzer) for collaboration and constructive discussion.
Published ahead of print on 20 April 2007.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Adékambi, T., P. Colson, and M. Drancourt. 2003. rpoB-based identification of nonpigmented and late pigmenting rapidly growing mycobacteria. J. Clin. Microbiol. 41:5699-5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adékambi, T., and M. Drancourt. 2004. Dissection of phylogenetic relationships among 19 rapidly growing Mycobacterium species by 16S rDNA, hsp65, sodA, recA and rpoB gene sequencing. Int. J. Syst. Evol. Microbiol. 54:2095-2105. [DOI] [PubMed] [Google Scholar]
- 3.Adékambi, T., P. Berger, D. Raoult, and M. Drancourt. 2006. rpoB gene sequence-based characterization of emerging non-tuberculous mycobacteria with descriptions of Mycobacterium bolletii sp. nov., Mycobacterium phocaicum sp. nov. and Mycobacterium aubagnense sp. nov. Int. J. Syst. Evol. Microbiol. 56:133-143. [DOI] [PubMed] [Google Scholar]
- 4.Blackwood, K. S., C. He, J. Gunton, C. Y. Turenne, J. Wolfe, and A. M. Kabani. 2000. Evaluation of recA sequences for identification of Mycobacterium species. J. Clin. Microbiol. 38:2846-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown-Elliott, B. A., and R. J. Wallace, Jr. 2002. Clinical and taxonomic status of pathogenic nonpigmented or late-pigmenting rapidly growing mycobacteria. Clin. Microbiol. Rev. 15:716-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carson, L. A., L. A. Bland, L. B. Cusick, M. S. Favero, G. A. Bolan, A. L. Reingold, and R. C. Good. 1988. Prevalence of nontuberculous mycobacteria in water supplies of hemodialysis centers. Appl. Environ. Microbiol. 54:3122-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cladera, A. M., A. Bennasar, M. Barceló, J. Lalucat, and E. García-Valdés. 2004. Comparative genetic diversity of Pseudomonas stutzeri genomovars, clonal structure, and phylogeny of the species. J. Bacteriol. 186:5239-5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Covert, T. C., M. R. Rodgers, A. L. Reyes, and G. N. Stelma, Jr. 1999. Occurrence of nontuberculous mycobacteria in environmental samples. Appl. Environ. Microbiol. 65:2492-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dailloux, M., C. Laurain, M. Weber, and P. Hartemann. 1999. Water and nontuberculous mycobacteria. Water Res. 33:2219-2228. [Google Scholar]
- 10.Dauendorffer, J. N., I. Guillemin, A. Aubry, C. Truffot-Pernot, W. Sougakoff, V. Jarlier, and E. Cambau. 2003. Identification of mycobacterial species by PCR sequencing of quinolone resistance-determining regions of DNA gyrase genes. J. Clin. Microbiol. 41:1311-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devulder, G., M. Pérouse de Montclos, and J. P. Flandrois. 2005. A multigene approach to phylogenetic analysis using the genus Mycobacterium as a model. Int. J. Syst. Evol. Microbiol. 55:293-302. [DOI] [PubMed] [Google Scholar]
- 12.Embil, J., P. Warren, M. Yakrus, R. Stark, S. Corne, D. Forrest, and E. Hershfield. 1997. Pulmonary illness associated with exposure to Mycobacterium avium complex in hot tub water—hypersensitivity pneumonitis or infection. Chest 111:813-816. [DOI] [PubMed] [Google Scholar]
- 13.Falkinham, J. O., III. 2002. Nontuberculous mycobacteria in the environment. Clin. Chest Med. 23:529-551. [DOI] [PubMed] [Google Scholar]
- 14.Felsenstein, J. 1989. PHYLIP—phylogeny inference package (version 3.0). Cladistics 5:164-166. [Google Scholar]
- 15.Gebo, K. A., A. Srinivasan, T. M. Perl, T. Ross, A. Groth, and W. G. Merz. 2002. Pseudo-outbreak of Mycobacterium fortuitum on a human immunodeficiency virus ward: transient respiratory tract colonization from a contaminated ice machine. Clin. Infect. Dis. 35:32-38. [DOI] [PubMed] [Google Scholar]
- 16.Gomila, M., J. Gascó, A. Busquets, J. Gil, R. Bernabeu, J. M. Buades, and J. Lalucat. 2005. Identification of culturable bacteria present in hemodialysis water and fluid. FEMS Microbiol. Ecol. 52:101-114. [DOI] [PubMed] [Google Scholar]
- 17.Hartmans, S., and J. A. M. DeBont. 1992. The genus Mycobacterium—nonmedical, p. 1214-1237. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, K.-H. Schleifer (ed.), The prokaryotes, 2nd edition. Springer Verlag, New York, NY.
- 18.Huson, D. H. 1998. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics 14:68-73. [DOI] [PubMed] [Google Scholar]
- 19.Iivanainen, E., J. Northrup, R. D. Arbeit, M. Ristola, M. L. Katila, and C. F. von Reyn. 1999. Isolation of mycobacteria from indoor swimming pools in Finland. APMIS 107:193-200. [DOI] [PubMed] [Google Scholar]
- 20.Iivanainen, E., P. J. Martikainen, P. Vaananen, and M. L. Katila. 1999. Environmental factors affecting the occurrence of mycobacteria in brook sediments. J. Appl. Microbiol. 86:673-681. [DOI] [PubMed] [Google Scholar]
- 21.Jolley, K. A., E. J. Feil, M. S. Chan, and M. C. Maiden. 2001. Sequence type analysis and recombinational tests (START). Bioinformatics 17:1230-1231. [DOI] [PubMed] [Google Scholar]
- 22.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, Inc., New York, NY.
- 23.Lane, D. J. 1991. 16S/23S rDNA sequencing, p. 115-175. In E. Stackebrand and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley, Chichester, United Kingdom.
- 24.Laussucq, S., A. L. Baltch, R. P. Smith, R. W. Smithwick, B. J. Davis, E. K. Desjardin, V. A. Silcox, A. B. Spellacy, R. T. Zeimis, H. M. Gruft, R. C. Good, and M. L. Cohen. 1988. Nosocomial Mycobacterium fortuitum colonization from a contaminated ice machine. Am. Rev. Respir. Dis. 138:891-894. [DOI] [PubMed] [Google Scholar]
- 25.Le Dantec, C., J. P. Duguet, A. Montiel, N. Dumoutier, S. Dubrou, and V. Vincent. 2002. Occurrence of mycobacteria in water treatment lines and in water distribution systems. Appl. Environ. Microbiol. 68:5318-5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leys, N. M., A. Ryngaert, L. Bastiaens, P. Wattiau, E. M. Top, W. Verstraete, and D. Springae. 2005. Occurrence and community composition of fast-growing Mycobacterium in soils contaminated with polycyclic aromatic hydrocarbons. FEMS Microbiol. Ecol. 51:375-388. [DOI] [PubMed] [Google Scholar]
- 27.Mangione, E. J., G. Huitt, D. Lenaway, J. Beebe, A. Bailey, M. Figoski, M. P. Rau, K. D. Albrecht, and M. A. Yakrus. 2001. Nontuberculous mycobacterial disease following hot tub exposure. Emerg. Infect. Dis. 7:1039-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore, J. S., M. Christensen, R. W. Wilson, R. J. Wallace, Y. S. Zhang, D. R. Nash, and B. Shelton. 2000. Mycobacterial contamination of metalworking fluids: involvement of a possible new taxon of rapidly growing mycobacteria. Am. Ind. Hyg. Assoc. J. 61:205-213. [DOI] [PubMed] [Google Scholar]
- 29.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 30.Reasoner, D. J., and E. E. Geldreich. 1985. A new medium for the enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 49:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ringuet, H., C. Akoua-Koffi, A. Honore, A. Varnerot, V. Vincent, P. Berche, J. Gaillard, and C. Pierre-Audigier. 1999. hsp65 sequencing for identification of rapidly growing mycobacteria. J. Clin. Microbiol. 37:852-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roth, A., U. Reischl, A. Streubel, L. Naumann, R. M. Kroppenstedt, M. Habicht, M. Fischer, and H. Mauch. 2000. Novel diagnostic algorithm for identification of mycobacteria using genus-specific amplification of the 16S-23S rRNA gene spacer and restriction endonucleases. J. Clin. Microbiol. 38:1094-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rozas, J., and R. Rozas. 1999. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15:174-175. [DOI] [PubMed] [Google Scholar]
- 34.Schulze-Röbbecke, R., C. Feldmann, R. Fischeder, B. Janning, M. Exner, and G. Wahl. 1995. Dental units: an environmental study of sources of potentially pathogenic mycobacteria. Tuber. Lung Dis. 76:318-323. [DOI] [PubMed] [Google Scholar]
- 35.September, S. M., V. S. Brözel, and S. N. Venter. 2004. Diversity of nontuberculoid Mycobacterium species in biofilms of urban and semiurban drinking water distribution systems. Appl. Environ. Microbiol. 70:7571-7573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shelton, B. G., W. D. Flanders, and G. K. Morris. 1999. Mycobacterium sp. as a possible cause of hypersensitivity pneumonitis in machine workers. Emerg. Infect. Dis. 5:270-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soini, H., E. C. Böttger, and M. K. Viljanen. 1994. Identification of mycobacteria by PCR-based determination of the 32-kilodalton protein gene. J. Clin. Microbiol. 32:2944-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steingrube, V. A., J. L. Gibson, B. A. Brown, Y. Zhang, R. W. Wilson, M. Rajagopalan, and R. J. Wallace, Jr. 1995. PCR amplification and restriction endonuclease analysis of a 65-kilodalton heat shock protein gene sequence for taxonomic separation of rapidly growing mycobacteria. J. Clin. Microbiol. 33:149-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takewaki, S. I., K. Okuzumi, H. Ishiko, K. I. Nakahara, A. Ohkubo, and R. Nagai. 1993. Genus-specific polymerase chain reaction for the mycobacterial dnaJ gene and species-specific oligonucleotide probes. J. Clin. Microbiol. 31:446-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The Clustal X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tortoli, E. 2003. Impact of genotypic studies on mycobacterial taxonomy: the new mycobacteria of the 1990s. Clin. Microbiol. Rev. 16:319-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tortoli, E. 2006. The new mycobacteria: an update. FEMS Immunol. Med. Microbiol. 48:159-178. [DOI] [PubMed] [Google Scholar]
- 43.Torvinen, E., T. Meklin, P. Torkko, S. Suomalainen, M. Reiman, M. L. Katila, L. Paulin, and A. Nevalainen. 2006. Mycobacteria and fungi in moisture-damaged building materials. Appl. Environ. Microbiol. 72:6822-6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vaerewijck, M. J. M., G. Huys, J. C. Palomino, J. Swings, and F. Portaels. 2005. Mycobacteria in drinking water distribution systems: ecology and significance for human health. FEMS Microbiol. Rev. 29:911-934. [DOI] [PubMed] [Google Scholar]
- 45.Wallace, R. J., Jr., B. A. Brown, and D. E. Griffith. 1998. Nosocomial outbreaks/pseudo-outbreaks caused by nontuberculous mycobacteria. Annu. Rev. Microbiol. 52:453-490. [DOI] [PubMed] [Google Scholar]
- 46.Wilson, K. 1987. Preparation of genomic DNA from bacteria, p. 241-242. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, Inc., New York, NY.
- 47.Zelazny, A. M., L. B. Calhoun, L. Li, Y. R. Shea, and S. H. Fischer. 2005. Identification of Mycobacterium species by secA1 sequences. J. Clin. Microbiol. 43:1051-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zolg, J. W., and S. Philippi-Schulz. 1994. The superoxide dismutase gene, a target for detection and identification of mycobacteria by PCR. J. Clin. Microbiol. 32:2801-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zumla, A., and J. Grange. 2002. Infection and disease caused by environmental mycobacteria. Curr. Opin. Pulm. Med. 8:166-172. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.