Abstract
We studied the diversity of Chloroflexus-like bacteria (CLB) in a hypersaline phototrophic microbial mat and assayed their near-infrared (NIR) light-dependent oxygen respiration rates. PCR with primers that were reported to specifically target the 16S rRNA gene from members of the phylum Chloroflexi resulted in the recovery of 49 sequences and 16 phylotypes (sequences of the same phylotype share more than 96% similarity), and 10 of the sequences (four phylotypes) appeared to be related to filamentous anoxygenic phototrophic members of the family Chloroflexaceae. Photopigment analysis revealed the presence of bacteriochlorophyll c (BChlc), BChld, and γ-carotene, pigments known to be produced by phototrophic CLB. Oxygen microsensor measurements for intact mats revealed a NIR (710 to 770 nm) light-dependent decrease in aerobic respiration, a phenomenon that we also observed in an axenic culture of Chloroflexus aurantiacus. The metabolic ability of phototrophic CLB to switch from anoxygenic photosynthesis under NIR illumination to aerobic respiration under non-NIR illumination was further used to estimate the contribution of these organisms to mat community respiration. Steady-state oxygen profiles under dark conditions and in the presence of visible (VIS) light (400 to 700 nm), NIR light (710 to 770 nm), and VIS light plus NIR light were compared. NIR light illumination led to a substantial increase in the oxygen concentration in the mat. The observed impact on oxygen dynamics shows that CLB play a significant role in the cycling of carbon in this hypersaline microbial mat ecosystem. This study further demonstrates that the method applied, a combination of microsensor techniques and VIS and NIR illumination, allows rapid establishment of the presence and significance of CLB in environmental samples.
Chloroflexus-like bacteria (CLB) have been reported to be conspicuously present in some microbial mats (4, 14, 16, 19, 23, 24, 29, 32, 34), while in other mats they seem to be virtually absent. CLB are multicellular filamentous anoxygenic phototrophs (13), which can move by gliding and are also characterized by the production of bacteriochlorophyll a (BChla) and in some species by the supplemental production of BChlc or BChld. All characterized CLB are members of the genera Chloroflexus, Chloronema, Oscillochloris, Roseiflexus, and Heliothrix in the family Chloroflexaceae in the phylum Chloroflexi (previously called the green nonsulfur bacteria). The phylum Chloroflexi accommodates additional genera, including filamentous but nonphototrophic species (Herpetosiphon) and even nonfilamentous nonphototrophic species (Thermoleophilum and Thermomicrobium) (13). Fluorescence microscopy with infrared detection has been used to visualize bacteriochlorophyll-containing CLB in hot spring microbial mats (27). We have used fluorescence microscopy with blue excitation to discriminate potential CLB from cyanobacteria, because the latter fluoresce in the red visible light range and the former do not fluoresce in the red visible light range. Chloroflexus aurantiacus, which was initially isolated and described by Pierson and Castenholz (28), is the most-studied species of the Chloroflexaceae and is characterized by a versatile metabolism as it can grow photohetero- and photoautotrophically, as well as chemotrophically by oxygen respiration. It produces BChlcs, which is characterized by an in vivo absorption maximum at 740 nm instead of the 745- to 755-nm absorption maximum of BChlc that is produced by the phylogenetically distantly related green sulfur bacteria (22). Despite the fact that CLB bacteriochlorophyll synthesis is repressed by oxygen (25, 28), CLB are often found to be abundant in the fully oxic photic zone of microbial mats (6, 14, 15, 19, 24), where they can utilize organic photosynthetic exudates from oxygenic phototrophs, probably their most preferred substrates (2, 13). Studies of BChla production in the purple sulfur bacterium (PSB) Thiocapsa roseopersicina have shown that an anoxic period of only a few hours (the dark period in microbial mats) during a 24-h period is sufficient for production of enough photopigment to enable phototrophic growth during the light, oxic period in mats (8, 31). The same may be true for CLB and may explain the observed abundance of CLB in the oxic zone of microbial mats; however, in situ studies of the function and metabolism of CLB are still limited (24, 29, 33, 34). The aim of the present study was to characterize the impact of CLB on the community carbon and oxygen metabolism of a hypersaline microbial mat, making use of the observation that intact mats, as well as a pure culture of C. aurantiacus, showed a significant decrease in aerobic respiration upon illumination with near-infrared (NIR) light. Therefore, we measured the contribution of CLB to microbial mat community oxygen respiration under different illumination regimens, including visible (VIS) light (400 to 700 nm), VIS light plus NIR light (400 to 700 plus 710 to 770 nm), and dark conditions. In this combined physiological and molecular study we obtained quantitative CLB pigment analysis data, as well as 16S rRNA gene-based evidence demonstrating the presence of CLB in the mat, and we characterized the phylogenetic diversity of the CLB.
MATERIALS AND METHODS
Sample collection.
Microbial mat samples were collected in October 2004 from near-shore mats of the hypersaline lake La Salada de Chiprana located in northeastern Spain (41°14′20″N, 0°10′55″W). Intact mat samples where taken for further studies to Bremen, Germany, where they were incubated in original lake water at 21°C with illumination cycles consisting of 16 h of light (300 μmol photons m−2 s−1) and 8 h of darkness in a glass aquarium.
Characterization of CLB in Lake Chiprana microbial mats. (i) Oxygen and hydrogen sulfide dynamics in intact mats.
In order to characterize the intact microbial mat, profiles of oxygen and hydrogen sulfide concentrations were determined at 100-μm intervals in the laboratory in a flowthrough chamber with artificial illumination (83 and 300 μmol photons m−2 s−1) using a fiber optic lamp (Schott KL 1500) which has a halogen lamp as the light source. Oxygen profiles were also determined under dark conditions and with 33 and 166 μmol photons m−2 s−1. Clark-type amperometric oxygen (tip diameter, 10 μm) and hydrogen sulfide (tip diameter, 15 μm) microsensors were used. Details concerning the use of oxygen and hydrogen sulfide sensors and calibration procedures have been described previously (17, 35). Microsensors were mounted on a motorized micromanipulator connected to a heavy stand. Positioning and data acquisition were performed automatically with a laptop computer. Before profiling, the microsensor tip was positioned on the sediment surface of the microbial mat with the aid of a binocular microscope.
(ii) Microscopic observations.
The topmost 2-mm green stratum (i.e., the photic zone of the mat) was suspended in filtered lake water, and aliquots were removed and used for fluorescence microscopy. Microscopic samples were excited with blue light, and in combination with a long-pass filter (>520 nm) filamentous bacteria that were fluorescent (cyanobacteria) or nonfluorescent in the visible part of the light spectrum could be distinguished. Nonfluorescent thin filaments without visible sulfur inclusions were considered CLB. The diversity of CLB morphotypes was determined on the basis of filament width and length. The ratio of nonfluorescent filaments to fluorescent filaments was determined visually using multiple sample preparations.
(iii) Photopigment analysis.
The photic zone (0- to 2-mm surface layer) was cut off frozen microbial mat samples and freeze-dried. Pigments were extracted after sonication in high-performance liquid chromatography (HPLC)-grade acetone. The extracts from two subsequent extractions were combined, and the carboxylic groups of extracted pigments were methylated with 2 or 3 drops of diazomethane dissolved in acetone using a modification of the method recommended by Sigma-Aldrich (catalog no. Z411736; Aldrich, Milwaukee, WI) using 1-methyl-3-nitro-1-nitrosoguanidine as the precursor. The solvent was evaporated using a speed vacuum machine for a few hours until the preparation was completely dry, leaving pigments in the tube. Pigments were redissolved in a 2-ml HPLC elution solution containing 45% acetonitrile, 50% methanol, and 5% water-based 50 mM ammonium-acetate buffer (pH 7.2). The liquid was filtered through a sterile 0.2-μm-pore-size filter, and a 100-μl aliquot was injected into an HPLC for pigment identification and quantification using a binary protocol as described previously (5).
(iv) CLB phylotype diversity: DNA extraction, PCR amplification, and cloning.
In order to estimate the diversity of CLB phylotypes in the mat studied, a clone library was constructed. Genomic DNA was extracted from 0.2-g mat samples as described previously (1) and purified with a Wizard DNA clean-up system (Promega). An approximately 400-bp fragment of the 16S rRNA gene was amplified from genomic DNA using two primers specific for bacteria of the phylum Chloroflexi (green nonsulfur bacteria) (11), GNSB-941F (5′AGCGGAGCGTGTGGTTT3′) and GNSB-1340R (5′CGCGGTTACTAGCAAC3′). Two microliters of template was added to a 50-μl reaction mixture containing 0.5 U Eppendorf Taq, 1× buffer, 4 mM of MgCl2, 4 mM of each deoxynucleoside triphosphate, and 1 μM of each primer. The reaction was performed in a Mastercycler thermocycler (Eppendorf, Hamburg, Germany) with the following cycling conditions: 95°C for 2 min and then 30 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min, followed by a final incubation at 72°C for 10 min. The PCR product was visualized on an agarose gel, and the 16S rRNA band was excised. The excised PCR product was then purified using a QIAquick gel extraction kit (QIAGEN, Hilden, Germany). Two microliters of purified product was then ligated into the pGEM-T Easy vector (Promega, Madison, WI) and then transformed into Escherichia coli TOP10 cells (Invitrogen, Carlsbad, CA) according to the manufacturer's recommendations. Overnight cultures were prepared from positive transformants in a 2-ml 96-well culture plate. Plasmid DNA was extracted and purified using a Montage 96 plasmid miniprep kit (Millipore, Billerica, MA).
(v) DNA sequencing and analysis.
Purified plasmids were sequenced in one direction with the M13F primer, using a BigDye Terminator v3.0 cycle sequencing kit (Applied Biosystems, Foster City, CA). Samples were run on an Applied Biosystems 3100 genetic analyzer (Foster City, CA). A phylogenetic tree was constructed with the ARB software package (http://www.arb-home.de) (21). First, the partial sequences retrieved and amplified from the mat were grouped into phylotypes based on the criterion that sequences of the same phylotype share more than 96% similarity. Phylogenetic trees were constructed with the maximum likelihood, maximum parsimony, and neighbor joining methods, using publicly available sequences that were at least 1,100 bp long. Representative sequences from each phylotype were then added to the phylogenic trees by parsimony.
Functional analysis of CLB in mats and culture. (i) Aerobic respiration of C. aurantiacus.
In order to determine the influence of different types of light (VIS and/or NIR illumination) on the metabolism of CLB (phototrophy versus aerobic respiration), an axenic culture of the hyperthermophilic strain C. aurantiacus DSM 635 was used as model for CLB in the natural environment (even though C. aurantiacus might be quite different from CLB present in hypersaline environments, pure cultures of hypersaline CLB species are not available). C. aurantiacus was cultured according to DSMZ recommendations in yeast extract medium amended with 1 mM sulfide. Cultures were incubated anaerobically at 55°C under a light-dark regimen consisting of 16 h of illumination with incandescent light (approximately 25 μmol photons m−2 s−1) and 8 h of darkness. Oxygen consumption under different light conditions was determined with sulfide-depleted cultures. Just before oxygen consumption measurement, culture aliquots were aerated and then incubated in 25-ml glass tubes in a water bath at 55°C. An oxygen microsensor, fitted in a butyl rubber stopper, was inserted into the culture (protein content, 0.58 ± 0.03 mg ml−1) without introduction of air bubbles and sealed to avoid contact with ambient air during the measurement. The change in the oxygen concentration over time was recorded. Cultures killed with formaldehyde (final concentration, 2%) were used to correct for abiotic oxygen consumption. Two sets of light-dark shift experiments were performed; the set of first experiments was performed with a combined VIS and NIR light source, and the second set of experiments was performed with only a VIS light source. In the first set of experiments light was provided by a 25-W incandescent (VIS plus NIR) light bulb in combination with two 40-mA NIR light-emitting diodes (LEDs) with a peak wavelength at 740 nm and a spectral full-width at half-maximum bandwidth of a 30-nm angle (LED-740-524; Roithner LaserTechnik, Austria). The light intensity as determined with a scalar irradiance light sensor (LI-250A; LI-COR Biosciences) was 15 μmol photons m−2 s−1 near the surface of the culture tube; however, the specific intensity of the NIR light was unknown as the light meter used is not sensitive in the NIR (>700-nm) light spectrum. The light source used in the second set of experiments was two warm white high-power LEDs (LXHL-MWGC; Lumileds, United States). The light range of these LEDs is restricted to the VIS part of the spectrum (400 to 700 nm). With this light source a light intensity of 60 μmol photons m−2 s−1 was measured at the surface of the inundated culture tube.
(ii) Aerobic respiration of intact mats under different illumination conditions.
In order to elucidate the effect of NIR light illumination on oxygen dynamics, intact mat pieces were incubated in a flowthrough chamber at room temperature and illuminated with LEDs with VIS radiance (400 to 700 nm: LXHL-MWGC; Lumileds) and/or with two 40-mA NIR LEDs (LED-740-524; Roithner LaserTechnik, Austria). The advantage of using separate LEDs instead of using VIS/NIR filters in combination with a full-spectrum light source is that the light intensity of the light source with one type of spectrum is not changed when the light source with the other spectral type is switched on or off. VIS LEDs were used to illuminate mats at 60 μmol photons m−2 s−1 with or without additional NIR light illumination, and oxygen profiles were recorded after steady-state conditions were reached. The microbial mat areal net oxygen production rates were calculated from the change in oxygen fluxes in the diffusion boundary layer using an oxygen diffusion coefficient of 2.1 × 10−5 (25°C, 75 ppt) (see reference 35 for details concerning the procedure used). In order to determine whether oxygen evolution occurred with NIR light illumination, the oxygen profiles in NIR light-illuminated mats were measured before and after the addition of 3-(3,4-dichlophenyl)-1-1-dimethylurea (DCMU), a specific inhibitor of the oxygen-evolving photosystem II. DCMU was added from a stock solution (1 mM dissolved in 70% ethanol) to a final concentration of 5 μM.
Nucleotide sequence accession numbers.
The partial 16S rRNA gene sequences determined in this study have been submitted to the GenBank database and assigned accession numbers DQ973818 to DQ 973833.
RESULTS
Characterization of CLB in Lake Chiprana microbial mats. (i) Microsensor characterization of the intact microbial mat.
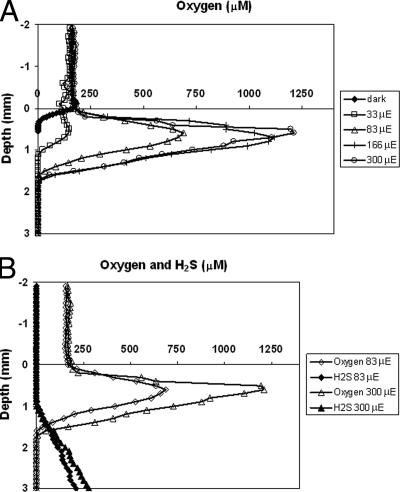
The oxygen concentration profiles for illuminated mats show that oxygen is produced mainly in the first 1 mm and that the concentration reaches more than six times air saturation when the mats are illuminated with 300 μmol photons m−2 s−1 (Fig. 1A), while in the dark oxygen penetrated to a maximum depth of only 0.4 mm. Oxygen and hydrogen sulfide cooccurred just above the oxic-anoxic interface in the 1- to 1.5-mm layer (Fig. 1B). The mat was entirely anoxic from 1.5 mm down, while the hydrogen sulfide concentration increased below this depth. These measurements thus show that CLB present in the photic zone of the mat (0- to 2-mm surface layer) encounter high oxygen concentrations during illumination in the topmost part and also sulfide in the lower part of the photic zone. Both components are relevant for CLB metabolism, as oxygen can be used as an electron acceptor in aerobic respiration, while sulfide can be used as an electron donor during photoautotrophic growth or, alternatively, as an electron donor for energy generation in aerobic respiration.
FIG. 1.
Steady-state oxygen and sulfide profiles in Lake Chiprana microbial mats at different light intensities. (A) Oxygen concentration profiles with 0, 33, 83, 166, and 300 μmol photons m−2 s−1. The oxygen concentration reaches up to six times the air saturation level at higher light intensities, indicating that CLB in the photic zone have to cope with high and fluctuating oxygen concentrations. (B) Oxygen and sulfide (H2S) concentration profiles at light intensities of 83 and 300 μmol photons m−2 s−1. Oxygen and sulfide cooccur in the zone between 1 and 1.5 mm deep, a zone where CLB are able to oxidize sulfide both phototrophically and chemotrophically, in the latter case using oxygen as the electron acceptor. μE, microeinsteins.
(ii) Microscopic observations.
The microbial mat comprised an association of filamentous cyanobacteria, mainly Oscillatoria-like (diameter, 2.5 μm), Microcoleus-like (diameter, up to 8 μm), and Pseudanabaena-like (diameter, about 3 μm) cyanobacteria, and had high densities of Chloroflexus-like filaments that did not exhibit red fluorescence. The last group had two different diameters (wider [>2 μm] and narrower [<1 μm]), with various lengths. The non-red-fluorescent filamentous bacteria were likely CLB, as they did not contain intracellular sulfur inclusions, typical of filamentous Beggiatoa strains. As CLB-specific NIR autofluorescence was not determined in this study, we cannot exclude the possibility that some of the filaments actually represented other bacteria, such as filamentous sulfate-reducing bacteria of the genus Desulfonema that were previously found to occur in hypersaline mats (10). The percentage of non-red-fluorescent filaments amounted to 20 to 30% of total filamentous bacteria in the photic zone.
(iii) Microbial mat photopigment analysis.
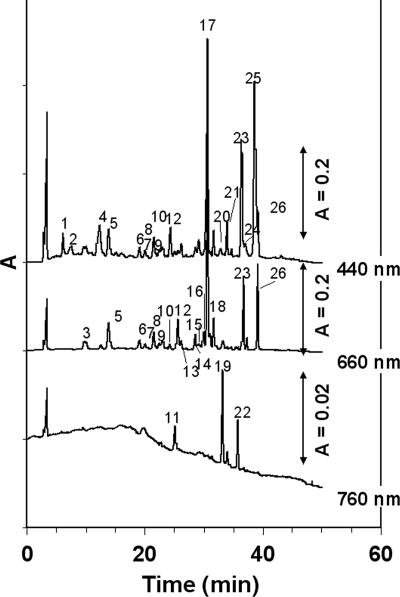
HPLC analysis of the pigments of the microbial mat studied revealed the presence of BChla, BChlc, and BChld, as well as γ-carotene, pigments that are known to be produced by photosynthetic members of the family Chloroflexaceae (12). Figure 2 shows the chromatograms at 440, 660, and 760 nm for a sample from the top layer of the mat, and the peaks are identified in Table 1. The pigment composition reflects a phototrophic community comprising cyanobacteria (chlorophyll a [Chla], zeaxanthin, and β-carotene) and CLB (BChlc/d, γ-carotene, and minor amounts of BChla). In addition, degradation products of these compounds were observed. The BChlc/d allomers eluted between 13 and 27 min (Fig. 2, peaks 5 to 10, 12, and 13). These retention times were longer than those observed for the farnesol-esterified BChlc homologs (BChlcF) found in a culture of Chlorobium tepidum. This indicates that the BChlc/d homologs were more hydrophobic than the BChlcF homologs and were thus esterified with another alcohol. C. aurantiacus contains different allomers of BChlc esterified with stearyl, phytyl, and geranylgeranyl alcohols. Further identification of the BChlc/d homologs requires HPLC-mass spectrometry. In addition, BChla (peak 11) was found in the mats, and we also observed two late-eluting BChld-like compounds. Different bacteriochlorophyll degradation products were identified as bacteriopheophorbide c and bacteriopheophytins a, c, and d. A number of peaks were quantified (Table 2). The ratio of BChlc to BChld to BChla was 72:26:1 on average.
FIG. 2.
HPLC chromatograms at 440, 660, and 760 nm for a sample from the top 5 mm of the Lake Chiprana microbial mat. Identified pigment molecules, indicated by numbers, are described in Table 1. The main pigments are Chla (peak 17) and BChlc allomers (peaks 5, 6, 8, 12 to 14, and 16).
TABLE 1.
Pigment identification, retention times, and λmax for the different peaks shown in the chromatograms in Fig. 2
| Peak | Retention time (min) | Compound | λmax |
|---|---|---|---|
| 1 | 6.13 | Unknown carotenoid | 448 |
| 2 | 7.50 | Bacteriopheaophorbide c, methyl ester | 438, 505, 662 |
| 3 | 9.66 | Phaeophorbide a, methyl ester | 411, 661 |
| 4 | 12.29 | Zeaxanthin | 451, 475 |
| 5 | 13.90 | BChlc | 434, 663. |
| 6 | 19.04 | BChlc | 430, 662 |
| 7 | 20.06 | BChld | 427, 652 |
| 8 | 21.49 | BChlc | 431, 668 |
| 9 | 22.40 | BChld | 428, 653 |
| 10 | 24.30 | BChld | 428, 653 |
| 11 | 25.00 | BChla | 373, 761 |
| 12 | 25.50 | BChlc | 434, 663 |
| 13 | 26.10 | BChlc | 435, 660 |
| 14 | 28.50 | Bacteriopheaophytine c | 412, 661 |
| 15 | 29.13 | Bacteriopheaophytine d | 409, 654 |
| 16 | 29.90 | Bacteriopheaophytine c | 411, 661 |
| 17 | 30.50 | Chla | 431, 660 |
| 18 | 31.70 | Allomer of Chla | 431, 659 |
| 19 | 33.20 | Bacteriopheaophytine a | 358, 525, 740 |
| 20 | 33.98 | BChld-like | 435, 652 |
| 21 | 34.75 | BChld-like | 433, 652 |
| 22 | 35.80 | Bacteriopheaophytine a | 359, 531, 666, 740 |
| 23 | 36.68 | Phaeophytin a | 409, 661 |
| 24 | 37.10 | γ-Carotene | 434, 460 |
| 25 | 38.70 | β-Carotene | 454, 480 |
| 26 | 39.10 | Pyrophaeophytin a | 409, 661 |
TABLE 2.
Quantification of pigments of the Lake Chiprana microbial mat in September 2004a
| Pigment(s) | Concn (μg cm−2)
|
|
|---|---|---|
| Mean | SD | |
| Total BChlc | 10.7 | 2.6 |
| Total BChld | 3.9 | 1.8 |
| Chla | 11.2 | 2.8 |
| Total phaeophytin a + pyrophaeophytin a | 9.5 | 0.8 |
| BChla | 0.15 | 0.06 |
| Zeaxanthin | 0.72 | 0.20 |
| γ-Carotene | 0.25 | 0.14 |
| β-Carotene | 1.14 | 0.26 |
The ratio of BChla, BChlc, and BChld indicates that CLB are abundant in this mat.
(iv) CLB phylotype diversity.
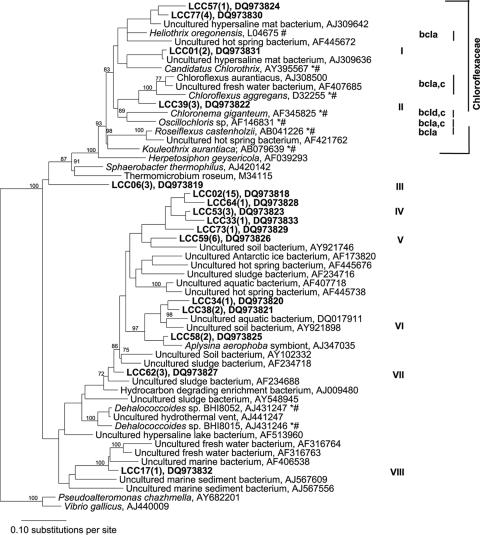
A total of 49 sequences were recovered by 16S rRNA PCR using Chloroflexi-specific primers designed by Gich et al. (11). This primer set targeted the 16S rRNA gene from various members of the phylum Chloroflexi, which includes isolated and characterized members of the anoxygenic phototrophic filamentous genera Chloroflexus, Chloronema, Oscillochloris, Heliothrix, and Roseiflexus. A total of eight major clusters (Fig. 3) of sequences were identified. There was significant diversity among the sequences that were recovered. Thus, we distinguished 16 different phylotypes, each comprising sequences that shared more than 96% sequence similarity. A representative sequence was selected from each phylotype that was considered in the phylogenetic analysis. Accordingly, most sequences were most closely related to the sequences from uncultured bacteria retrieved from various microbial mats, soils, sediments, and sludges. In general, the sequences recovered in this study were about 75 to 86% identical to sequences recovered in other studies. Ten of 49 sequences from this study clustered with sequences derived from members of the family Chloroflexaceae (clusters I to II); the remaining 39 sequences clustered with sequences from various other uncultured groups of bacteria distantly related to the family Chloroflexaceae (clusters III to VIII) but still within the phylum Chloroflexi. Some of these sequences appear to be distantly related to Dehalococcoides (phylum Chloroflexi, class Dehalococcoides). Seven sequences (cluster I) represented by LCC01 (two sequences), LCC77 (four sequences), and LCC57 were 90 to 99% identical to sequences recovered from a hypersaline microbial mat in Guerrero Negro, Mexico (23). Three sequences represented by LCC39 (cluster II) shared 88% identity with C. aurantiacus. They were the sequences most closely related to any 16S rRNA gene from a cultured organism.
FIG. 3.
16S rRNA gene maximum parsimony tree for representative sequences obtained in this study (bold type) and sequences retrieved from the database. Sequences from this study in the tree represent groups of sequences that shared more than 96% identity; the numbers in parentheses indicate the numbers of sequences in the groups. Asterisks indicate known phototrophic species, and number signs indicate previously isolated and described species. The bootstrap values at the nodes are percentages based on 1,000 replications. Sequences from this study, as well as LO4675, AJ09636, and AJ309642, were excluded from the bootstrap analysis. Group I represents sequences recovered in this study, which formed a separate clade that included H. oregonesis (a chlorosome-less, BChla- but not BChlc- or BChld-producing species) and “Candidatus Chlorothrix” (chlorosome-containing, BChla- and BChlc-producing species); group II represents sequences obtained in this study that formed a clade with Chloroflexus, Chloronema, and Oscillochloris (chlorosome-containing, BChla- plus BChlc- or BChld-producing species). Groups III to VIII represent sequences from this study which cluster with sequences distantly related to the family Chloroflexaceae but are still in the phylum Chloroflexi.
Functional analysis of the CLB impact on microbial mat oxygen respiration. (i) Aerobic respiration of C. aurantiacus.
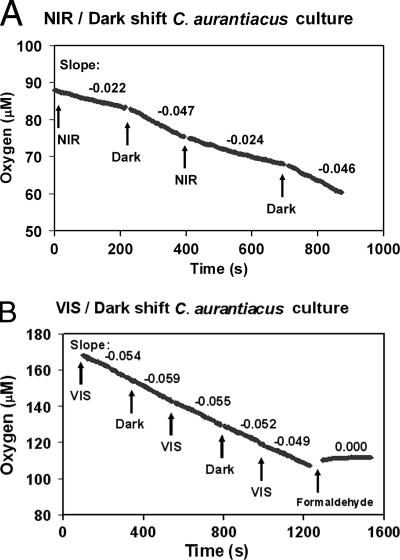
C. aurantiacus pregrown under anoxic conditions in the presence of light was subjected to aerobic conditions by aerating culture aliquots just before aerobic respiration rates were measured. Examples of oxygen consumption measurements obtained during incandescent light bulb-NIR LED/dark or VIS LED/dark shifts are presented in Fig. 4. Calculated oxygen consumption rates under different illumination conditions are presented in Table 3. The oxygen consumption rate with NIR light illumination was significantly lower (ca. 50%) than that in the dark or with only VIS light illumination. No significant difference in the oxygen consumption rates between dark conditions and illumination with only VIS light was observed. The level of respiration in the dark calculated in the first experimental series was somewhat lower than that calculated in the second series (Table 3). This may have been due to the lower initial oxygen concentration in the first series. The formaldehyde-killed culture showed no measurable oxygen uptake over time. The NIR light applied to the C. aurantiacus culture was the maximum intensity that could be obtained with the NIR LED configuration used in this study. It may well be that this intensity was not high enough to saturate anoxygenic photosynthesis of the C. aurantiacus culture.
FIG. 4.
Oxygen consumption in aerated axenic cultures of C. aurantiacus under different illumination conditions. (A) Alternating illumination with NIR light (25-W incandescent light bulb plus two 40-mA NIR LEDs [710 to 770 nm]) and darkness. (B) Alternating illumination with VIS light (two VIS LEDs [400 to 700 nm]) and darkness, as well as the formaldehyde-killed control. See Table 3 for calculated specific respiration rates.
TABLE 3.
C. aurantiacus aerobic respiration rates in aerated axenic culture aliquots (n = 4) under different illumination conditions
| Expt series | Light conditions | Respiration rate (μmol O2 g protein−1 min−1)a |
|---|---|---|
| 1 | Bulb + NIR LEDs (15)b | 2.48 ± 0.17 |
| Dark | 4.47 ± 0.59 | |
| 2 | VIS LEDs (60)b | 5.44 ± 0.29 |
| Dark | 5.75 ± 0.50 |
The respiration rates under dark conditions and with VIS light illumination are not significantly different. However, the respiration rates with NIR light illumination (25-W incandescent light bulb plus two 40-mA NIR LEDs) and dark conditions are significantly different.
The values in parentheses are light intensities at 400 to 700 nm expressed in μmol photons m−2 s−1.
(ii) Effect of NIR light illumination on microbial mat community respiration.
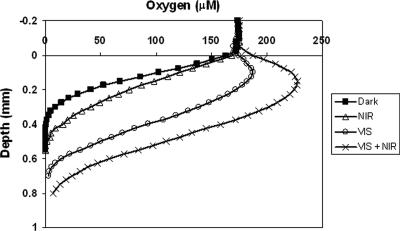
The effect of NIR light illumination on microbial mat community oxygen consumption was inferred from the steady-state oxygen profiles acquired under the different light conditions. Upon illumination with only NIR light, oxygen penetration significantly increased compared to that under dark conditions, from 0.40 to 0.55 mm (Fig. 5). A comparable phenomenon was observed when VIS light and NIR light were combined; the oxygen penetration depths were 0.7 and 0.9 mm when VIS light was used alone and when VIS light and NIR light were combined, respectively. With VIS light illumination alone (60 μmol photons m−2 s−1) the oxygen concentration reached a maximum of 190 μM at a depth of 0.1 mm. When NIR light was subsequently added to the white light, the maximum oxygen concentration increased to 230 μM and oxygen penetrated to a depth of 0.175 mm. The calculated microbial mat areal net oxygen production or consumption values determined from the oxygen profiles measured under the different light conditions are presented in Table 4. Controls containing mats illuminated with only NIR light in which oxygen profiles were measured before and after DCMU addition showed no difference in oxygen concentration and penetration depth. These observations indicate that NIR illumination did not result in oxygenic photosynthesis in this specific hypersaline microbial mat. Otherwise, in DCMU-inhibited mats the oxygen concentration would have been decreased due to continued respiration and ceased oxygenic photosynthesis. The unchanged oxygen concentration profile after DCMU addition also shows that the addition of ethanol (part of the DCMU stock solution) to the mat did not result in increased aerobic respiration rates in the short term. The latter conclusion also supports the finding of Ludwig et al. (20) that aerobic respiration in Lake Chiprana microbial mats is not limited by organic carbon compounds.
FIG. 5.
Steady-state oxygen concentration profiles in the Lake Chiprana microbial mat under four different light conditions: (i) darkness, (ii) NIR light (two 40-mA NIR LEDs [710 to 770 nm]), (iii) VIS light (two VIS LEDs [400 to 700 nm]; 60 μmol photons m−2 s−1), and (iv) two NIR LEDs plus two VIS LEDs.
TABLE 4.
Net areal oxygen production rates calculated from oxygen concentration profiles in Lake Chiprana microbial mats under different illumination conditionsa
| Light conditions | Net areal O2 production (nmol O2 cm−2 s−1) |
|---|---|
| Dark | −0.13 |
| NIR | −0.10 |
| VIS | 0.03 |
| VIS + NIR | 0.07 |
See Fig. 4. Negative values indicate net consumption.
DISCUSSION
The investigated microbial mat from hypersaline Lake Chiprana in Spain appeared to be rich in CLB in the photic zone, as demonstrated by microscopy, pigment analysis, and the diversity of CLB-related partial 16S rRNA gene sequences. The fact that CLB occur not only in hot spring microbial mats but also in hypersaline mats was also determined in previous studies (14, 23, 19, 32); however, the in situ metabolic function of CLB in carbon cycling remains to be clarified. In this study we developed a method that allows us to evaluate the proportion of the oxygen and carbon metabolism that is directly attributed to the metabolic activity of CLB in the mat. For this we used the finding that CLB can switch their metabolism from anoxygenic photosynthesis with NIR illumination to aerobic respiration in the absence of NIR illumination. We corroborated this finding for a pure culture of C. aurantiacus. As the electromagnetic spectrum of sunlight comprises both VIS and NIR light, we decided to use artificial illumination with specific LEDs and not filters, which were used originally (9). Filters may decrease the light intensity or otherwise affect the nontarget light field and thus may influence, e.g., the VIS light intensity and simultaneously the oxygenic photosynthesis rates, which would influence the oxygen concentration in mats and thus hamper the quantification of CLB respiration rates. In this study illumination with NIR light with a spectrum of 740 ± 30 nm in addition to darkness or illumination with only VIS light (with a spectrum of 400 to 700 nm) led to a substantial increase in the net areal mat oxygen production rate. As 740 nm is the specific absorption wavelength of BChlcs, which is known to be produced by C. aurantiacus, and to a lesser extent of BChlc and BChld, which are also produced by CLB species, we concluded that the observed effect on mat community oxygen dynamics was due to the metabolic activity of CLB species. Like CLB, in the presence of oxygen PSB (7) and purple nonsulfur bacteria may immediately switch their metabolism from anoxygenic photosynthesis to aerobic respiration when NIR illumination is suddenly stopped. However, these bacteria have a very low absorption cross section at 740 nm and, therefore, virtually do not use the light of the LEDs used in this study. Moreover, microscopic observations revealed only very small quantities of typical PSB morphotypes, and, in general, the specific respiration rates of Chromatiaceae (PSB) (7, 26) are 1 to 2 orders of magnitude lower than those that we found for C. aurantiacus. Green sulfur bacteria that also contain BChla, BChlc, and BChld have been found in some microbial mats. However, these bacteria are strict anaerobes that are confined to the permanent anoxic layers of the mats and cannot contribute to oxygen respiration. Recently, a cyanobacterium that produces Chld and absorbs light in the NIR light spectrum was isolated (18). Whether such presumably oxygen-producing organisms comprise a substantial part of the microbial community in the mat remains to be investigated, although no Chld was detected in this study. However, we concluded that if present, such organisms could not be responsible for the observed increase in oxygen concentration upon additional NIR light illumination as control experiments with DCMU, a specific inhibitor of photosystem II and thus oxygen production, did not result in a decrease in the oxygen concentration when mats illuminated with only NIR light were used.
The role that CLB play in the local carbon cycle can be inferred from their respiration rate under non-NIR light conditions. Axenic culture experiments with C. aurantiacus showed that the oxygen respiration decreased by about 50% upon illumination with NIR light. CLB can switch from anoxygenic phototrophy, which provides energy due to cyclic electron transport in photosystem I, to aerobic respiration to sustain energy generation under dark conditions (13). It can thus be assumed that the amount of oxygen respired under non-NIR light conditions at least equals the equivalent energy that was produced by anoxygenic photosynthesis by active CLB illuminated with NIR light. Therefore, one way to estimate the role of CLB in the local carbon cycle is to compare their non-NIR light respiration to the total community respiration. In the dark, the total community respiration, as inferred from microbial mat areal oxygen uptake, decreased from 0.13 to 0.10 nmol O2 cm−2 s−1, a decrease of 25%, upon illumination with NIR light. When NIR light was supplied along with VIS light, the net community areal oxygen production increased from 0.03 to 0.07 nmol O2 cm−2 s−1; thus, the apparent net primary production more than doubled, likely as a result of a decrease in CLB respiration due to a switch to anoxygenic phototrophy for energy generation.
The measurable change in oxygen dynamics upon illumination with NIR light (740 nm) of intact mats can thus be attributed to CLB activity. However, as this metabolic shift is directly linked to the mode of energy generation, no conclusions can be drawn as to whether a shift in the electron donor for autotrophic growth (e.g., reduced sulfur compounds) or a shift in the carbon source (organic or inorganic) for hetero- or autotrophic growth also occurs. CLB are known to be able to use various inorganic compounds as electron donors during autotrophic growth, as well as inorganic and various organic compounds as carbon sources during phototrophic or heterotrophic growth (13, 34). The fact that CLB can be found at high densities in microbial mats is likely due to this versatile metabolism. In mats light intensity and oxygen, reduced sulfur compound, and dissolved inorganic and organic carbon concentrations change rapidly during a full 24-h diel cycle, conditions to which CLB are maximally adapted. In such systems these bacteria would have a competitive advantage over more specialized but metabolically more restricted organisms.
The capacity to switch from anoxygenic photosynthesis to respiration has thus been clearly demonstrated for C. aurantiacus in culture experiments and has been inferred for the CLB in a microbial mat from oxygen profiles. This ability, which is apparently widespread among CLB, has important consequences for the quantification of gross photosynthesis in mats using the traditional light/dark shift method, which was introduced by Revsbech et al. in 1981 (30) and has been used in numerous studies since. This method is based on the assumption (among others) that in a microbial mat under steady-state (stable oxygen profile) conditions, the community oxygen consumption rate is unchanged initially (for at least a few seconds) when the mat is switched from light to darkness. The rate of oxygen disappearance at the start of the dark period then equals the rate of oxygen production in the light (30). This assumption, however, does not hold when C. aurantiacus or other CLB make up a significant part of the microbial community, as these organisms may switch within seconds from anoxygenic photosynthesis in the light to aerobic respiration in the dark. The fact that this effect can be substantial was shown in this study, where the apparent net photosynthesis more than doubled when NIR light was supplied along with VIS light. Hence, using the traditional light/dark shift method, gross photosynthesis rates are substantially overestimated in CLB-rich microbial mats. However, this effect can be compensated for, if only VIS light (400 to 700 nm) is used as a light source (CLB behave as aerobic bacteria [i.e., continuously respire]) or, alternatively, an additional NIR light source remains on continuously (CLB continue anoxygenic phototrophy and do not switch to aerobic respiration when VIS light is switched off) during measurements.
The high diversity of 16S rRNA gene phylotypes affiliated with the phylum Chloroflexi indicates that besides a number of sequences that group in the family Chloroflexaceae, nonfilamentous and even nonphototrophic Chloroflexi may be present in the mat examined. It is difficult to infer the type of chlorophyll that a bacterium possesses based on our 16S rRNA data given the paucity of 16S rRNA gene sequences available for cultured members of the Chloroflexaceae. However, the three sequences represented by LCC39 (cluster II) group with sequences from two characterized Chloroflexus species and are therefore likely to represent BChlc-producing CLB active in this system. The six sequences in cluster I that were recovered in this study are most closely related to the 16S rRNA gene from Heliothrix oregonensis, a CLB that does not, like Chloroflexus species, produce BChlc or BChld in addition to BChla. The organisms responsible for these sequences may thus not have contributed to the observed changes in community oxygen respiration when illumination with 740-nm NIR light was manipulated. The three sequences represented by LCC39 from the mat are the sequences that are most closely related to the 16S rRNA gene from C. aurantiacus which originated from a hot spring microbial mat and which so far is the most intensely studied and characterized species in the family Chloroflexaceae. However, they share sequence identity of only 88%. The highest sequence identity obtained for the hypersaline mat studied here (cluster I) was the sequence identity with sequences that originated from a mat in Guerrero Negro, Mexico (accession no. AJ309642 and AJ309636) (23), which was also a hypersaline microbial mat but was on a different continent. The sequence similarity between these sequences was up to 99%, which indicates that at least some hypersaline Chloroflexaceae may have a cosmopolitan distribution.
This study focused on CLB, which are filamentous phototrophic bacteria that are members of the family Chloroflexaceae. Although the clone library in this study retrieved 10 of 49 sequences that grouped in the family Chloroflexaceae, it is not known whether all these sequences represent filamentous phototrophic bacteria, as only a few sequences from this group represent well-characterized strains. Furthermore, the clone library shows that sequences retrieved in this study that fall outside the family Chloroflexaceae but still cluster within the phylum Chloroflexi are all most closely related to sequences from uncultured organisms. The apparently high but uncharacterized diversity of Chloroflexi-related sequences retrieved from the specific hypersaline mat studied seems to be typical, as other studies of various hot spring mats (24, 29) and hypersaline mats (19, 23), as well as other ecosystems (3, 11), reported the same diversity. An intriguing question is what kind of species these sequences belong to and what ecological role they play in these ecosystems. Future cultivation and ecophysiological characterization studies must resolve this question.
Acknowledgments
We thank Alfredo Legaz (COMENA, Caspe) and the local authorities in Chiprana for granting permission to access Lake Chiprana and take microbial mat samples. We are grateful to Dirk de Beer and Lubos Polerecky for discussions and technical help.
A. Bachar was supported by grant DFG JO-412 from the German Research Foundation. R. de Wit acknowledges support from the Deutscher Akademischer Austausch Dienst (DAAD) and the Agence National de la Recherche (program CYANOCARBO).
Footnotes
Published ahead of print on 20 April 2007.
REFERENCES
- 1.Abed, R. M. M., and F. Garcia-Pichel. 2001. Long-term compositional changes after transplant in a microbial mat cyanobacterial community revealed using a polyphasic approach. Environ. Microbiol. 3:53-62. [DOI] [PubMed] [Google Scholar]
- 2.Bateson, M. M., and D. M. Ward. 1988. Photoexcretion and fate of glycolate in a hot-spring cyanobacterial mat. Appl. Environ. Microbiol. 54:1738-1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bjornsson, L., P. Hugenholtz, G. W. Tyson, and L. L. Blackall. 2002. Filamentous Chloroflexi (green non-sulfur bacteria) are abundant in wastewater treatment processes with biological nutrient removal. Microbiology 149:2309-2318. [DOI] [PubMed] [Google Scholar]
- 4.Boomer, S. M., D. P. Lodge, B. E. Dutton, and B. Pierson. 2002. Molecular characterization of novel red green nonsulfur bacteria from five distinct hot spring communities in Yellowstone National Park. Appl. Environ. Microbiol. 68:346-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buffan-Dubau, E., O. Pringault, and R. DeWit. 2001. Artificial cold-adapted microbial mats cultured from Antarctic lake samples. 1. Formation and structure. Aquat. Microb. Ecol. 26:115-125. [Google Scholar]
- 6.D'Amelio, E. D., Y. Cohen, and D. J. DesMarais. 1987. Association of a new type of gliding, filamentous, purple phototrophic bacterium inside bundles of Microcoleus chthonoplastes in hypersaline microbial mats. Arch. Microbiol. 147:213-220. [DOI] [PubMed] [Google Scholar]
- 7.DeWit, R., and H. VanGemerden. 1987. Chemolithotrophic growth of the phototrophic sulfur bacterium Thiocapsa roseopersicina, FEMS Microbiol. Ecol. 45:117-126. [Google Scholar]
- 8.DeWit, R., and H. VanGemerden. 1990. Growth and metabolism of the purple sulfur bacterium Thiocapsa roseopersicina under combined light dark and oxic anoxic regimens. Arch. Microbiol. 154:459-464. [Google Scholar]
- 9.DeWit, R., L. Falcón, and C. Charpy-Roubaud. 2005. Heterotrophic dinitrogen fixation (acetylene reduction) in phosphate-fertilised Microcoleus chthonoplastes microbial mat from the hypersaline inland lake “la Salada de Chiprana” (NE Spain). Hydrobiology 534:245-253. [Google Scholar]
- 10.Fukui, M., A. Teske, B. Assmus, G. Muyzer, and F. Widdel. 1999. Physiology, phylogenetic relationships, and ecology of filamentous sulfate-reducing bacteria (genus Desulfonema). Arch. Microbiol. 172:193-203. [DOI] [PubMed] [Google Scholar]
- 11.Gich, F., J. Garcia-Gil, and J. Overmann. 2001. Previously unknown and phylogenetically diverse members of the green nonsulfur bacteria are indigenous to freshwater lakes. Arch. Microbiol. 177:1-10. [DOI] [PubMed] [Google Scholar]
- 12.Gich, F., R. L. Airs, M. Danielsen, B. J. Keely, C. A. Abella, J. Garcia-Gil, M. Miller, and C. M. Borrego. 2003. Characterization of the chlorosome antenna of the filamentous anoxygenic phototrophic bacterium Chloronema sp. strain UdG9001. Arch. Microbiol. 180:417-426. [DOI] [PubMed] [Google Scholar]
- 13.Hanada, S., and B. K. Pierson. November 2002, posting date. The family Chloroflexaceae. In M. Dworkin et al. (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed., release 3.11. Springer-Verlag, New York, NY. http://link.springer-ny.com/link/service/books/10125. Accessed 28 August 2006.
- 14.Jonkers, H. M., R. Ludwig, R. DeWit, O. Pringault, G. Muyzer, H. Niemann, N. Finke, and D. DeBeer. 2003. Structural and functional analysis of a microbial mat ecosystem from a unique permanent hypersaline inland lake: ‘La Salada de Chiprana’ (NE Spain). FEMS Microbiol. Ecol. 44:175-189. [DOI] [PubMed] [Google Scholar]
- 15.Jørgensen, B. B., and D. C. Nelson. 1988. Bacterial zonation, photosynthesis, and spectral light-distribution in hot-spring microbial mats of Iceland. Microb. Ecol. 16:133-147. [DOI] [PubMed] [Google Scholar]
- 16.Klappenbach, J. A., and B. K. Pierson. 2004. Phylogenetic and physiological characterization of a filamentous anoxygenic photoautotrophic bacterium ‘Candidatus Chlorothrix halophila’ gen. nov., sp. nov. recovered from hypersaline microbial mats. Arch. Microbiol. 181:17-25. [DOI] [PubMed] [Google Scholar]
- 17.Kühl, M., C. Steuckart, G. Eickert, and P. Jeroschewski. 1998. A H2S microsensor for profiling biofilms and sediments: application in an acidic lake sediment. Aquat. Microb. Ecol. 15:201-209. [Google Scholar]
- 18.Kühl, M., M. Chen, P. J. Ralph, U. Schreiber, and A. W. D. Larkum. 2005. A niche for cyanobacteria containing chlorophyll d. Nature 433:820. [DOI] [PubMed] [Google Scholar]
- 19.Ley, R. E., J. K. Harris, J. Wilcox, J. R. Spear, S. R. Miller, B. M. Bebout, J. A. Maresca, D. A. Bryant, M. L. Sogin, and N. R. Pace. 2006. Unexpected diversity and complexity of the Guerrero Negro hypersaline microbial mat. Appl. Environ. Microbiol. 72:3685-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ludwig, R., O. Pringault, R. DeWit, D. DeBeer, and H. M. Jonkers. 2006. Limitation of oxygenic photosynthesis and oxygen consumption by phosphate and organic nitrogen in a hypersaline microbial mat: a microsensor study. FEMS Microbiol. Ecol. 57:9-17. [DOI] [PubMed] [Google Scholar]
- 21.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madigan, M. T., J. M. Martinko, and J. Parker. 2000. Brock—biology of microorganisms, 9th ed. Prentice-Hall, Inc., Englewood Cliffs, NJ.
- 23.Nübel, U., M. M. Bateson, M. T. Madigan, M. Kühl, and D. M. Ward. 2001. Diversity and distribution in hypersaline microbial mats of bacteria related to Chloroflexus spp. Appl. Environ. Microbiol. 67:4365-4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nübel, U., M. M. Bateson, V. VanDieken, A. Wieland, M. Kühl, and D. M. Ward. 2002. Microscopic examination of distribution and phenotypic properties of phylogenetically diverse Chloroflexaceae-related bacteria in hot spring microbial mats. Appl. Environ. Microbiol. 68:4593-4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oelze, J. 1992. Light and oxygen regulation of the synthesis of bacteriochlorophylls a and c in Chloroflexus aurantiacus. J. Bacteriol. 174:5021-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Overmann, J., and N. Pfennig. 1992. Continuous growth and respiration of Chromatiaceae species at low oxygen concentrations. Arch. Microbiol. 158:59-67. [Google Scholar]
- 27.Pierson, B. K., and H. M. Howard. 1972. Detection of bacteriochlorophyll containing microorganisms by infrared fluorescence photomicrography. J. Gen. Microbiol. 73:359-363. [Google Scholar]
- 28.Pierson, B. K., and R. W. Castenholz. 1974. Studies of pigments and growth in Chloroflexus aurantiacus, a phototrophic filamentous bacterium. Arch. Microbiol. 100:283-305. [DOI] [PubMed] [Google Scholar]
- 29.Pierson, B. K., D. Valdez, M. Larsen, E. Morgan, and E. E. Mack. 1994. Chloroflexus-like organisms from marine and hypersaline environments—distribution and diversity. Photosynth. Res. 41:35-52. [DOI] [PubMed] [Google Scholar]
- 30.Revsbech, N. P., B. B. Jørgensen, and O. Brix. 1981. Primary production of microalgae in sediments measured by oxygen microprofile, H-CO-14(3)-fixation, and oxygen-exchange methods. Limnol. Oceanogr. 26:717-730. [Google Scholar]
- 31.Schaub, B. E. M., and H. VanGemerden. 1994. Simultaneous phototrophic and chemotropic growth in the purple sulfur bacterium Thiocapsa roseopersicina M1. FEMS Microbiol. Ecol. 13:185-195. [Google Scholar]
- 32.Sorensen, K. B., D. E. Canfield, A. P. Teske, and A. Oren. 2005. Community composition of a hypersaline endoevaporitic microbial mat. Appl. Environ. Microbiol. 71:7352-7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van der Meer, M. T. J., S. Schouten, J. S. S. Damste, J. W. DeLeeuw, and D. M. Ward. 2003. Compound-specific isotopic fractionation patterns suggest different carbon metabolisms among Chloroflexus-like bacteria in hot-spring microbial mats. Appl. Environ. Microbiol. 69:6000-6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Meer, M. T. J., S. Schouten, M. M. Bateson, U. Nübel, A. Wieland, M. Kühl, J. W. De Leeuw, J. S. S. Damste, and D. M. Ward. 2005. Diel variations in carbon metabolism by green nonsulfur-like bacteria in alkaline siliceous hot spring microbial mats from Yellowstone National Park. Appl. Environ. Microbiol. 71:3978-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wieland, A., and M. Kühl. 2000. Short-term temperature effects on oxygen and sulfide cycling in a hypersaline cyanobacterial mat (Solar Lake, Egypt). Mar. Ecol. Prog. Ser. 196:87-102. [Google Scholar]