Abstract
Blood Falls is the surface manifestation of brine released from below the Taylor Glacier, McMurdo Dry Valleys, Antarctica. Geochemical analyses of Blood Falls show that this brine is of a marine origin. The discovery that 74% of clones and isolates from Blood Falls share high 16S rRNA gene sequence homology with phylotypes from marine systems supports this contention. The bacterial 16S rRNA gene clone library was dominated by a phylotype that had 99% sequence identity with Thiomicrospira arctica (46% of the library), a psychrophilic marine autotrophic sulfur oxidizer. The remainder of the library contained phylotypes related to the classes Betaproteobacteria, Deltaproteobacteria, and Gammaproteobacteria and the division Bacteroidetes and included clones whose closest cultured relatives metabolize iron and sulfur compounds. These findings are consistent with the high iron and sulfate concentrations detected in Blood Falls, which are likely due to the interactions of the subglacial brine with the underlying iron-rich bedrock. Our results, together with previous reports, suggest that the brine below the Taylor Glacier hosts a viable ecosystem with microorganisms capable of growth, supported by chemical energy present in reduced iron and sulfur compounds. The metabolic and phylogenetic structure of this subglacial microbial assemblage appears to be controlled by glacier hydrology, bedrock lithology, and the preglacial ecosystem.
Recent changes in our understanding of subglacial environments have led scientists to investigate the role of microorganisms in subglacial processes (65). Though these processes were once thought to be devoid of microbially mediated reactions (51), it is now clear that microorganisms contribute to subglacial weathering and carbon cycling (e.g., references 24, 47, 58, 60, 61, and 68). A growing consensus among these studies is that there is a strong link between geochemical signatures in subglacial materials and the metabolic processes present. The physical isolation, constant low temperatures, and permanent darkness of the subglacial environment make subglacial systems ideal sites for studying the relationship between microbial diversity and ecosystem function.
Measurements of diversity in a particular habitat establish the community structure in which the functional or ecological niches of the system are filled (5). Emerging themes in the ecology of the subglacial ecosystem are that glacier hydrology and bedrock lithology control microbial community structure (60, 65). The niche space generated by these controlling factors would clearly affect metabolic community structure by influencing electron acceptor and donor availability in the subglacial setting. Our study of the Taylor Glacier demonstrates that the preglacial ecosystem is an additional factor controlling subglacial microbial community structure.
Here, we present data on the microbial diversity associated with the subglacial discharge from the Taylor Glacier in the McMurdo Dry Valleys, Antarctica. Hypersaline, iron-rich, liquid brine episodically drains from Taylor Glacier at Blood Falls, which is the end of a conduit where this subglacial brine emerges to meet the atmosphere. The ionic compositions and 36Cl values for outflow at Blood Falls are consistent with those for marine waters that have experienced cryogenic concentration (41, 43). The subglacial brine is thought to be a remnant of seawater originating from the last marine incursion into the McMurdo Dry Valley network during the Pliocene Epoch (∼5 million years ago), when the Dry Valleys were fjord lands (17). Although geochemical studies of the subglacial outflow have been conducted sporadically since the early 1960s (4, 6, 35, 43), our study is the first to investigate the microbial diversity associated with Blood Falls.
MATERIALS AND METHODS
Site description.
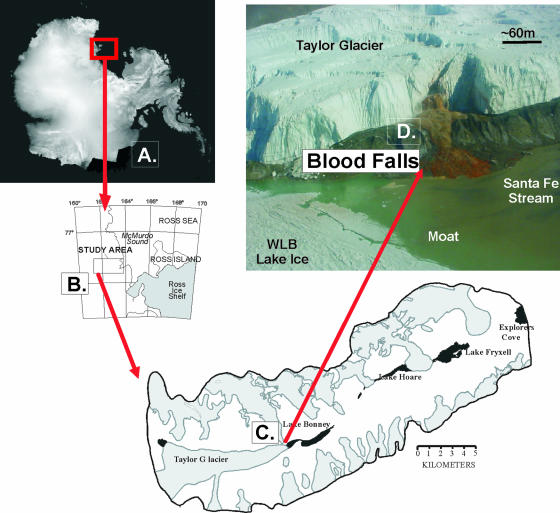
The McMurdo Dry Valleys (Fig. 1) form the largest ice-free region in Antarctica (∼4,000 km2) and are characterized by strong winds, low precipitation (<6-mm water equivalents), and an average annual air temperature near −20°C (50). Dry polar desert conditions are believed to have persisted in the region for the past 15 million years (62, 63). This polar desert contains a mosaic of permanently ice-covered lakes, ephemeral streams, arid soils, exposed bedrock, and both alpine and outlet glaciers. Taylor Glacier, located in the Taylor Valley, is an outlet glacier that drains the Polar Plateau (23). The Taylor Glacier is unique among the Dry Valley glaciers in that the presence of subglacial brine near its terminus results in geomorphic behavior more like that of a temperate or polythermal glacier (54). Ice-penetrating-radar data indicate water or slush below the glacier corresponding to an 80-m depression in the bedrock topology at ∼4 km up-glacier from the terminus (28). This depression is below sea level and forms what is believed to have been a third lobe of Lake Bonney (43). When the chemically reduced subglacial brine flows from below the glacier and is exposed to the atmosphere, it becomes oxidized and a red salt cone, known as Blood Falls, precipitates at the northern end of the glacier terminus (Fig. 1) (6, 35).
FIG. 1.
Location of Blood Falls. The McMurdo Dry Valleys are located in East Antarctica (A) west of the McMurdo Sound (B). The Taylor Glacier is located in the western end of the Taylor Valley (C). In the map of the Taylor Valley, lakes appear black and glaciers are gray. A photograph shows the subglacial outflow at Blood Falls, which occurs at the northern end of the Taylor Glacier terminus (D).
Total direct bacterial counts.
Environmental samples (10 ml) were fixed with prefiltered (0.2 μm) formalin to a final concentration of 2% (vol/vol). The samples were filtered onto polycarbonate filters (0.2-μm pore size) and stained with SYBR gold DNA stain (Molecular Probes) as described by Lisle and Priscu (39). More than 300 bacteria per sample were counted in randomly selected fields with a Zeiss Axioskop 50 epifluorescence microscope.
Microbial cultivation.
We used a matrix of heterotrophic and autotrophic media designed to selectively enrich organisms from the subglacial brine. Enrichment strategies changed with each sampling season as we learned more about the geochemistry of undiluted outflow. It is currently unknown what triggers release of subglacial brine from the Taylor Glacier; therefore, it was not possible to predict the extent of outflow that would be present each season. Commercially available media, R2A agar (Difco) (53) and marine agar (Bacto), were used to assess heterotrophic growth from outflow samples. Chemoautotrophic growth was tested for by using a medium made with CO2 as the carbon source, Fe(III) or O2 as the electron acceptor, and H2 or thiosulfate as the electron donor. These media were based on the known geochemical composition of outflow and are described in detail below. Samples of outflow for culture work were collected directly into autoclaved 74-ml serum vials by submerging the vials in the flow and crimp sealing them with sterile butyl rubber stoppers with no headspace. The serum vials were kept in the dark below 4°C until the return to McMurdo Station for incubation in selective media (approximately 5 to 10 d). Research efforts were focused on those enrichments in which colonies appeared after several months of incubation. Outflow samples (100 μl; ∼500 cells) were streaked onto prechilled plates and incubated at 2 to 4°C until colonies appeared (about 3 to 4 weeks). Supraglacial melt was tested for growth on Marine agar plates as a control; samples (200 μl; ∼75 cells) were streaked onto prechilled plates and incubated at 2 to 4°C. No growth was observed in supraglacial melt marine agar plates after 4 weeks of incubation.
Thiosulfate-oxidizer (S-Ox) agar plates were prepared for enrichment of organisms that use thiosulfate as an electron donor. It should be noted that organisms capable of using agar for growth could also be enriched with these plates. S-Ox medium ingredients were added per liter of distilled water as follows: ammonium chloride (0.2 g), potassium dibasic phosphate trihydrate (0.4 g), potassium dihydrogen phosphate (0.1 g), magnesium chloride hexahydrate (0.2 g), calcium chloride dehydrate (0.04 g), and agar (15 g). One milliliter each of vitamin solution (ATCC catalog no. MD-VS) and trace minerals (ATCC catalog no. MD-TMS) was added after the autoclaving. Thiosulfate (3 mM final concentration) was added (10 ml) from a 300 mM stock solution. The pH of the medium was adjusted to 7 with NaOH (0.5 M), autoclaved, and aseptically transferred into sterile petri dishes.
The iron-reducing (Fe-Rd) medium was adapted from a mineral salts medium (called MS) described in Boone et al. (7) with the addition of Fe-OOH sludge (2 ml). Amorphous iron(III) oxide was prepared by neutralizing a 0.4 M solution of FeCl3·6H2O with 1 M NaOH and washing it with distilled water (40). The Fe(III) gel was stored cold (4°C) and added to the bulk medium by use of a sterile pipette. The sodium hydroxide was dissolved and equilibrated with CO2 gas. Media were dispensed anaerobically (31) into CO2-sparged serum vials, crimp sealed with butyl rubber stoppers, and autoclaved (30 min). Enrichments for iron reduction were prepared by inoculating 0.5 ml (∼2,500 cells) of outflow, collected as described above, into 4.5 ml Fe-Rd medium and then serially diluting the enrichment 1:100 (or ∼25 cells). The headspace was then pressurized with H2 gas (30 lb/in2).
All media were prechilled (2°C) before inoculation, and all enrichments were incubated at 1 to 4°C. Most marine and R2A agar plates showed signs of growth within 2 to 4 weeks, while autotrophic enrichments required longer incubations (>2 months). When colonies appeared, a single colony was selected and restreaked on a fresh agar plate to ensure purity. One enrichment from Blood Falls outflow showed reduction of the iron in the Fe-Rd broth after ∼9 months of incubation. Aliquots of this enrichment broth were transferred to marine agar plates, streaked for isolation, and restreaked for purity. No contaminants appeared after microscopic observation of pure colonies, and no contaminants appeared in the uninoculated medium.
DNA extraction.
Samples for environmental DNA extraction were collected directly from Blood Falls outflow during the 1999-2000 austral summer (Table 1). Samples (1 liter) were maintained between 0 and 4°C and filtered within ∼4 h. Cells were concentrated onto a 90-mm-diameter polyethersulfone membrane filter (0.2-μm pore size), placed in a sterile plastic bag, heat sealed, and stored at −20°C until extraction. DNA was extracted from environmental samples and pure cultures using an UltraClean soil DNA isolation kit (Mo Bio Laboratories, Inc.) according to the manufacturer's protocol. The alternative protocol for maximum yields was followed. The filter containing the environmental sample was cut into pieces by using flame sterilized scissors and tweezers and then processed with the Mo Bio kit. DNA from each isolate was collected by picking a single colony from pure culture streak plates with a sterile pipette tip, which was then processed with the Mo Bio kit.
TABLE 1.
Summary of geochemical parameters, types of flow, and medium strategies for samples collected at Blood Fallsa
| Sample group | Collection date | Isolation sample identification code | Concn (no. of cells ml−1) | pH | Avg value (mM) for:
|
Blood Falls flow type | Medium strategy | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Cl− | SO42− | DOC | DIC | |||||||
| Clone library | 18 January 2000 | NA | 6.4 | 1,420 | 49.0 | 0.45 | NA | Active outflow | None | |
| Culture | 11 November 2001 | DV-X | NA | 6.0 | 1,070 | 44.0 | NA | NA | Diluted flow | R2A agar (heterotroph) |
| Culture | 19 November 2002 | S-X | 1.0 × 104 | 6.2 | 22 | 8.30 | NA | NA | Diluted flow | Marine agar; FeRd (chemoautotroph and heterotroph) |
| Culture | 17 November 2003 | ThoX | 7.6 × 105 | 8.9 | 2 | 0.26 | 0.08 | 0.9 | Diluted flow | S-Ox (chemoautotroph) |
| Culture | 14 November 2004 | BF05_X | 5.0 × 104 | 6.2 | 1,430 | 50.7 | 0.42 | 55 | Active outflow | Marine agar (heterotroph) |
| Taylor Glacier ice | 9 December 2006 | 2.3 × 103 | 8.6 | 1.6 × 10−3 | 9.2 × 10−4 | 0.06 | NA | |||
The isolation sample identification code refers to strain designations for isolates obtained from the listed sample. X represents the strain number. The Cl− and SO42− values for Taylor Glacier ice are from Lyons et al. (42). DOC, dissolved organic carbon; DIC, dissolved inorganic carbon; NA, not available.
16S rRNA gene clone library construction.
The microbial 16S rRNA genes were amplified from extracted DNA by using oligonucleotide primers 4F and 9F with 1492R (Table 2). Primers were purchased from Macromolecular Resources (Colorado State University). Each 50-μl PCR mixture contained 5 μl DNA template, deoxynucleoside triphosphate nucleotide mix (200 μM final concentration; Fisher Scientific), bovine serum albumin (0.5 μg/μl), 0.5 μM (final concentration) of each primer, 10× Taq buffer (1.5 mM Mg2+), 5× TaqMaster (a proprietary PCR enhancer; Eppendorf), and 1.25 units of Taq DNA polymerase (Eppendorf MasterTaq kit). The reaction mixture was PCR amplified in an Eppendorf Mastercycler gradient thermal cycler under conditions that included 15 s of denaturation at 95°C, 45 s of annealing at 55°C, and 1.5 min of primer extension at 72°C. These steps were repeated for 30 cycles. PCR products were purified using a QIAquick PCR purification kit (QIAGEN) to remove primers, nucleotides, polymerases, and salts. Purified PCR products were cloned using a pGEM-T Easy Vector System cloning kit with the pGEM-T Easy vector (Promega) according to the manufacturer's protocol.
TABLE 2.
Oligonucleotide primers used for PCR amplification and sequencing reactions of 16S rRNA gene from Blood Falls genomic DNA, clones, and isolates
| Primer use(s) and namea | Sequence (5′→3′) |
|---|---|
| PCR and sequencing | |
| 4F | TCCGGTTGATCCTGCCRG |
| 9F | GAGTTTGATCTGGCTCAG |
| 1492R | GGTTACCTTGTTACGACTT |
| 534R | ATTACCGCGGCTGCTGG |
| Sequencing | |
| 27F | AGAGTTTGATCCTGGCTCAG |
| 515F | GTGCCAGCMGCCGCGGTAA |
| 926R | ACCGCTTGTGCGGGCCC |
| 1068F | GCATGGCYGYCGTCAG |
| 1391R | GACGGGCGGTGTGTRCA |
F, forward primer; R, reverse primer.
16S rRNA gene screening and sequencing.
The clone library constructed from an environmental sample was screened by restriction fragment length polymorphism (RFLP) analysis with restriction enzymes HinPI and MspI (New England Biolabs). Each 10-μl RFLP reaction mixture contained 2 μl of a supplied enzyme buffer (New England Biolabs) with two units of each enzyme and 8.7 μl of a PCR product amplified directly from an individual clone colony as described above and incubated at 37°C for 2 h. Representatives (two or three clones) for each unique RFLP pattern were selected for sequencing. Plasmid DNA was prepared for sequencing from clones by using a QIAprep mini prep kit (QIAGEN) according the manufacturer's protocol. The PCR product from the pure cultures was precipitated prior to sequencing by using ethanol as described by Shapiro (57). DNA and primers (Table 2) were sent to the Translational Genomics Research Institute (TGen) (www.tgen.org) for sequencing. Sequences of the DNA of each clone and isolate were obtained in the forward and reverse directions with a minimum twofold coverage of the 16S rRNA gene. TGen uses Applied Biosystems automated sequencers.
Phylogenetic analysis.
Nucleotide sequences were assembled with BioEdit (26). Any ambiguous or incorrectly aligned positions were checked and aligned manually on the basis of the conserved primary sequence and secondary structures. Sequences were aligned using the ClustalX function in BioEdit and then further aligned manually to related sequences obtained from GenBank by using BLAST version 2.0 (3). The nearest neighbors for which full-length 16S rRNA gene data were available in GenBank and cultured isolates and clones with available ecological data were selected for analysis. All assembled sequences were checked for chimeras by using the Ribosomal Database Project II chimera check program (14) and the Bellerophon Sever run with two different correction models (Kimura and Jukes-Cantor) and with no correction model (29). No chimeras were detected by these methods. Aligned sequences were imported into the Mega 2.1 program (38) or BioEdit for phylogenetic analysis. A neighbor-joining phylogenetic tree was constructed using a Kimura two-parameter model for estimating evolutionary distances between sequences without a mask. A maximum likelihood phylogenetic tree was estimated from the nucleotide sequences by using fastDNAml version 3.3 (20, 49). Neighbor-joining bootstrap tests of phylogeny were run with 100 replicates.
Estimations of richness and diversity.
The richness and diversity of 16S rRNA genes were estimated from the clone library data. Numbers and proportional abundances of unique phylotypes were calculated based on the occurrence of unique RFLP patterns. Two or more representatives for each unique RFLP pattern were selected for sequence analysis. Sequence results were used to confirm RFLP pattern assignments as unique phylotypes. Unique phylotypes were defined as having a final 16S rRNA gene sequence similarity of 97% or greater compared to the other sequences in the clone library. Similarity between related sequences was determined using the software program DNADist (version 3.5c) (19). The relative abundances of these unique phylotypes were determined based on matching of RFLP patterns to the sequenced clone(s). Phylotype richness (S) and abundance (N) values were determined from clone library data. These measurements (S and N) were used for subsequent Shannon diversity (H′) calculation. A log series alpha value (α) was calculated for the clone library data and used to predict expected log abundance values for each phylotype (44). Chao1 estimates were calculated using the web interface provided by Kemp and Aller (34) (www.asol.org/lomethods/free/2004/0114a.html).
Nucleotide sequence accession numbers.
Sequences were deposited in GenBank under accession numbers DQ677840 through DQ677876.
RESULTS
The results presented in this study are based on samples collected during the austral summers (October to January) between 1999 and 2004. Blood Falls is a hydrologically dynamic feature that is subject to potential sample bias depending upon the date, time of day, and specific area of sample collection. Geochemical analysis of samples collected at Blood Falls allows us to infer the “purity” of our samples from the subglacial source. The chemistry of subglacial brine released at Blood Falls is markedly different from the chemistry of Taylor Glacier ice and the other streams that derive from supraglacial melt during the austral summer (42, 43). The chemistry of Blood Falls can vary with the rate of subglacial discharge, with low-discharge waters being diluted with contemporary glacial surface melt. Chloride and sulfate concentrations (Table 1) from Blood Falls, for example, are consistently high during periods of active subglacial outflow (November 2004 outflow concentrations: for Cl−, 1,440 mM; for SO42−, 50 mM) versus those during periods of diluted flow (November 2003 concentrations: for Cl−, 2 mM; for SO42−, 0.26 mM). Regardless of the fractional dilution of the outflow, measurements at Blood Falls are clearly distinct from those for supraglacial melt from Taylor Glacier surface ice, which contained concentrations of Cl− and SO42− measured at 1.6 μM and 0.92 μM, respectively (42). Blood Falls discharge is also iron rich (total iron concentration = 3.8 mM) and anoxic (no oxygen detected by Winkler titration) at its source, whereas melt from the Taylor Glacier is highly oxygenated (∼0.5 mM O2) (J. Mikucki, unpublished data). Bacterial cell numbers were lower in Taylor Glacier ice (2.3 × 103 ml−1) than in samples of diluted or concentrated subglacial outflow (range = 1 × 104 to 7.6 × 105 cells ml−1).
The clone library was constructed from a single sample collected in January 2000 when Blood Falls outflow was active based on geochemical data and did not appear to be diluted with supraglacial melt. Isolates were obtained from samples collected over 4 years (2001 to 2005), representing years of active subglacial outflow (2004 to 2005) as well as years in which outflow appears to have been diluted to some extent with supraglacial melt (2001 to 2004). Here, we present all the available diversity data to provide a more accurate description of the microbial community structure of the dynamic Blood Falls system. We focus our discussion on results obtained from the two samples collected during undiluted or concentrated outflow (18 January 2000 and 14 November 2005).
Cultured isolates.
The cultivation of microorganisms from natural systems is subject to bias from the moment a sample is removed from the environment. Our culture efforts were designed to survey organisms that could be readily cultured on synthetic media as well as media designed to reflect in situ conditions. This effort was by no means exhaustive, but the isolates obtained by our methods do provide insight into the ecology of the Blood Falls system.
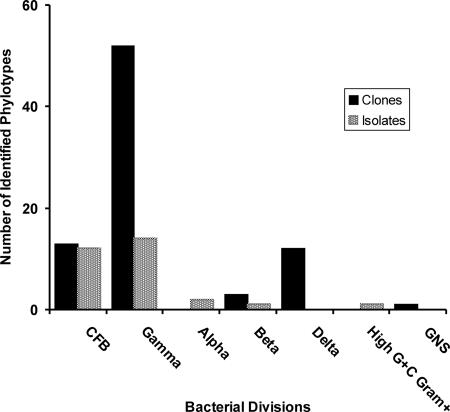
Seven distinct phylotypes were detected by culturing. The majority of bacterial isolates obtained from Blood Falls environmental samples were related to members of the Proteobacteria and Bacteroidetes divisions (Fig. 2). The most common isolates were from the genera Flavobacterium (represented by isolates DV3, S2, S13, S15, Tho1, Tho3, Tho8, BF05_1, BF05_2, and BF05_16), Psychrobacter (represented by isolates DV8, DV13, S5, S6, S8, S9, S11, S12, and S16), and Marinobacter (represented by isolates BF05-4, BF05_5, and BF05_16). Culture work on samples collected during diluted outflow events revealed several organisms not detected in the clone library, including isolate S10, which was closely related to an Arthrobacter isolate from sea ice (33), and an Alphaproteobacteria isolate (Tho7) that clustered within the genus Caulobacter (1). We were most interested in isolates BF05_4, 5, and 16, which were obtained during active subglacial flow (November 2004) and showed high sequence similarity to clones for our clone library. Isolate BF05_4 had 99.35% sequence similarity (based on 1,402 bp) to clone BF Clone_65 (which clusters with the Marinobacter). Phylotypes similar to BF05_4 have been previously isolated from both lobes of Lake Bonney. Strain ELB-17 (99.28% sequence similarity to BF05_4) was isolated from a 17-m depth in the east lobe (GenBank accession number AY518678) (69). Strain WLB16-007 (99.86% sequence similarity to BF05_4) was isolated from a 16-m depth in the west lobe of Lake Bonney (GenBank accession number DQ015849) (25).
FIG. 2.
Distribution of represented bacterial divisions detected in the Blood Falls clone library and cultures. Clones that were not sequenced were identified based on RFLP pattern. A total of 111 phylotypes were identified. CFB, Cytophaga-Flavobacteria-Bacteroides phylum; GNS, green nonsulfur.
Autotrophic enrichments yielded no strict chemolithotrophs; however, the ferrihydrite level in the Fe-Rd enrichment medium was reduced after approximately 9 months of incubation. When an aliquot of enrichment culture was transferred to marine agar for isolation, colonies appeared within 3 days. Phylogenetic analysis of the isolate (named BF_FeRd-1) confirmed a close relation to Shewanella frigidimarina (strain ACAM 591) (99% sequence similarity based on 1,400 bp) (9).
Clone library.
Our clone library was constructed from a sample collected during concentrated subglacial outflow (18 January 2000). Clones (81 total) were screened by RFLP analysis, and 40 were selected for complete sequencing. Unique phylotypes were distinguished based on a conservative definition (clones with 97% and greater sequence similarity were considered to be the same phylotype). Based on this definition, 16 distinct phylotypes were detected. The Blood Falls clone library was dominated by representatives of the phylum Proteobacteria (67 clones, or 83% of the library) (Fig. 2). The Gamma-, Delta- and Betaproteobacteria classes were represented by 52, 12, and 3 clones, respectively. Phylogenetic analysis using both neighbor-joining and maximum likelihood analysis of the 16S rRNA gene sequences yielded similar tree topologies; the neighbor-joining trees generated from the sequence data are presented (Fig. 3 and 4).
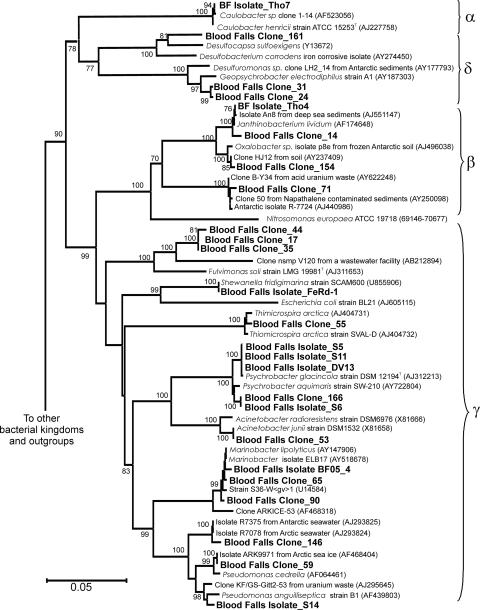
FIG. 3.
Neighbor-joining phylogenetic tree of Proteobacteria 16S rRNA gene sequences constructed with a Kimura two-parameter model. Selected sequences from the major bacterial lineages were used to the root the tree. Results of bootstrap analysis with 100 replications are noted. The scale bar represents 0.05 nucleotide substitutions per sequence position. Isolates and clones from Blood Falls are in bold. GenBank accession numbers are listed in parentheses.
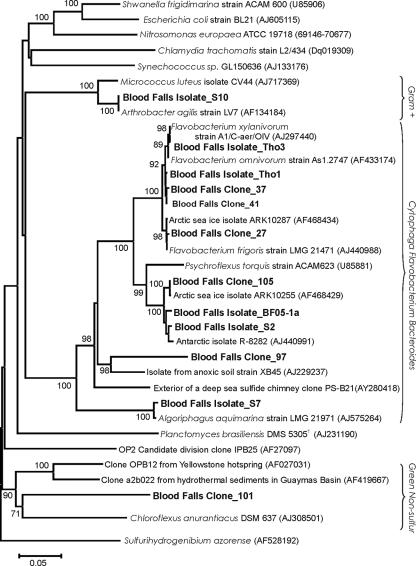
FIG. 4.
Neighbor-joining phylogenetic tree of 16S rRNA gene sequences from selected isolates and clones constructed with a Kimura two-parameter model. Results of bootstrap analysis with 100 replications are noted. The scale bar represents 0.05 nucleotide substitutions per sequence position. Isolates and clones from Blood Falls are in bold. GenBank accession numbers are listed in parentheses.
The most abundant clone in our library (BF Clone_55) was closely related (99% sequence similarity based on 1,420 bp) to an obligate chemolithoautotrophic sulfur-oxidizing bacterium, Thiomicrospira arctica, which was originally isolated from Arctic marine sediments (37). T. arctica is a true psychrophile and is capable of growth under high Na+ concentrations (as high as 1,240 mM). These values are consistent with in situ Na+ concentrations in Blood Falls. Clones belonging to the class Deltaproteobacteria constituted 15% of the library (12 clones). The closest sequence match (98% identity; 1,402 bp) for nine of these clones (BF Clone_24 and 31) was Geopsychrobacter electrodiphilus, a psychrotolerant bacterium capable of growth using Fe(III), Mn(IV), and elemental sulfur as electron acceptors (27). The remaining Deltaproteobacteria phylotype (BF Clone_161) was more distantly related to its nearest relatives in the GenBank database. This phylotype had 93.2% sequence similarity (based on 1,412 bp) to an isolate (strain STP 23) from anaerobic lake sediments that can disproportionate sulfite, ferment lactose, and use sulfite and thiosulfate as electron donors and acceptors (56) and 92.2% similarity (based on 1,412 bp) to Desulfocapsa sulfoexigens, an anaerobe isolated from marine surface sediments that disproportionates elemental sulfur and thiosulfate (21). Thirteen of the 81 clones were grouped in the Bacteroidetes division (sometimes referred to as the Cytophaga-Flavobacteria-Bacteroides or CFB division in the literature). CFB-type isolates obtained on S-Ox agar were related (95.1% sequence similarity) to deep sea isolates that are capable of oxidizing thiosulfate (64), suggesting that other phylotypes in Blood Falls may be involved in sulfur oxidation. A single sequence that was distantly related to other uncultured clones in the Chloroflexi (green nonsulfur) division, including an “uncultured Chloroflexi” from pasture soil (∼29%; 100 bp) and OPB 11 and OPB 12 (∼34%; 1,000 bp) from Obsidian Pool in Yellowstone (30), was detected. No Archaeal DNA was amplified using primers 4F and 1492R.
Diversity.
The use of 16S rRNA gene analysis for determining microbial diversity includes bias from DNA extraction, PCR, cloning, and gene copy number (18). Given these potential problems, the frequency of the 16S rRNA gene is not necessarily a reflection of the true in situ microbial community structure (67). Although sequenced microbial genomes of Blood Falls phylotypes currently do not exist, several phylogenetically related organisms have been analyzed (http://img.jgi.doe.gov and reference 36). Genomes available for the Pseudomonas and Acinetobacter genera, for example, have high rRNA gene copy numbers, but these phylotypes were not among the abundant clones retrieved from the Blood Falls library. There appears to be no correlation between the 16S rRNA gene copy number for the nearest relative with a genome sequence and the frequency of our environmental phylotypes detected (r = −0.24; P = 0.42), implying that copy number is not a factor in the frequency of clones from Blood Falls.
The use of several different microbial diversity measurements allows us to explore the physical and biological factors that may regulate community structure. The Shannon index is a frequently used measure of diversity and therefore provides a relative statistic for comparing studies of different environmental systems. H′ is not without biases, as it assumes that all species are represented in an infinitely large sample. Since there is no estimate of error for this statistic, the error increases as the proportion of species represented in the sample declines (44). Despite this potential for bias, diversity descriptions based on 16S rRNA genes provide a useful means of comparison with other systems and this approach has been used in many recent microbial diversity studies (e.g., references 16, 32, 48, and 55).
The bacterial 16S rRNA gene Shannon diversity value calculated for Blood Falls (H′ = 1.75) is comparable to measurements from other icy and glaciated environments (Table 3), including that of its proglacial lake, Lake Bonney. H′ was calculated for clone libraries from the west lobe of Lake Bonney at 13-m (H′ = 2.1) and 16-m (H′ = 2.4) depths (25). A microbial diversity study of the forefields of two receding glaciers (59) reported decreased Shannon diversities with increasing distances from both glaciers (from 2.31 and 2.83 at the glacier termini to 1.97 and 2.25 at 150-m distances from the termini). Both forefields showed higher diversities (∼24%; standard deviation = ±11%) than Blood Falls. However, Blood Falls has twice the diversity found in Antarctic pack ice (H′ = 0.81 to 0.84) and ∼60% greater diversity than that of the Artic pack ice (H′ = 1.06 to 1.09), despite the similarity in phylotype composition found in these locations. The similar phylotypes detected in both Blood Falls and polar pack ice included Shewanella fridigimarina and members of the genera Psychrobacter, Marinobacter, Pseudomonas, and Acinetobacter (13). Blood Falls is a hypersaline system, yet it shows diversity values lower than those reported for other hypersaline brines, for example, the deep hypersaline anoxic brines in the Mediterranean Sea (H′ = 2.17 to 3.88) (66).
TABLE 3.
Shannon diversity values for Blood Falls and other ecological systemsa
| Habitat sampled | N | H′ | Latitude | Phylotype detection method | Source or reference |
|---|---|---|---|---|---|
| Blood Falls | 81 | 1.75 | 77°S | RFLP and cloning | This study |
| WLB (13 m) | 28 | 2.1 | 77°S | RFLP and cloning | 25 |
| WLB (16 m) | 57 | 2.4 | 77°S | RFLP and cloning | 25 |
| Antarctic pack ice | 95 | 0.81 | 70°S | RFLP and cloning | 13 |
| 103 | 0.84 | 70°S | RFLP and cloning | 13 | |
| Arctic pack ice | 86 | 1.06 | 75°N | RFLP and cloning | 13 |
| 106 | 1.09 | 80°N | RFLP and cloning | 13 | |
| Glacier terminus | 25b | 2.31b | 46°N | DGGE | 59 |
| 11c | 2.83c | 46°N | DGGE | 59 | |
| 150 m from glacier | 15b | 1.97b | 46°N | DGGE | 59 |
| 9c | 2.25c | 46°N | DGGE | 59 | |
| Arizona arid soils | 190 | 6.6-7.1 | 36°N | RFLP and cloning | 16 |
Two glaciers were studied in the Sigler and Zeyer report (59), Damma Glacier and Rotfirn Glacier. N, abundance; WLB, west lobe of Lake Bonney.
Result for Damma Glacier.
Result for Rotfirn Glacier.
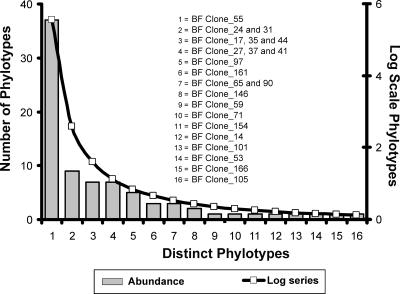
A phylotype distribution curve (i.e., a relative abundance curve) for Blood Falls (Fig. 5) revealed a few abundant clones (represented by BF Clone_55, 24, 31, 17, 35, 44, 27, 37, 41, and 97), with most sequences being rare and generating a long right-handed tail. The clone library data fit a log series distribution (Fig. 5). Log series α (5.97) was calculated from the clone library data and superimposed on the phylotype distribution curve (44). The log series describes the species abundance found in the Blood Falls clone library (P was >95% based on the goodness of fit test, where χ2 was 6.14, with 15 degrees of freedom).
FIG. 5.
Phylotype distribution curve based on bacterial 16S rRNA gene clone library data from an environmental sample of Blood Falls outflow. Phylotypes were determined based on RFLP pattern and confirmed with sequence analysis, and the log series curve is fitted to the distribution curve. The clone identification is listed for each phylotype and corresponds to the clones presented in the phylogenetic trees (Fig. 3 and 4).
The Shannon diversity (H′) value, which takes both evenness and abundance into account, was 1.75 for the Blood Falls clone library. Values for Shannon diversity typically range between 1.5 and 3.5 and rarely surpass 4.5 (44, 45). However, complex systems, such as soils, tend to have high phylotype diversity; for example, Arizona soil samples had reported Shannon diversity values between 6.6 and 7.1 (16). Phylotype richness generally decreases when one or a few populations attain a high density, signifying successful dominance by a single population (5), which may explain the low diversity observed in Blood Falls. Chao1 uses the number of singletons (phylotypes that appear once) and doubletons (phylotypes that appear twice) to estimate the diversity found in a given sample. Observed/predicted values for Chao1 indicated that only 53% of the total diversity in Blood Falls was sampled, implying that the measurements presented here may be conservative estimates of the bacterial diversity present in Blood Falls.
DISCUSSION
Microbial diversity in Blood Falls outflow. (i) Microbial diversity in subglacial environments.
Although diversity measurements can provide information on the richness of a particular system, these metrics do not provide specific information on the genetic diversity present. The phylotype diversity reported for subglacial systems from temperate latitudes differs considerably from that for Blood Falls. This difference is likely due to the physical hydrology of the glacier as well as the preglacial ecosystem. In temperate glaciers, there is direct communication between supraglacial melt and the subglacial bed, providing oxygen and the class Betaproteobacteria has been found to dominate alpine snowpack (2) and is the most abundant bacterial class reported for temperate subglacial systems. No Betaproteobacteria were detected in Blood Falls. Foght et al. (22) used cultivation to examine the diversity of basal sediments from the Franz Josef and Fox Glaciers, New Zealand. The majority of isolates detected using cultivation were members of the class Betaproteobacteria. A molecular survey based on sequencing of denaturing gradient gel electrophoresis (DGGE) fragments from samples collected from a subglacial volcanic crater lake in Iceland also revealed a system dominated by members of the Betaproteobacteria (24). A comparative study of the 16S rRNA gene diversity from the John Evens Glacier in Canada and the Bench Glacier in Alaska was dominated by sequences from the Betaproteobacteria as well (60). The New Zealand, Alaskan, and Canadian glaciers mentioned above are wet-based glaciers with meltwater from glacier ice reaching the basal environment. Despite the similar physical setting and potential metabolic niches of subglacial systems, the 16S rRNA gene diversity detected in Blood Falls is unique among reported subglacial systems. The majority of the phylotypes described for Blood Falls samples were related to other previously described marine organisms (10, 11, 52), reflecting its marine preglacial heritage. Below, we describe the distinct microbial niches implied by our data (Table 4).
TABLE 4.
Description of cultivated relatives nearest to selected Blood Falls clones and isolates obtained from concentrated subglacial outflow samples during December 1999 and December 2004a
| Putative ecological role | Blood Falls clone or isolate | Closest cultivated relative | % Similarity | % of library | Comments | Reference |
|---|---|---|---|---|---|---|
| Primary producer | C55 | Thiomicrospira arctica | 98.8 | 45.7 | Chemoautotrophic sulfur oxidizer; psychrophilic arctic marine sediments | 37 |
| Sulfur and iron reducer | C24, C31 | Geopsychrobacter electrodiphilus | 98.0 | 11.1 | Anaerobic, psychrotolerant, from marine sediments; chemoorganotroph using Fe(III), S°, Mn(IV), or AQDS as the electron acceptor | 27 |
| C161 | Desulfocapsa sulfoexigens | 92.7 | 3.7 | Anaerobic, psychrotolerant, from marine sediments; disproportionates elemental sulfur, sulfite, and thiosulfate using Fe(III) as a scavenging agent | 21 | |
| Heterotroph | C65, C90, isolate BF05_4a | Marinobacter lipolyticus | 96.1 | 3.7 | Moderate halophile with lipolytic activity | 46 |
| C154 | Antarctic soil isolate | 98.3 | 1.2 | Protease-producing isolate | 70 | |
| C105 | Psychroflexus tropicus | 95.5 | 1.2 | Hypersaline lake isolate | 15 | |
| Isolates BF05-1 and -2 | Abyssal strain AII3 Cytophaga-Flavobacterium | 95.1 | NA | Thiosulfate-oxidizing strain isolated from marine sediments from a 4,500-m depth | 64 |
AQDS, anthraquinone-2,6-disulfonate; NA, not applicable.
(ii) Sulfur oxidation and autotrophy.
There is growing evidence that biological sulfur oxidation is an important subglacial process. Significant biological sulfate production has been measured in subglacial sediment and water samples by incubation experiments (58) and stable isotopic measurements (8). Bottrell and Tranter (8) found that biological sulfur oxidation occurred under partially anoxic conditions, with Fe(III) derived from Fe-OH minerals as the oxidant. Our phylogenetic data support these previous studies and lead us to contend that Fe and S have an important role in the metabolism of the Taylor Glacier subglacial system. The majority of clones in the Blood Falls library (46%; 37 clones) were related to an obligate chemoautotroph (Thiomicrospira arctica), implying the capacity for subglacial chemosynthetic primary production. This notion is further supported by the dark CO2 fixation that we measured in Blood Falls concentrated outflow samples (J. Mikucki, unpublished data). The rates calculated for Blood Falls (1.2 nmol C liter−1 d−1) were statistically similar (P = 0.11) to rates measured in samples from a subglacial volcanic lake (3.3 nmol C liter−1 d−1) (24). The genus Thiomicrospira is an important group in hydrothermal vent communities and is widely distributed in marine environments, suggesting that this genus may play an important role in global carbon and sulfur cycles (12). It is possible that in situ primary production by chemoautotrophs like Thiomicrospira may provide new organic carbon to the subglacial environment below the Taylor Glacier in the total absence of sunlight and at permanently low temperatures.
(iii) Iron and sulfur reduction.
Several lines of evidence suggest that microbially mediated iron and sulfur reduction occurs in Blood Falls. The medium containing oxidized iron as the sole electron acceptor was reduced when inoculated with Blood Falls outflow (and H2 gas) after several months of incubation. From this enrichment, a close relative to Shewanella frigidimarina, a known iron-reducing organotroph, was isolated. Nine clones from the Blood Falls clone library clustered with members of the sulfur- and iron-reducing Geobacteraceae family with high 16S rRNA gene homology (98% similarity). Our findings are also consistent with geomicrobial studies from other subglacial environments. Sulfate reduction was measured in the subglacial environments of two Arctic glaciers by sulfate loss in incubations of basal ice from the John Evans Glacier in Canada (61) and stable-isotope analyses of subglacial outflow from Finsterwalderbreen in Svalbard (68).
(iv) Organic matter degradation.
Heterotrophic activity has been measured in Blood Falls outflow by labeled thymidine incorporation and extracellular enzyme activity assays (47). The isolates in the present study were not tested for their carbon utilization abilities, but the existing literature on the cultivated organisms nearest to the clones and isolates from Blood Falls implies the ability to degrade an array of carbon compounds. Members of the division Bacteroidetes were well represented in our clone library (13 clones; 16% of the library), and its members were readily cultivated from Blood Falls samples (12 isolates).
The ecology of the Taylor Glacier subglacial habitat.
The phylotype abundance data for Blood Falls fit a log series distribution (Fig. 5) which describes a habitat where the biological assemblage is composed of a small number of abundant phylotypes and a larger proportion of rare phylotypes that are present in relatively low numbers. This is typical of biological communities where a few dominant species account for the majority of energy inputs while the less abundant species determine the overall diversity of the system (5). A log series distribution typically occurs where one or only a few factors control the ecology of the community (for example, the availability of oxygen or sunlight). Glacier hydrology imparts strong feedbacks on the availability of oxygen as an electron acceptor and may be a robust regulator in subglacial community structure. The ability to gain energy efficiently at low temperatures from iron and sulfur minerals or available organic carbon would be a selective advantage below the Taylor Glacier and in other subglacial environments. The phylotypes in Blood Falls that were related to organisms that metabolize iron and sulfur compounds were the most abundant in the Blood Falls log series curve. The low diversity (based on the Shannon index) coupled with a log series distribution of phylotypes indicates that Blood Falls is a physically controlled ecosystem where adaptations to the physiochemical stress of subglacial conditions contribute to community structure control. Collectively, these data indicate that the environmental factors creating the subglacial environment (the preglacial ecosystem, the glacier bed substratum, and glacier hydrology) all influence the genetic structure of subglacial microbial diversity.
Conclusions.
Microbial growth in the subglacial setting must be able to proceed in the absence of sunlight and under permanently cold conditions. In the hydrologic regime of the Taylor Glacier, anoxia also is likely to be an important regulator of microbial energetics. Organisms below the Taylor Glacier must contend with elevated salinities and high iron concentrations. The biogeochemical data presented here imply that the bacterial assemblage below the Taylor Glacier can grow chemoautotrophically or chemoorganotrophically by harvesting energy from bedrock minerals or the assemblage may grow heterotrophically on ancient marine organics by respiring Fe(III) or SO42−. Given the length of time that this marine system has been isolated from phototrophic production (∼2 million years), the ability to degrade and consume increasingly recalcitrant organic carbon would also be important. The detection of putatively chemoautotrophic phylotypes suggests that subglacial systems can be sustained independent of new carbon fluxes by in situ CO2 fixation. Collectively, our data indicate that the subglacial community structure is constrained by a combination of bedrock lithology, glacier hydrology, and the preglacial history of the covered terrain.
Acknowledgments
We are grateful to all our McMurdo LTER and MO colleagues for support and suggestions, especially to Kathy Welch, W. Berry Lyons, and Martyn Tranter for critical discussions about the geochemistry of Blood Falls and to Christine Foreman and the “limno” team for assistance in sample collection. John Lisle collected the sample used for our clone library. We appreciate the thoughtful comments of Ann Pearson and two anonymous reviewers whose input greatly improved the organization and accuracy of the manuscript.
This research was supported by the following grants: NSF-OPP 00926250, NSF-OPP 0085400, NSF-OPP 0440943, and MCB 0237335 to J.C.P. and NASA GSRP NG5-50481 and NSF-ANT-0528710 to J.A.M.
Footnotes
Published ahead of print on 27 April 2007.
REFERENCES
- 1.Abraham, W. R., C. Strompl, H. Meyer, S. Lindholst, E. R. B. Moore, R. Christ, M. Vancanneyt, B. J. Tindall, A. Bennasar, J. Smit, and M. Tesar. 1999. Phylogeny and polyphasic taxonomy of Caulobacter species. Proposal of Maricaulis gen. nov. with Maricaulis maris (Poindexter) comb. nov. as the type species, and emended description of the genera Brevundimonas and Caulobacter. Int. J. Syst. Bacteriol. 49:1053-1073. [DOI] [PubMed] [Google Scholar]
- 2.Alfreider, A., J. Pernthaler, R. Amann, B. Sattler, F. O. Glöckner, A. Wille, and R. Psenner. 1996. Community analysis of the bacterial assemblages in the winter cover and pelagic layer of a high mountain lake by in situ hybridization. Appl. Environ. Microbiol. 62:2138-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Angino, E. E., K. B. Armitage, and J. C. Tash. 1964. Physiochemical limnology of Lake Bonney, Antarctica. Limnol. Oceanogr. 9:207-217. [Google Scholar]
- 5.Atlas, R. M. 1984. Diversity of microbial communities. Adv. Microb. Ecol. 7:1-47. [Google Scholar]
- 6.Black, R. F., M. L. Jackson, and T. E. Berg. 1965. Saline discharge from Taylor Glacier, Victoria Land, Antarctica. J. Geol. 74:175-181. [Google Scholar]
- 7.Boone, D. R., R. L. Johnson, and Y. Liu. 1989. Diffusion of interspecies electrons carriers H2 and formate in methanogenic ecosystems and its implications in the measurements of Km for H2 or formate uptake. Appl. Environ. Microbiol. 55:1735-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bottrell, S. H., and M. Tranter. 2002. Sulphide oxidation under partially anoxic conditions at the bed of the Haut Glacier d'Arolla, Switzerland. Hydrol. Process. 16:2363-2368. [Google Scholar]
- 9.Bowman, J. P., S. A. McCammon, D. S. Nichols, J. H. Skerratt, S. M. Rea, P. D. Nicols, and T. A. McMeekin. 1997. Shewanella gelidimarina sp. nov. and Shewanella firgidimarina sp. nov., novel Antarctic species with the ability to produce eicosapentaenoic acid (20:5w3) and grow anaerobically by dissimilatory Fe(III) reduction. Int. J. Syst. Bacteriol. 47:1040-1047. [DOI] [PubMed] [Google Scholar]
- 10.Bowman, J. P., and R. D. McCuaig. 2003. Biodiversity, community structural shifts, and biogeography of prokaryotes within Antarctic continental shelf sediments. Appl. Environ. Microbiol. 69:2463-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bowman, J. P., S. M. Rea, S. A. McCammon, and T. A. McMeekin. 2000. Diversity and community structure within anoxic sediment from marine salinity meromictic lakes and a coastal meromictic marine basin, Vestfold Hills, Eastern Antarctica. Environ. Microbiol. 2:227-237. [DOI] [PubMed] [Google Scholar]
- 12.Brinkhoff, T., and G. Muyzer. 1997. Increased species diversity and extended habitat range of sulfur oxidizing Thiomicrospira spp. Appl. Environ. Microbiol. 63:3789-3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brinkmeyer, R., K. Knittel, J. Jurgens, H. Weyland, R. Amann, and E. Helmke. 2003. Diversity and structure of bacterial communities in Arctic versus Antarctic pack ice. Appl. Environ. Microbiol. 69:6610-6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 1:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donachie, S. P., J. P. Bowman, and M. Alam. 2004. Psychroflexus tropicus sp. nov., an obligately halophilic Cytophaga-Flavobacterium-Bacterioides group bacterium from an Hawaiian hypersaline lake. Int. J. Syst. Evol. Microbiol. 54:935-940. [DOI] [PubMed] [Google Scholar]
- 16.Dunbar, J., S. Takala, S. M. Barns, J. A. Davis, and C. R. Kuske. 1999. Levels of bacterial community diversity in four arid soils compared by cultivation and 16S rRNA gene cloning. Appl. Environ. Microbiol. 65:1662-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elston, D. P., and S. L. Bressler. 1981. Magnetic stratigraphy of DVDP drill cores and late Cenozoic history of Taylor Valley, Transantarctic Mountains, Antarctica, p. 413-426. In L. D. McGinnis (ed.), Dry Valley drilling project. Antarctic Research Series, vol. 33. American Geophysical Union, Washington, DC. [Google Scholar]
- 18.Farrelly, V., F. A. Rainey, and E. Stackebrandt. 1995. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl. Environ. Microbiol. 61:2798-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Felsenstein, J. 1993. DNADIST, 3.5c ed. University of Washington, Seattle, WA.
- 20.Felsenstein, J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17:368-376. [DOI] [PubMed] [Google Scholar]
- 21.Finster, K., W. Liesack, and B. Thamdrup. 1998. Elemental sulfur and thiosulfate disproportionation by Desulfocapsa sulfoexigens sp. nov., a new anaerobic bacterium isolated from marine surface sediment. Appl. Environ. Microbiol. 3:119-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Foght, J., J. Aislabie, S. Turner, C. E. Brown, J. Ryburn, D. J. Saul, and W. Lawson. 2004. Culturable bacteria in subglacial sediment and ice from two southern hemisphere glaciers. Microb. Ecol. 47:329-340. [DOI] [PubMed] [Google Scholar]
- 23.Fountain, A. G., W. B. Lyons, M. B. Burkins, G. L. Dana, P. T. Doran, K. J. Lewis, D. M. McKnight, D. L. Moorhead, A. N. Parsons, J. C. Priscu, D. H. Wall, R. A. Wharton, Jr., and R. A. Virginia. 1999. Physical controls on the Taylor Valley ecosystem, Antarctica. Bioscience 49:961-971. [Google Scholar]
- 24.Gaidos, E. J., B. Lanoil, T. Thorsteinsson, A. Graham, M. Skidmore, S.-K. Han, T. Rust, and B. Popp. 2004. A viable microbial community in a subglacial volcanic crater lake, Iceland. Astrobiology 4:327-344. [DOI] [PubMed] [Google Scholar]
- 25.Glatz, R. E., P. W. Lepp, B. B. Ward, and C. A. Francis. 2006. Phanktonic microbial community composition across steep physical/chemical gradients in permanently ice-covered Lake Bonney, Antarctica. Geobiology 4:53-67. [Google Scholar]
- 26.Hall, T. A. 1999. BioEdit: a user friendly biological sequence alignment editor and analysis program for Windows 85/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 27.Holmes, D. E., J. S. Nicoll, D. R. Bond, and D. R. Lovley. 2004. Potential role of a novel psychrotolerant member of the family Geobacteraceae, Geopsychrobacter electrodiphilus gen. nov., sp. nov., in electricity production by a marine sediment fuel cell. Appl. Environ. Microbiol. 70:6023-6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hubbard, A., W. Lawson, B. Anderson, B. Hubbard, and H. Blatter. 2004. Evidence for subglacial ponding across Taylor Glacier, Dry Valleys, Antarctica. Ann. Glaciol. 39:79-84. [Google Scholar]
- 29.Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319. [DOI] [PubMed] [Google Scholar]
- 30.Hugenholtz, P., C. Pitulle, K. L. Hershberger, and N. R. Pace. 1998. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180:366-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hungate, R. E. 1969. A roll tube method for cultivation of strict anaerobes, p. 117-132. In J. R. Norris and D. W. Ribbons (ed.), Methods in Microbiology, vol. 3B. Academic Press, New York, NY. [Google Scholar]
- 32.Inagaki, F., M. Suzuki, K. Takai, H. Oida, T. Sakamoto, K. Aoki, K. H. Nealson, and K. Horikoshi. 2003. Microbial communities associated with geological horizons in coastal subseafloor sediment from the Sea of Okhotsk. Appl. Environ. Microbiol. 69:7224-7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Junge, K., J. J. Gosink, H. G. Hoppe, and J. T. Staley. 1998. Arthrobacter, Brachybacteriu and Planococcus, isolates identified from Antarctic sea ice brine. Description of Planococcus mcmeekenii, sp. nov. Syst. Appl. Microbiol. 21:306-314. [DOI] [PubMed] [Google Scholar]
- 34.Kemp, P. F., and J. Y. Aller. 2004. Estimating prokaryotic diversity: when are 16S rDNA libraries large enough? Limnol. Oceanogr. Methods 2:114-125. [Google Scholar]
- 35.Keys, J. R. 1979. Saline discharge at the terminus of the Taylor Glacier. Antarctic J. US 14:82-85. [Google Scholar]
- 36.Klappenbach, J. A., P. R. Saxman, J. R. Cole, and T. M. Schmidt. 2001. rrndb: the Ribosomal RNA Operon Copy Number Database. Nucleic Acids Res. 29:181-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knittel, K., J. Kuever, A. Meyerdierks, R. Meinke, R. Amann, and T. Brinkhoff. 2005. Thiomicrospira arctica sp. nov. and Thiomicrospira psychrophila sp. nov. psychrophilic obligately chemolithoautotrophic sulfur-oxidizing bacteria isolated from marine Arctic sediments. Int. J. Syst. Evol. Microbiol. 55:781-786. [DOI] [PubMed] [Google Scholar]
- 38.Kumar, S. K., K. Tamura, and M. Nei. 1993. MEGA: Molecular Evolutionary Genetics Analysis, 2.1 ed. Pennsylvania State University, University Park, PA.
- 39.Lisle, J. T., and J. C. Priscu. 2004. The occurrence of lysogenic bacteria and microbial aggregates in the lakes of the McMurdo Dry Valleys, Antarctica Microb. Ecol. 47:427-439. [DOI] [PubMed] [Google Scholar]
- 40.Lovley, D. R., and E. J. P. Phillips. 1986. Organic matter mineralization with reduction of ferric iron in anaerobic sediments. Appl. Environ. Microbiol. 51:683-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lyons, W. B., K. A. Welch, and P. Sharma. 1998. Chlorine-36 in the waters of the McMurdo Dry Valley lakes, southern Victoria Land, Antarctica: revisited. Geochim. Cosmochim. Acta 62:185-191. [Google Scholar]
- 42.Lyons, W. B., K. A. Welch, K. Neumann, J. K. Toxey, R. McArthur, C. Williams, D. M. McKnight, and D. Moorhead. 1998. Geochemical linkages among glaciers, streams and lakes within the Taylor Valley, Antarctica. In J. C. Priscu (ed.), Ecosystem dynamics in a polar desert, vol. 72. American Geophysical Union, Washington, DC.
- 43.Lyons, W. B., K. A. Welch, G. Snyder, J. Olesik, E. Y. Graham, G. M. Marion, and R. J. Poreda. 2005. Halogen geochemistry of the McMurdo Dry Valleys lakes, Antarctica: clues to the origin of solutes and lake evolution. Geochim. Cosmochim. Acta 69:305-323. [Google Scholar]
- 44.Magurran, A. 1988. Ecological diversity and its measurement. Princeton University Press, Princeton, NJ.
- 45.Margalef, R. 1972. Interpretation not strictly statistical of representation of biological entities in multifactorial space. Investig. Pesquera 36:183-190. [Google Scholar]
- 46.Martin, S., M. C. Marquez, C. Sanchez-Porro, E. Mellado, D. R. Arahal, and A. Ventosa. 2003. Marinobacter lipolyticus sp. nov., a novel moderate halophile with lipolytic activity. Int. J. Syst. Evol. Microbiol. 53:1383-1387. [DOI] [PubMed] [Google Scholar]
- 47.Mikucki, J. A., C. M. Foreman, B. Sattler, W. B. Lyons, and J. C. Priscu. 2004. Geomicrobiology of Blood Falls: an iron-rich saline discharge at the terminus of the Taylor Glacier, Antarctica. Aquat. Geochem. 10:199-220. [Google Scholar]
- 48.Moyer, C. L., F. C. Dobbs, and D. M. Karl. 1994. Estimation of diversity and community structure through restriction fragment length polymophism distribution and analysis of bacterial 16S rRNA genes from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii. Appl. Environ. Microbiol. 60:871-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olsen, G. J., H. Matsuda, R. Hagstrom, and R. Overbeek. 1994. fastDNAml: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comp. Appl. Biosci. 10:41-48. [DOI] [PubMed] [Google Scholar]
- 50.Priscu, J. C. 1998. Ecosystem dynamics in a polar desert, vol. 72. American Geophysical Union, Washington, DC.
- 51.Raiswell, R. 1984. Chemical models of solute acquisition in glacial meltwaters. J. Glaciol. 30:49-57. [Google Scholar]
- 52.Ravenschlag, K., K. Sahm, J. Pernthaler, and R. Amann. 1999. High bacterial diversity in permanently cold marine sediments. Appl. Environ. Microbiol. 65:3982-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reasoner, D. J., and E. E. Geldreich. 1979. A new medium for the enumeration and subculture of bacteria from potable water, abstr. N7. Abstr. 79th Annu. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC. [DOI] [PMC free article] [PubMed]
- 54.Robinson, P. H. 1984. Ice dynamics and thermal regime of Taylor Glacier South Victoria Land, Antarctica. J. Glaciol. 30:153-159. [Google Scholar]
- 55.Rosenzweig, M. L. 1995. Species diversity in space and time. Cambridge University Press, New York, NY.
- 56.Sass, H., E. Wieringa, H. Cypionka, H. D. Babenzien, and J. Overmann. 1998. High genetic and physiological diversity of sulfate-reducing bacteria isolated from an oligotrophic lake sediment. Arch. Microbiol. 170:243-251. [DOI] [PubMed] [Google Scholar]
- 57.Shapiro, D. J. 1981. Quantitative ethanol precipitation of nanogram quantities of DNA and RNA. Ann. Biochem. 110:229-231. [DOI] [PubMed] [Google Scholar]
- 58.Sharp, M., J. Parkes, B. Cragg, I. J. Fairchild, H. Lamb, and M. Tranter. 1999. Widespread bacterial populations at glacier beds and their relationship to rock weathering and carbon cycling. Geology 27:107-110. [Google Scholar]
- 59.Sigler, W. V., and J. Zeyer. 2002. Microbial diversity and activity along the forefields of two receding glaciers. Microb. Ecol. 43:397-407. [DOI] [PubMed] [Google Scholar]
- 60.Skidmore, M., S. P. Anderson, M. Sharp, J. Foght, and B. D. Lanoil. 2005. Comparison of microbial community compositions of two subglacial environments reveals a possible role for microbes in chemical weathering processes. Appl. Environ. Microbiol. 71:6986-6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Skidmore, M. L., J. M. Foght, and M. J. Sharp. 2000. Microbial life beneath a high Arctic glacier. Appl. Environ. Microbiol. 66:3214-3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sugden, D. E., D. R. Marchant, and G. H. Denton. 1993. The case for a stable East Antarctic ice sheet: the background. Geograf. Annaler. 75A:151-154. [Google Scholar]
- 63.Summerfield, M. A., F. M. Stuart, H. A. P. Cockburn, D. E. Sugden, G. H. Denton, T. Dunai, and D. R. Marchant. 1999. Long-term rates of denudation in the Dry Valleys, Transantarctic Mountains, Southern Victoria Land, Antarctica, based on in-situ-produced cosmogenic 21Ne. Geomorphology 27:113-129. [Google Scholar]
- 64.Teske, A., T. Brinkhoff, G. Muyzer, D. P. Moser, J. Rethmeier, and H. W. Jannasch. 2000. Diversity of thiosulfate-oxidizing bacteria from marine sediments and hydrothermal vents. Appl. Environ. Microbiol. 66:3125-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tranter, M., M. Skidmore, and J. Wadham. 2005. Hydrological controls on microbial communities in subglacial environments. Hydrol. Process. 19:995-998. [Google Scholar]
- 66.van der Wielen, P. W. J. J., H. Bohuis, S. Borin, D. Daffonchio, C. Corselli, L. Giuliano, G. D'Auria, G. J. de Lange, A. Huebner, S. P. Varnavas, J. Thomson, C. Tamburini, D. Marty, T. J. McGenity, K. N. Timmis, and the BioDeep Scientific Party. 2005. The enigma of prokaryotic life in deep hypersaline anoxic basins. Science 307:121-123. [DOI] [PubMed] [Google Scholar]
- 67.von Wintzingerode, F., U. B. Gobel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 68.Wadham, J. L., S. Bottrell, M. Tranter, and R. Raiswell. 2004. Stable isotope evidence for microbial sulphate reduction at the bed of a polythermal high Arctic glacier. Earth Planet. Sci. Lett. 219:341-355. [Google Scholar]
- 69.Ward, B. B., and J. C. Priscu. 1997. Detection and characterization of denitrifying bacteria form a permanently ice-covered Antarctic lake. Hydrobiologia 347:57-68. [Google Scholar]
- 70.Wery, N., U. Gerike, A. Sharman, J. B. Chaudhuri, D. W. Hough, and M. J. Danson. 2003. Use of a packed-column bioreactor for isolation of diverse protease-producing bacteria from Antarctic soil. Appl. Environ. Microbiol. 69:1457-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]