Abstract
Vineyards of southern France and northern Italy are affected by the flavescence dorée (FD) phytoplasma, a quarantine pathogen transmitted by the leafhopper of Nearctic origin Scaphoideus titanus. To better trace propagation of FD strains and identify possible passage between the vineyard and wild plant compartments, molecular typing of phytoplasma strains was applied. The sequences of the two genetic loci map and uvrB-degV, along with the sequence of the secY gene, were determined among a collection of FD and FD-related phytoplasmas infecting grapevine, alder, elm, blackberry, and Spanish broom in Europe. Sequence comparisons and phylogenetic analyses consistently indicated the existence of three FD phytoplasma strain clusters. Strain cluster FD1 (comprising isolate FD70) displayed low variability and represented 17% of the disease cases in the French vineyard, with a higher incidence of the cases in southwestern France. Strain cluster FD2 (comprising isolates FD92 and FD-D) displayed no variability and was detected both in France (83% of the cases) and in Italy, whereas the more-variable strain cluster FD3 (comprising isolate FD-C) was detected only in Italy. The clonal property of FD2 and its wide distribution are consistent with diffusion through propagation of infected-plant material. German Palatinate grapevine yellows phytoplasmas (PGY) appeared variable and were often related to some of the alder phytoplasmas (AldY) detected in Italy and France. Finally, phylogenetic analyses concluded that FD, PGY, and AldY were members of the same phylogenetic subclade, which may have originated in Europe.
Phytoplasmas are phloem-restricted wall-less bacteria pathogenic to many plant species worldwide (37, 52). Phytoplasmas can be spread both by hemipteran insect vectors (63) and by vegetative multiplication of infected-plant material. Controlling phytoplasma-induced diseases in perennial crops depends on field surveys and implementation of prophylactic sanitary measures requiring sensitive and specific detection of phytoplasmas in plants. Genetically different phytoplasmas can infect the same plant species; therefore, precise identification and typing of phytoplasma strains are necessary to ascertain the causes and origin of new outbreaks and predict the route of disease spread.
Vineyards in southern France, northern Italy, and Spain are affected by the flavescence dorée (FD) phytoplasma, a quarantine pathogen of grapevine (7, 8, 16, 24). The classification of phytoplasmas, which are uncultivable and currently described under the provisional genus “Candidatus Phytoplasma,” is mainly based on 16S rRNA gene phylogeny, genomic diversity, and plant and insect host ranges (32, 36, 59). The FD phytoplasma belongs to the 16SrV taxonomic group (36). Members of this group share high 16S rRNA gene sequence similarity (34, 38), but the group consists of phytoplasmas with an important variety of specific biological niches restricted to woody perennial hosts. “Ca. Phytoplasma ulmi” is responsible for yellows of elm species in North America and Europe (38) and “Ca. Phytoplasma ziziphi” is the agent of jujube witches'-broom and cherry lethal yellows in Asia (34, 38). In Europe, other phytoplasmas of group 16SrV are mainly infecting grapevine (23, 43), alder (46, 51), blackberry (26, 50), Spartium, and eucalyptus (44, 45). Most of the insect vectors naturally disseminating group 16SrV phytoplasmas have been identified. The elm yellows phytoplasmas are transmitted in North America by Scaphoideus luteolus (Van Duzee) (5) and in Europe by Macropsis mendax (Fieber) (15), whereas FD phytoplasmas are specifically transmitted by Scaphoideus titanus (Ball) (53, 58) and rubus stunt phytoplasma by Macropsis fuscula (Zetterstedt) (26). Phytoplasmas associated with Palatinate grapevine yellows (PGY) and alder yellows (AldY) are both transmitted by the alder leafhopper Oncopsis alni (Schrank) (41, 42) and were classified as members of the group 16SrV on the basis of their high 16S rRNA gene and secY sequence similarity to the corresponding genes of FD phytoplasmas (2, 3).
The genomic diversity in this phytoplasma group was recently examined. Sequence and restriction fragment length polymorphism (RFLP) analysis of the 16S rRNA genes and the 16S-23S intergenic spacer allowed differentiation of two different FD phytoplasma isolates (48). Variability analysis of two nonribosomal genetic loci, namely, secY (2, 3) and rpsC (47), showed that FD phytoplasma variants detected in France and Italy belonged to three different strain clusters and seemed closely related to phytoplasmas infecting European alder or grapevine in German Palatinate (2, 3).
To further document the genetic diversity of FD phytoplasmas in France and evaluate the genetic relationship with other 16SrV group phytoplasmas in Europe, the variability of two newly characterized FD phytoplasma genetic loci was determined. We present in the current paper the description of map and degV genes isolated by subtractive suppression hybridization (SSH) and characterized through genome walking. Diversity of the map and degV genetic loci and secY among a collection of 16SrV group phytoplasma isolates is described and discussed regarding alder and grapevine phytoplasmosis epidemiology. Comments on the consequences for phytoplasma taxonomy in the 16SrV group are also presented.
MATERIALS AND METHODS
Phytoplasma reference strains.
Phytoplasma reference isolates listed in Table 1 that had previously been transmitted to Catharanthus roseus periwinkle cv. Cooler were maintained in this host by grafting. Periwinkle plants were grown at 20 to 25°C with a photoperiod of 16 h. The FD92 isolate was continuously propagated on broad bean (Vicia faba cv. Agua dulce) through transmission by Euscelidius variegatus (Kirschbaum), reared on oat and broad bean at 20 to 25°C (19).
TABLE 1.
Phytoplasma reference strains propagated on herbaceous hosts
| Strain | Disease | Original host-exptl host | Geographic origin (yr) |
|---|---|---|---|
| EY1 | Elm yellows | Ulmus americana-Catharanthus roseus | New York State |
| ULW | Elm yellows | Ulmus carpinifolia-C roseus | France |
| FD70 | Flavescence dorée | Scaphoideus titanus captured on Vitis vinifera-C roseus | Landes, France (1970) |
| FD92 | Flavescence dorée | S titanus captured on V vinifera-Euscelidius variegatus and Vicia faba | Landes, France (1992) |
| RuS | Rubus stunt | Rubus fruticosus-C roseus | Southern Italy |
| ALY | Alder yellows | Alnus glutinosa-C roseus | Basilicata, Italy |
| EY17-49 | PGY,a A type | V vinifera-C roseus | Rheinland-Pfalz, Germany |
| EY38 | PGY, C type | V vinifera-C roseus | Rheinland-Pfalz, Germany |
| HD1 | Hemp dogbane | Apocynum cannabinum | New York State |
Palatinate grapevine yellows.
Phytoplasma isolates and nucleic acid extraction.
Grapevines exhibiting yellows were sampled from French, German, and Italian vineyards. Other phytoplasma isolates were collected from yellows-diseased elms and alders, stunting brambles, or proliferating broom-bushes in France and in Italy. Plant host and geographical origin of phytoplasma isolates are indicated in Table 2. Nucleic acids were extracted from 1 g of leaf midribs by the method of Maixner et al. (39) or Angelini et al. (2). The resulting nucleic acid pellets were resuspended in 60 μl of 10 mM Tris-HCl-1 mM EDTA, pH 7.8.
TABLE 2.
Phytoplasma isolates collected from grapevines, trees, and bushes
| Isolate(s) | Disease-host | Geographic origin |
|---|---|---|
| V00-SP5 and V00-SP9 | FD-Vitis vinifera | Gironde, France |
| V01-9 and V02-101 | FD-V vinifera | Gironde, France |
| V03-1-1, 9-4, 9-8, and 9-20 | FD-V vinifera | Gironde, France |
| V03-4-3, 4-4, 5-1, 5-2, and 5-3 | FD-V vinifera | Lot-et-Garonne, France |
| V03-2-2 and 9-1 | FD-V vinifera | Dordogne, France |
| V03-9-2, 9-16, and 9-17 | FD-V vinifera | Pyrénées Atlantiques, France |
| V03-9-21 | FD-V vinifera | Landes, France |
| V04-11-01 | Yellows-V vinifera | Haut-Rhin, France |
| V04-11-02 and 11-03 | FD-V vinifera | Saône-et-Loire, France |
| V04-11-04 and 11-05 | FD-V vinifera | Corrèze, France |
| V04-11-06 | FD-V vinifera | Vendée, France |
| V04-11-07, 11-08, 11-09, 11-10, and 11-11 | FD-V vinifera | Charente, France |
| V04-11-13, 11-14, and 11-15 | FD-V vinifera | Aveyron, France |
| V04-11-16, 11-17, and 11-18 | FD-V vinifera | Gers, France |
| V04-11-19, 11-21, and 11-53 | FD-V vinifera | Lot, France |
| V04-11-22, 11-23, and 11-24 | FD-V vinifera | Tarn, France |
| V04-11-25, 11-26, 11-27, and 11-29 | FD-V vinifera | Tarn-et-Garonne, France |
| V04-11-30 and 11-31 | FD-V vinifera | Aude, France |
| V04-11-35, 11-37, and 11-38 | FD-V vinifera | Hérault, France |
| V04-11-39, 11-40, 11-41, and 11-43 | FD-V vinifera | Vaucluse, France |
| V04-11-44, 11-45, and 11-46 | FD-V vinifera | Drôme, France |
| V04-11-49, 11-50, 11-51, and 11-52 | FD-V vinifera | Savoie, France |
| V04-11-54, 11-55, and 11-56 | FD-V vinifera | Charente, France |
| VI04-C28 and C29 | FD-V vinifera | Veneto, Italy |
| VI04-D004-03 | FD-V vinifera | Veneto, Italy |
| VI04-Lig1 and Lig2 | FD-V vinifera | Liguria, Italy |
| VI04-Toscana1 | FD-V vinifera | Toscana, Italy |
| VI04-248-04 and 188-04 | FD-V vinifera | Piemonte, Italy |
| PGY-B | Yellows (B type)-V vinifera | Rheinland-Pfalz, Germany |
| SI04-2160 and SI04-S4 | Witches'-broom-Spartium junceum | Campania, Italy |
| AI04-3-7 and AI04-3-13 | Yellows-Alnus glutinosa | Basilicata, Italy |
| WJ1444-32 | Yellows-A glutinosa | Pyrénées Orientales, France |
| AI04-2-4 | Yellows-A glutinosa | Friuli Venezia Giulia, Italy |
| RI04-2-6 | Stunting-Rubus fruticosus | Friuli Venezia Giulia, Italy |
| RI04-3-26 and RI04-2157 | Stunting-R fruticosus | Basilicata, Italy |
| WJ1295-78 | Stunting-Rosa sp. | Pyrénées Orientales, France |
| WJ1295-21 and WJ1295-31 | Yellows-Rosa canina | Pyrénées Orientales, France |
| WJ1274-81 and WJ1296-30 | Yellows-Ulmus carpinifolia | Pyrénées Orientales, France |
| EI04-2-2 | Yellows-Ulmus minor | Friuli Venezia Giulia, Italy |
| EI04-3-3 | Yellows-U minor | Campania, Italy |
| E04-D482 and D708 | Yellows-Ulmus glabra | Haute-Vienne, France |
| E04-D438 | Yellows-U minor | Loire Atlantique, France |
| E04-D714 | Yellows-U glabra | Haute-Vienne, France |
SSH library and genome walking.
The SSH protocol was performed according to the PCR-Select bacterial genome subtraction kit (Clontech) with some modifications reported by Cimerman et al. (21). Total DNA (4 μg) from healthy periwinkle and FD70-infected periwinkle was digested by RsaI endonuclease (MBI Fermentas) to constitute driver DNA and tester DNA, respectively. Tester DNA (100 ng) from infected periwinkle was ligated in two separate reactions of 10 μl: one with adaptor 1 and the other with adaptor 2R. Then, 1 μl of each ligation product was heat denatured and separately hybridized to an excess of driver (600 ng of RsaI-digested healthy periwinkle DNA) for 1.5 h at 63°C. The two hybridization mixtures were then mixed together in the presence of 300 ng of heat-denatured driver and hybridized overnight at 63°C. Hybrids carrying both adaptors 1 and 2R were amplified by nested PCR according to the manufacturer's instructions using Taq Advantage cDNA polymerase mix (BD Biosciences-Clontech). PCR amplification was performed in 25-μl reaction mixture volume with 0.4 μM of primer P1. The templates were first heated for 2 min at 72°C to fill the ends and then denatured for 25 s at 94°C. Thermal PCR conditions consisted of 25 cycles (10 s at 94°C, 30 s at 66°C, and 1 min 30 s at 72°C) with a single final extension of 7 min at 72°C. One microliter of a 1:40 dilution of the primary PCR mixture was submitted to a nested amplification of 18 thermal cycles with primers NP1 and NP2R using the same parameters as described above, except for the annealing temperature, which was 68°C. The PCR products were cloned into pGEMt-easy (Promega Corp). In order to select plasmids carrying DNA of the FD70 phytoplasma, plasmid inserts were labeled by PCR with incorporation of digoxigenin-11-dUTP (DIG Labeling Mix Plus; Roche). Probes were used to hybridize dot blots consisting of NaOH-denatured (0.4 M) healthy or infected-plant DNA (10 μg) spotted on Nytran Super Charge nylon N+ transfer membrane (0.45 μm) (Schleicher & Schuell). Dot blots were hybridized and washed according to standard procedures (56). Hybridized probes were revealed using anti-digoxigenin Fab fragments and CDP-Star as substrates according to the instructions of the manufacturer of the DIG DNA labeling and detection kit (Roche).
Four genome walking libraries were prepared according to the instructions with the Genomewalker kit (Clontech) by digesting total DNA (4 μg) from FD70-infected periwinkle by EcoRV, PvuII, HincII, and SwaI and ligating digested DNA to the genome walking adaptor. Genome walking nested PCR was carried out according to the manufacturer's instructions using primer pairs MFD9g1-AP1 (first PCR) and MFD9g1N-AP2 (second PCR) (Table 3) for the region bordering secY, and using primer pairs MFD32g1-AP1 (first PCR) and MFD32g1N-AP2 (second PCR) (Table 3) for the region bordering uvrB. PCR products were cloned into pGEMt-easy (Promega Corp.).
TABLE 3.
Primers used for genome walking and secY, secY-map, and uvrB-degV amplification and sequencing
| Genetic locus | Method | Primer | Sequence (5′-3′) |
|---|---|---|---|
| uvrB | Genome walking | MFD32g1 | TAGGAATTAAAGTAGCTTATCTTCATAGTG |
| MFD32gN1 | CTGGTGTTTATGATTGTTTAGTTGGA | ||
| secY | Genome walking | MFD9g1 | TTGTAAGATGACGATCAGAATTAGGA |
| MFD9g1N | GCAAAGATGTAGCGGAACATTTGTC | ||
| PCR | FD9r | TTTGCTTTCATATCTTGTRTCG | |
| FD9f2L | GTTTTAGCTAAAGGTGATTTAAC | ||
| Nested PCR | FD9r2L | TAAAAGACTAGTCCCRCCAAAAG | |
| FD9f3L | AATAAGGTAGTTTTATATGACAAG | ||
| Sequencing | FD9ri | CTATTTATAGCGTAATTAATAGG | |
| secY-map | PCR | FD9f5 | CAAAAAATTACTTTTGGCGGGAC |
| MAPr1 | TGCTCAAAATGAGCGCTTAAAC | ||
| Nested PCR | FD9f6 | GTCGCTTTAGAATCGACACA | |
| MAPr2 | TCGGAAGTAACAGCAGTCCA | ||
| uvrB-degV | PCR | UVRBf1 | GAAGGTCTAGATTTGCCTGAAGT |
| DEGVr4 | CTCCATTTTTGTTAACCTGT | ||
| Nested PCR | UVRBf3 | TTAATCCAAACTATCGGAAGA | |
| DEGVr3 | CCTTTTGTTTTGTTTAAACGTCC | ||
| Sequencing | DegVf1 | GCTTCGACATCAACTAGTTG |
PCR amplification and RFLP.
Amplification of secY by nested PCR was adapted from previous publications (2, 3) (Table 3). Primer sequences are given in Table 3, and primers are shown schematically in Fig. 1. PCR and nested PCR amplifications of secY, secY-map, and uvrB-degV genetic loci were performed in 25 μl for the first PCR and 50 μl for nested amplification with 1 μM of each PCR primer, 2 mM MgCl2, 5% dimethyl sulfoxide (DMSO), and 0.04 unit/μl of Taq polymerase (Promega). One microliter of plant nucleic acid extract diluted 10 times in water was used for the first PCR, and 0.5 μl of the first amplification was directly used as a template for nested amplification. For secY-map and uvrB-degV loci, PCR conditions were 1 min at 92°C and 35 cycles, with 1 cycle consisting of 30 s at 92°C, 30 s at 52°C, and 1 min 30 s at 66°C. For secY, the PCR conditions were 40 cycles, with 1 cycle consisting of 1 min at 92°C, 1 min at 55°C, and 1 min 30 s at 66°C, followed by a final extension of 5 min at 66°C. For routine typing of FD isolates, 10 μl of amplified product was double digested with 10 units of AluI and Eco72I restriction enzymes (MBI-Fermentas) according to the manufacturer's instructions. Digested products were analyzed on 8% polyacrylamide gels.
FIG. 1.
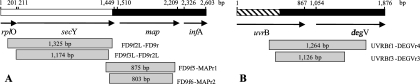
Genetic organization of secY-map (A) and uvrB-degV (B) genetic loci in the grapevine flavescence dorée phytoplasma FD70 and schematic representation of PCR products (gray boxes). White and hatched areas indicate previously characterized and SSH sequences, respectively. Black areas depict sequences determined through genome walking.
Sequencing and analysis.
Sequencing reactions were performed by Genome-Express (Grenoble, France) on MegaBACE capillary sequencing instruments. secY, secY-map, and uvrB-degV PCR products were sequenced using primers FD9r2L and FD9ri, primers FD9-F6 and MAP-R2, and primers UVRB-F3, and DEGV-F1, respectively. The raw sequence chromatograms were assembled and edited using Phred, Phrap, and Consed software (27, 28, 30). As most sequences were determined on a single strand, all sequences were edited by two experts in addition to base calling with Phred software. Only bases with Phred quality above 30 (error probability lower than 0.1%) were selected for analysis. This led to the exclusion of sequence extremities and finally retained sequences shorter than the PCR products. In detail and according to strains, sequence lengths were 674 to 676 bp for secY-map (positions 21 to 694 in the FD9f6-MAPr2 PCR fragment of the FD70 isolate), 1007 to 1031 bp for secY (positions 62 to 1074 in the FD9r2L-FD9f3L PCR fragment of the FD70 isolate), and 1017 to 1037 bp for uvrB-degV (positions 96 to 1112 in the UVRBf3-DEGVr3 PCR fragment of the FD70 isolate). Database searches were performed using BLAST programs (1) on the NCBI server (http://www.ncbi.nlm.nih.gov/BLAST/). Multiple-sequence alignments were performed using the CLUSTAL W program (61). Phylogenetic reconstructions using maximum parsimony were performed using MEGA2 (35) with randomized bootstrapping evaluation of branching validity.
Nucleotide sequence accession numbers.
The complete sequences of the secY-map and uvrB-degV loci of the phytoplasma isolate FD70 were deposited at EMBL under the accession numbers AM238512 and AM238511, respectively. For the other phytoplasma isolates, the accession numbers are AM384884 to AM384902 for secY-map, AM396411 to AM396433 for uvrB-degV, and AM397285 to AM397300 for secY sequences.
RESULTS
Characterization of map and degV genes of FD70 phytoplasma.
In order to isolate FD phytoplasma genes, SSH was carried out by subtracting RsaI-digested healthy periwinkle DNA from the RsaI-digested DNA of a periwinkle infected with the FD70 isolate. The resulting SSH product was cloned into pGEMt-easy. Out of 72 RsaI-SSH plasmidic inserts used as a hybridization probe, the insert FD32 was the only insert to hybridize the total DNA of FD70-infected periwinkle without hybridizing the total DNA of healthy periwinkle. The 544-bp sequence of the FD32 insert contained a partial open reading frame whose translation product shared 77% identity with the UvrB protein of “Ca. Phytoplasma asteris” (OY-M) (55). This FD70 partial gene sequence was further extended by chromosome walking, and a final sequence of 1,876 nucleotides was characterized. According to similarity with sequences in gene databases, this sequence corresponds to the second half of the uvrB gene encoding the subunit B of excinuclease ABC (positions 1 to 870) and the nearly complete gene encoding a protein of the DegV family (positions 1054 to 1876 [Fig. 1]). The same genome walking strategy was used to characterize a 2,603-bp-long genetic locus by determining the region downstream of the previously characterized FD9 phytoplasma DNA fragment (24). According to similarity with sequences in gene databases, this sequence contains the 3′ end of the rplO gene encoding the 50S ribosomal protein L15 (positions 1 to 204), the full-length secY gene which encodes the SecY protein of the protein secretion machinery (positions 211 to 1452), the entire map gene encoding the methionine aminopeptidase (positions 1511 to 2254), followed by the infA gene, which encodes the translation initiation factor IF-1 (Fig. 1).
Polyvalent amplification of map and uvrB-degV genetic loci of group 16SrV phytoplasmas.
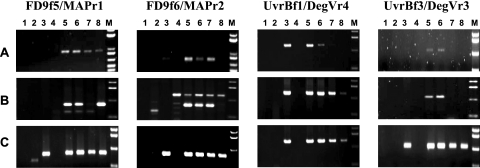
PCR detection of phytoplasmas in woody hosts necessitates nested amplification in most cases. In order to set up a nested amplification protocol for map and uvrB-degV, two primer pairs were designed for the FD70 DNA sequence (Fig. 1). Efficiency and polyvalence of primer pairs were evaluated for a panel of five 16SrV group phytoplasmas propagated on C. roseus or V. faba (Table 1). Surprisingly, an elongation temperature of 72°C, classically used for PCR amplification, did not lead to efficient amplification of both loci for all phytoplasmas tested (Fig. 2A). Changing annealing temperatures did not achieve higher yield of amplification (data not shown). Addition of 5% DMSO as a chemical enhancer for PCR (6) improved amplification with UvrBf1-DegVr4 primers but resulted in nonspecific amplification with primer pairs targeted to the map gene (Fig. 2B). However, combining DMSO and a lower elongation temperature of 66°C led to the efficient amplification of map and uvrB-degV for FD92 and FD70 FD isolates (Fig. 2C, lanes 3 and 5), the alder yellows isolate ALY (lane 6), the rubus stunt isolate RuS, and the elm yellows isolate EY1 (lanes 7 and 8). The improvement resulting from the use of a lower elongation temperature could partially be explained by the very low G+C content of secY-map and uvrB-degV genes (26% and 22%, respectively) especially in intergenic sequences where it falls to 6.5% and 9%, respectively. The DNA strands being synthesized certainly melt when the temperature is raised to 72°C, preventing exponential amplification of the targeted genes.
FIG. 2.
PCR amplifications of map and uvrB-degV genetic loci for phytoplasma reference strains of group 16SrV under various conditions. (A) Elongation temperature of 72°C without DMSO; (B) elongation temperature of 72°C with 5% DMSO; and (C) elongation temperature of 66°C with 5% DMSO. Lanes 1, H2O; lanes 2, healthy broad bean, lanes 3, FD92-infected broad bean; lanes 4, healthy periwinkle, lanes 5 to 8, periwinkle infected by FD70, ALY, RuS, and EY1 phytoplasmas, respectively; M, molecular size markers (1-kbp DNA ladder).
Nested amplification protocols for the map and uvrB-degV genes (genes drawn in Fig. 1) were applied to a panel of woody plants described in Table 3, and amplified products were submitted to DNA sequencing.
Genetic variability of FD phytoplasma isolates.
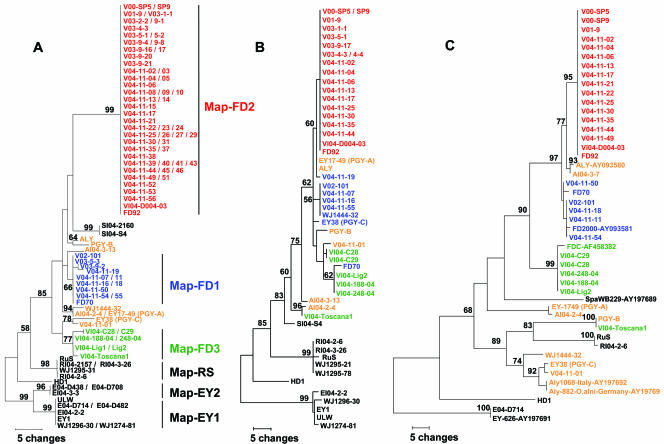
The diversity of FD phytoplasma isolates was assessed by determining the variability of map, uvrB-degV, and secY genetic loci. The sequences of the map gene (674 bp) were determined for 64 French and 8 Italian grapevine samples that have been diagnosed as FD phytoplasma infected by expert laboratories using one of the standard PCR detection techniques (22, 31, 49, 57). Multiple-sequence alignment and maximum parsimony analysis of map sequences indicated the clustering of FD isolates into three sequence clusters (Fig. 3A). Fifty-two grapevine samples from France (83%) and an Italian isolate VI04-D004-003 collected in Veneto (FD-D type according to Angelini et al. [2]) had the same map sequence, identical to the sequence of the reference strain FD92 isolated in southwestern France in 1992. This group of isolates will be referred as strain cluster FD2. Eleven French grapevine samples (17%) had map sequences differing from the sequence of strain cluster FD2 by 12 base substitutions. Most of the strains in strain cluster FD1 originated from southwestern France and had a map sequence identical to the sequence of the reference strain FD70 isolated in southwestern France in 1970 (18). Strain cluster FD1 was more variable than strain cluster FD2, as the two FD1 members V03-9-2 and V04-11-19 could be distinguished by one and two single-nucleotide polymorphisms (SNPs), respectively. Finally, strain cluster FD3 was made up of only Italian FD isolates, and the map sequence of strain cluster FD3 differed from the sequences of strain clusters FD1 and FD2 by at least 8 and 12 base substitutions, respectively. The variability of strain cluster FD3 was in the same range as that of strain cluster FD1 with three isolates presenting one (VI04-Toscana1) or two SNPs (VI04-C28 and C29 corresponding to FD-C types according to Angelini et al. [2]). No insertion or deletion was observed between map sequences of FD isolates.
FIG. 3.
Phylogenetic trees constructed by parsimony analysis of sequences of map (A), uvrB-degV (B), and secY (C) genetic loci. “Ca. Phytoplasma ulmi” isolates were taken as the outgroup. Branch lengths are proportional to the number of inferred character state transformations. For clarity, the scales of the trees are proportional to the lengths of alignments. The strain clusters identified according to map are shown to the right of the tree in Fig. 1A. Bootstrap values for 100 replicates are shown on the branches. Phytoplasma isolates are described in Tables 1 and 2. Members of the FD clusters and AldY and PGY isolates are colored blue (cluster FD1), red (cluster FD2), green (cluster FD3), and orange (AldY and PGY).
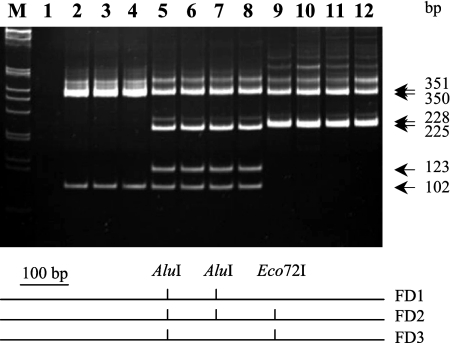
As an application for routine typing, the three FD strain clusters could easily be differentiated by RFLP of FD9f6-MAPr2 PCR products (Fig. 4) due to the lack of an AluI restriction site at position 452 in strain cluster FD3 and the lack of an Eco72I site at position 575 in the map sequence of strain cluster FD1. As expected, a double AluI and Eco72I digestion of map PCR products generated three fragments of 102, 350, and 351 bp for grapevine isolates of strain cluster FD1 (Fig. 4, lanes 2 to 4), four fragments of 102, 123, 225, and 351 bp for grapevine isolates of strain cluster FD2 (Fig. 4, lanes 5 to 8), and three fragments of 225, 228, and 350 bp for grapevine isolates of strain cluster FD3 (Fig. 4, lanes 9 to 12).
FIG. 4.
Analysis of AluI-Eco72I digestion of FD9f6-MAPr2 PCR products on 8% polyacrylamide gels and restriction map of the different FD strain clusters. Lane 1, healthy grapevine; lanes 2 to 12, grapevine samples V03-5-3, V04-11-18, V04-11-11, V00-SP9, V04-11-17, V04-11-30, V04-11-39, VI04-C29, VI04-Lig1, VI04-Toscana1, and VI04-248-04, respectively. Lane M, 1-kb ladder from Invitrogen.
Globally, comparison and parsimony analysis of uvrB-degV sequences from FD isolates resulted in three clusters corresponding to the strain clusters described from map sequences (Fig. 3B). The uvrB-degV locus nevertheless appeared less variable than map; the maximum number of base substitutions between strain cluster FD2 and FD3 was reduced to 8 over 1,037 nucleotides compared to 14 over 674 nucleotides for map. Seventeen French FD isolates and again the Italian isolate VI04-D004-03 from strain cluster FD2 had the same uvrB-degV sequence identical to the sequence of the reference strain FD92. Four French FD isolates from strain cluster FD1 had the same sequence, but it differed from that of the reference strain FD70. As observed for map, the French isolate V04-11-19 exhibited five and two SNPs compared to other members of strain cluster FD1 and FD2, respectively. Five of the Italian FD isolates from strain cluster FD3 grouped together. In this strain cluster, Italian isolates from Liguria and Piemonte showed a 7-bp deletion in the uvrB-degV intergenic sequence, a feature also observed for the reference isolate FD70 and the German PGY-C. The Italian FD isolate VI04-Toscana1 displayed a uvrB-degV sequence (Fig. 3B) as well as a secY sequence (Fig. 3C) very different from the three FD strain cluster, making this isolate a remarkably divergent variant. For example, its secY sequence had a CAAACG insertion at position 105, and a AAA triplet was inserted at position 533, two mutations also found in the German PGY-B isolate. Divergent genetic variants of FD phytoplasma have also been detected in Tuscany, Italy, by other authors (9). Taking apart this exception, secY sequence typing consistently grouped FD isolates in three clusters (Fig. 3C), whose composition was in agreement with the clusters found by map and uvrB-degV sequence typing. The whole strain cluster FD2 again displayed identical sequences. In strain cluster FD1, isolate V04-11-50 presented a single SNP variation and no base substitutions were observed for the five Italian FD isolates of strain cluster FD3. Finally, a French isolate (V04-11-01) collected in northeastern France had map and secY sequences closely related to those of PGY-C isolated in German Palatinate and could therefore be considered a PGY-C variant.
Genetic variability of alder yellows, PGY, and Spartium witches'-broom isolates.
Three German isolates of PGY and four Italian and French isolates of alder yellows (AldY) were submitted to sequence typing. The three genetic loci analyzed gave different clustering of PGY and AldY sequences, but all three showed important variability and no monophyletic origin for these phytoplasmas transmitted by the same alder leafhopper vector O. alni (41, 42). For example, map sequences distinguished four clusters: (i) ALY and PGY-B (differing by three SNPs); (ii) AI04-3-13, AI04-2-4 and PGY-A (differing by two SNPs from WJ1444-32); and finally (iii) PGY-C and (iv) V04-11-01. The variability between clusters ranged between 7 and 15 SNPs. Surprisingly, the cluster constituted by PGY-A, AI04-2-4, and WJ1444-32 differed by only four to six SNPs from strain cluster FD1. In the same way, the cluster made of ALY and PGY-B differed from strain cluster FD1 by five to seven SNPs. The cluster made of PGY-C and V04-11-01 displayed 8 SNPs compared to FD3 strain cluster, whereas 12 to 15 SNPs distinguished PGY-C from any of the other PGY-AldY clusters. For the uvrB-degV locus, only ALY and PGY-A had an identical sequence, and the variability of the other AldY and PGY isolates ranged from 4 SNPs (ALY versus WJ1444-32) to 13 SNPs (PGY-B versus AI04-2-4). Again, most of the strains of the AldY and PGY cluster showed higher uvrB-degV sequence similarity with strain cluster FD1. For instance, ALY/PGY-A and strain cluster FD1 differed by only one SNP; WJ1444-32 (AldY) and strain cluster FD1 had identical sequences, and AI04-2-4 (AldY) differed from the FD isolate VI04-Toscana1 by one SNP. These two isolates also shared a unique deletion of a T at position 439. Similarly, PGY-C had a 7-bp deletion at position 319 which was found only in uvrB-degV intergenic sequence of strain cluster FD3 (except VI04-Toscana1) and of isolate FD70. Comparison of secY sequences allowed differentiation of four clusters of AldY and PGY isolates. The Italian isolates ALY and AI04-3-7 differed by only 4 or 5 SNPs from strain cluster FD1 and FD2 but displayed 38 SNPs compared to the French isolate WJ1444-32, 44 and 46 SNPs compared to PGY-A and PGY-C, respectively, and 58 SNPs compared to PGY-B. Diversity was also important between PGY isolates which did not cluster together according to phylogenetic analysis of secY sequences. The only Spartium witches'-broom isolate analyzed clustered, for the three genetic loci, with FD, alder yellows, and PGY isolates.
Taken together, parsimony analyses of map, uvrB-degV, and to a lower extent, secY sequences indicated a common monophyletic origin for FD, AldY, PGY, and Spartium witches'-broom phytoplasmas, as all isolates clustered on a common phylogenetic branch supported by bootstrap values of 85%, 83%, and 68%, respectively. However, unlike the congruent evolution of the three genetic loci for the FD strain cluster and isolate ALY, the phylogenetic analyses of the three genetic markers often indicated different phylogenetic branching for AldY and PGY isolates.
Genetic diversity of rubus stunt and elm yellows phytoplasmas in Europe.
Variability of map and uvrB-degV was determined for 15 phytoplasma isolates associated either with stunting of Rubus or yellowing of elm and dog rose. These isolates had previously been characterized by 16S RFLP typing as members of the 16SrV phylogenetic group (9, 12, 30, 42). The elm yellows phytoplasma isolate EY1 and the hemp dogbane phytoplasma isolate HD1 from the United States were also tested. For Rubus and dog rose phytoplasmas (strain cluster RS), no sequence variability in the map locus was found, and only one SNP was detected in uvrB-degV locus (Fig. 3A and B). All isolates of strain cluster RS had a monophyletic origin, as they clustered in a single group supported by bootstrap values of 98% and 99%. They were all characterized by a specific ATT insertion at position 344 in the uvrB-degV intergenic sequence. Nucleotide sequence similarity between strain cluster RS and other phytoplasmas ranged between 96% and 98% for map and between 95% and 97% for uvrB-degV.
“Ca. Phytoplasma ulmi” isolates split into homogenous strain cluster EY1 and EY2 according to map sequences, which differed by 8 SNPs over 674 bp. Isolates from Italy and France were found in both groups. The variability of uvrB-degV was lower and only reached 3 SNPs over 1,025 bp between isolates WJ1296-30 and EI04-2-2. Sequence similarities with the other phytoplasmas tested were in the range of 96 to 97% for map and about 95% for uvrB-degV. Parsimony analyses of both genetic loci indicated a single monophyletic origin as all “Ca. Phytoplasma ulmi” isolates clustered on a branch supported by high bootstrap values of 99% (Fig. 3A and B).
DISCUSSION
Molecular typing to improve epidemiological knowledge of FD strain clusters and variants.
Sequence typing of the housekeeping gene map and the degV gene encoding a protein of unknown function allowed us to consistently distinguish three major FD groups of isolates occurring in France or Italy. Other typing approaches targeting secY and rpsC have previously concluded to the existence of three distinct groups of isolates (2, 3, 47), whereas only two distinct strain clusters were established according to 16S rRNA gene and internal transcribed spacer typing (2, 48). The present variability study of map and uvrB-degV confirmed that the three groups of FD phytoplasmas clearly constitute consistent lineages. Variants could be distinguished on the basis of a few SNPs in map and uvrB-degV genetic loci in two of the three strain clusters: FD1 and FD3. The FD1 strain cluster is genetically close to the reference strain FD70 isolated in southwestern France in 1970 (18). It had an incidence of about 17% in the French samples examined and was mainly restricted to the southwestern part of the country where map SNP variants could also be identified. This strain is also present in Italy but only in samples from the Piemonte and Lombardia regions (47). The FD2 strain cluster was the most widespread, and it was also prevalent in France (83% of the disease cases according to our data) and Italy (about half of the isolates tested) (47). The FD3 strain cluster (reference isolate FD-C) was restricted to Italy, where it was first detected in Veneto (48). The clonal property of strain cluster FD2 (reference strains FD88, FD92, and FD-D) and its wide distribution can be explained by the long-distance propagation of infected-plant material. However, differences in biological properties between FD strains resulting in better dynamics of grapevine colonization and/or insect transmission of the FD2 strain cluster cannot be excluded. It was recently demonstrated that strains of both FD1 and FD2 strain clusters, i.e., strains FD2000 and FD92, respectively, decreased in the same way the longevity of the experimental vector E. variegatus (12). The same reduced longevity, which could influence FD diffusion, was also reported in the natural vector S. titanus infected with FD92 (13), but other FD strains have not yet been checked in that respect on the natural vector. All three strain clusters described in this work can be transmitted to grapevine by S titanus (14, 53, 58), but comparison of their kinetics of insect transmission and multiplication in plants remains to be achieved. Routine typing of FD outbreaks by plant protection services will be implemented if fast and affordable methods can be developed. Simple PCR-RFLP analysis can be applied for an initial screening, but it remains time-consuming compared to sequencing, which is still too costly for widespread implementation by plant protection services. In order to better trace diffusion of FD strains, a good compromise could be to submit each year to sequence typing a representative set of isolates which should include new outbreaks. Qualitative survey of FD epidemics is of great value as it provides identification of new variants and confirmation of strain identity when propagation through nurseries is suspected. It would be worthy to study peculiar grapevine yellows isolates with similarities to PGY or AldY, such as V04-11-01 from northeastern France or VI04-Toscana1 from Italy, to understand their particular epidemiological properties.
Consequence of a probable common origin of group 16SrV phytoplasmas from grapevine and alder on the epidemiology of grapevine yellows in Europe.
Because S titanus, the specific insect vector of FD, was introduced from North America, it has been assumed that FD phytoplasma might have been introduced in Europe when phylloxera-resistant rootstocks were imported from North America (17, 40). Actually, positive enzyme-linked immunosorbent assays were obtained with antibodies to FD phytoplasma on S titanus specimens from vineyards affected with grapevine yellows in New York (40) in 1989 to 1990, but no additional data have been produced since. High tolerance to FD of rootstock varieties that were bred from American Vitis species (20) is evidence favoring the latter hypothesis. In contrast, more recent data tend to support a recent association of a Nearctic leafhopper and a European pathogen (3, 4, 13). Our data bring strong additional genetic evidence of a common origin between alder yellows and FD phytoplasmas and are more in favor of a European origin of this 16SrV subclade. The diversity and phylogenetic analyses presently based on three nonribosomal genes pointed out FD, PGY, and AldY as being members of the same phylogenetic subclade. Some alder isolates appeared to be more related to FD isolates than to other PGY or AldY phytoplasmas. Alder yellows phytoplasma is widespread in European alders, and it was reported in Italy, France, Switzerland, Austria, Germany, and the eastern Baltic region, but its presence was never reported in America (33, 51, 62). It is transmitted by O alni to alder that may be tolerant and remain symptomless (41) but also to grapevine in which it was reported as PGY (42). As proposed previously (10), some strains of FD-related phytoplasmas could have been erratically transmitted to grapevine by occasional vine-feeding vector species, such as O alni. Then phytoplasmas could have been transmitted to neighboring vines and vineyards by competent S titanus and spread further by trading of plant material (16, 20) to vineyards of southwestern France and northern Italy inhabited by S titanus populations. Success of such a series of events should have happened at least three times to give rise to the three genetically distinct FD strain clusters that we describe in this work. However, PGY occurs in a viticulture area to which S titanus has not yet expanded (10, 42, 43), and until now experimental transmission of PGY phytoplasmas by S titanus specimens has remained unsuccessful (M. Maixner and E. Boudon-Padieu, unpublished results). As for FD-C (strain cluster FD3), grapevine-to-grapevine transmission by S titanus has been demonstrated (53). Nevertheless, although FD-C isolates have been consistently (4) and frequently (E. Angelini, personal communication) detected in wild Clematis vitalba growing in the underbrush near affected vineyards, insect transmission from Clematis to grapevine could not be obtained (E. Angelini, personal communication; E. Boudon-Padieu, unpublished results). Without experimental confirmation of the ability of S titanus to transmit some PGY phytoplasma or of vector transmission of Clematis phytoplasma to grapevine, our hypothesis will remain unproven.
Taxonomic implications of group 16SrV phytoplasma variability.
Phytoplasmas have been classified primarily on the basis of the 16S rRNA gene sequence and ecological properties (36, 59). Following the proposal of Murray and Schleifer for recording putative taxa (54), “Candidatus Phytoplasma” species have been described and rules for phytoplasma taxonomy proposed (29, 32). Regarding the level of 16S sequence similarity between subtaxons, it is recognized that below a level of 97.5%, it is unlikely that two organisms have more than 60 to 70% DNA similarity and hence that they constitute different species (60). However, some phytoplasma groups with 16S rRNA gene sequence similarity above 97.5% include phytoplasma strains with very different biological properties, such as insect vector, host plant specificity, and symptomatology, and with genomic variability. For such cases, description of two different species is recommended only when all parameters listed above are verified (32). Phytoplasma group 16SrV is one of these, in which “Ca. Phytoplasma ziziphi” (strain cluster 16SrV-B) and “Ca. Phytoplasma ulmi” (strain cluster 16SrV-A) were described despite their high 16S sequence similarity (34, 38). Diversity and phylogenetic analyses based on three genes (16S rRNA gene, rpsC, and secY) clearly indicated that the 16SrV-A RFLP strain cluster consisting of elm yellows strains represents a distinct lineage divergent from the 16SrV-B RFLP strain cluster. “Ca. Phytoplasma ulmi” showed 98.2% 16S rRNA gene sequence similarity, 93% similarity in the ribosomal protein L22 and S3 genes, and 81.0% similarity in the secY gene with the corresponding sequences of “Ca. Phytoplasma ziziphi” (38). As there were no plant hosts or insect vectors common to 16SrV-A and 16SrV-B phytoplasmas, both candidate species could be designated. All the other members of this group remained as strain cluster 16SrV-C, D, and E. In the present study, diversity of map and uvrB-degV is in total agreement with the taxonomic status of “Ca. Phytoplasma ulmi” and confirms the previous description of genetic diversity among “Ca. Phytoplasma ulmi” isolates (11). Sequence similarity between “Ca. Phytoplasma ulmi” isolates and members of RFLP strain cluster 16SrV-C (isolates FD-C, ALY, FD70, and SpaWB), 16SrV-D (isolate FD-D), and 16SrV-E (RuS) ranged from 95% to 97% according to the gene and the strain cluster. Part of our data is also consistent with a possible distinction of rubus stunt phytoplasma as a new “Candidatus” species, especially as its specific insect vector and plant host range are well determined (25, 26, 33, 50). Recently, the working group on phytoplasma taxonomy suggested the provisional name of “Ca. Phytoplasma vitis” for FD phytoplasma (32). To take into account the genetic proximity of FD, AldY, and PGY phytoplasmas, this new candidate species should include these three groups of phytoplasmas with some members infecting grapevine as the preferred host and being transmitted by S titanus and the other members infecting alder as the preferred host and grapevine as secondary host and being transmitted by O alni. However, in contrast to rpsC (34, 43), map, and uvrB-degV (present paper), the phylogeny of secY does not give a phylogenetic grouping of the isolates totally in agreement with these two taxons, and therefore, genomic diversity in this group will need further documentation before describing new species.
Acknowledgments
This work was supported by research grants from the Conseil Interprofessionnel du Vin de Bordeaux (contract 6099-2004) and from the Conseil Régional d'Aquitaine (contract B05977). Support for G. Arnaud was provided by the Ministère de l'Enseignement Supérieur et de la Recherche.
We thank our colleagues P. Pracros, J. L. Danet, S. Baron, and F. Ferrer for growing plants and insects, maintaining phytoplasma-infected periwinkles, and their support in sampling diseased grapevines. We gratefully acknowledge the French national and regional “Services de Protection des Plantes,” Elisa Angelini, Assunta Bertaccini, Luigi Carraro, Denis Clair, and Cristina Marzachi for providing phytoplasma-infected plant samples or extracts.
Footnotes
Published ahead of print on 27 April 2007.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST (Basic Local Alignment Search Tool) and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angelini, E., D. Clair, M. Borgo, A. Bertaccini, and E. Boudon-Padieu. 2001. Flavescence dorée in France and Italy: occurrence of closely related phytoplasma isolates and their near relationships to palatinate grapevine yellows and an alder yellows phytoplasma. Vitis 40:79-86. [Google Scholar]
- 3.Angelini, E., E. Negrisolo, D. Clair, M. Borgo, and E. Boudon-Padieu. 2003. Phylogenetic relationships among Flavescence doree strains and related phytoplasmas determined by heteroduplex mobility assay and sequence of ribosomal and nonribosomal DNA. Plant Pathol. 52:663-672. [Google Scholar]
- 4.Angelini, E., F. Squizzato, G. Luchetta, and M. Borgo. 2004. Detection of a phytoplasma associated with grapevine Flavescence dorée in Clematis vitalba. Eur. J. Plant Pathol. 110:193-201. [Google Scholar]
- 5.Baker, W. L. 1949. Notes on the transmission of the virus causing phloem necrosis of American elm, with notes on the biology of its insect vector. J. Econ. Entomol. 42:729-732. [Google Scholar]
- 6.Baskaran, N., R. P. Kandpal, A. K. Bhargava, M. W. Glynn, A. Bale, and S. M. Weissman. 1996. Uniform amplification of a mixture of deoxyribonucleic acids with varying GC content. Genome Res. 6:633-638. [DOI] [PubMed] [Google Scholar]
- 7.Batlle, A., A. Laviña, C. Kuszala, D. Clair, J. Larrue, and E. Boudon-Padieu. 1997. Detection of flavescence doree phytoplasma in grapevine in northern Spain. Vitis 36:211-212. [Google Scholar]
- 8.Belli, G., A. Fortusini, and D. Rui. 1985. Recent spread of flavescence dorée and its vector in vineyards of northern Italy. Phytopathol. Mediterr. 24:189-191. [Google Scholar]
- 9.Botti, S., and A. Bertaccini. 2003. Molecular variability in Flavescence dorée phytoplasmas as marker for the disease outbreaks in vineyards, p. 62-63. In Proceedings of the 14th Meeting of the International Council for the Study of Virus and Virus-like Disease of the Grapevine. Locorotondo (BARI), Italy, 12 to 17 September 2003.
- 10.Boudon-Padieu, E. 2002. Flavescence dorée of the grapevine: knowledge and new developments in epidemiology, etiology and diagnosis, p. 15-34. Atti Giornate Fitopatologiche, Baselga di Piné (Trento), Italy.
- 11.Boudon-Padieu, E., J. Larrue, D. Clair, J. Hourdel, A. Jeanneau, R. Sforza, and E. Collin. 2004. Detection and prophylaxis of elm yellows phytoplasma in France. Investig. Agrar. Sist. Recur. For. 13:71-80. [Google Scholar]
- 12.Bressan, A., D. Clair, O. Semetey, and E. Boudon-Padieu. 2005. Effect of two strains of Flavescence dorée phytoplasma on the survival and fecundity of the experimental leafhopper vector Euscelidius variegatus Kirschbaum. J. Invertebr. Pathol. 89:144-149. [DOI] [PubMed] [Google Scholar]
- 13.Bressan, A., V. Girolami, and E. Boudon-Padieu. 2005. Reduced fitness of the leafhopper vector Scaphoideus titanus exposed to Flavescence dorée phytoplasma. Entomol. Exp. Appl. 115:283-290. [Google Scholar]
- 14.Bressan, A., J. Larrue, and E. Boudon Padieu. 2006. Patterns of phytoplasma-infected and -infective Scaphoideus titanus leafhoppers in vineyards with high incidence of Flavescence dorée. Entomol. Exp. Appl. 119:61-69. [Google Scholar]
- 15.Carraro, L., F. Ferrini, P. Ermacora, N. Loi, M. Martini, and R. Osler. 2004. Macropsis mendax as a vector of elm yellows phytoplasma of Ulmus species. Plant Pathol. 53:90-95. [Google Scholar]
- 16.Caudwell, A. 1957. Deux années d'étude sur la flavescence dorée, nouvelle maladie grave de la vigne. Ann. Amélior. Plantes 4:359-363. [Google Scholar]
- 17.Caudwell, A. 1983. L'origine des jaunisses à Mycoplasmes (MLO) des plantes et l'exemple des jaunisses de la vigne. Agronomie 2:103-111. [Google Scholar]
- 18.Caudwell, A., C. Kuszala, J. C. Bachelier, and J. Larrue. 1970. Transmission de la Flavescence dorée de la vigne aux plantes herbacées par l'allongement du temps d'utilisation de la cicadelle Scaphoideus littoralis Ball et l'étude de sa survie sur un grand nombre d'espèces végétales. Ann. Phytopathol. 2:415-428. [Google Scholar]
- 19.Caudwell, A., C. Kuszala, J. Larrue, and J. C. Bachelier. 1972. Transmission de la Flavescence dorée de la Fève à la Fève par des cicadelles des genres Euscelis et Euscelidius. Ann. Phytopathol. 1572:181-189. [Google Scholar]
- 20.Caudwell, A., J. Larrue, V. Tassart, R. Boidron, S. Grenan, M. Leguay, and P. Bernard. 1994. Caractère porteur de la Flavescence dorée chez les vignes porte-greffes en particulier le 3309 C et le Fercal. Agronomie 14:83-94. [Google Scholar]
- 21.Cimerman, A., G. Arnaud, and X. Foissac. 2006. Stolbur phytoplasma genome survey achieved using a suppression subtractive hybridization approach with high specificity. Appl. Environ. Microbiol. 72:3274-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clair, D., J. Larrue, G. Aubert, J. Gillet, G. Cloquemin, and E. Boudon-Padieu. 2003. A multiplex nested-PCR assay for sensitive and simultaneous detection and direct identification of phytoplasma in the elm yellows group and Stolbur group and its use in survey of grapevine yellows in France. Vitis 42:151-157. [Google Scholar]
- 23.Daire, X., D. Clair, J. Larrue, E. Boudon-Padieu, A. Alma, A. Arzone, L. Carraro, R. Osler, E. Refatti, G. Granata, R. Credi, E. Tanne, R. Pearson, and A. Caudwell. 1993. Occurrence of diverse MLOs in tissues of grapevine affected by grapevine yellows in different countries. Vitis 32:247-248. [Google Scholar]
- 24.Daire, X., D. Clair, W. Reinert, and E. Boudon-Padieu. 1997. Detection and differentiation of grapevine yellows phytoplasmas belonging to the elm yellows group and to the stolbur strain cluster by PCR amplification of non-ribosomal DNA. Eur. J. Plant Pathol. 103:507-514. [Google Scholar]
- 25.Davies, D. L. 2000. The occurrence of two phytoplasmas associated with stunted Rubus species in the UK. Plant Pathol. 49:86-88. [Google Scholar]
- 26.de Fluiter, H. J., and F. A. van der Meer. 1953. Rubus stunt, a leafhopper borne virus disease. Tijdschr. Plantenziekten 59:195-197. (In German.) [Google Scholar]
- 27.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 28.Ewing, B., L. Hillier, M. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 29.Firrao, G., K. Gibb, and C. Streten. 2005. Short taxonomic guide to the genus ‘Candidatus Phytoplasma’. J. Plant Pathol. 87:249-263. [Google Scholar]
- 30.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 31.Gundersen, D. E., and I.-M. Lee. 1996. Ultrasensitive detection of phytoplasmas by nested-PCR assays using two universal primer pairs. Phytopathol. Mediterr. 35:114-151. [Google Scholar]
- 32.The IRPCM Phytoplasma/Spiroplasma Working Team-Phytoplasma Taxonomy Group. 2004. ‘Candidatus Phytoplasma’, a taxon for the wall-less, non-helical prokaryotes that colonize plant phloem and insects. Int. J. Syst. Evol. Microbiol. 54:1243-1255. [DOI] [PubMed] [Google Scholar]
- 33.Jarausch, W., B. Jarausch-Wehrheim, J. L. Danet, J. M. Broquaire, F. Dosba, C. Saillard, and M. Garnier. 2001. Detection and identification of European stone fruit yellows and other phytoplasmas in wild plants in the surroundings of apricot chlorotic leaf roll-affected orchards in southern France. Eur. J. Plant Pathol. 107:209-217. [Google Scholar]
- 34.Jung, H. Y., T. Sawayanagi, S. Kakizawa, H. Nishigawa, W. Wei, K. Oshima, S. Miyata, M. Ugaki, T. Hibi, and S. Namba. 2003. ‘Candidatus Phytoplasma ziziphi’, a novel phytoplasma taxon associated with jujube witches'-broom disease. Int. J. Syst. Evol. Microbiol. 53:1037-1041. [DOI] [PubMed] [Google Scholar]
- 35.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis (MEGA) software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 36.Lee, I. M., R. E. Davis, and D. E. Gundersen-Rindal. 2000. Phytoplasma: phytopathogenic mollicutes. Annu. Rev. Microbiol. 54:221-255. [DOI] [PubMed] [Google Scholar]
- 37.Lee, I. M., D. E. Gundersen-Rindal, and A. Bertaccini. 1998. Phytoplasma: ecology and genomic diversity. Phytopathology 88:1359-1366. [DOI] [PubMed] [Google Scholar]
- 38.Lee, I. M., M. Martini, C. Marcone, and S. F. Zhu. 2004. Classification of phytoplasma strains in the elm yellows group (16SrV) and proposal of ‘Candidatus Phytoplasma ulmi’ for the phytoplasma associated with elm yellows. Int. J. Syst. Evol. Microbiol. 54:337-347. [DOI] [PubMed] [Google Scholar]
- 39.Maixner, M., U. Ahrens, and E. Seemüller. 1995. Detection of the German grapevine yellows (Vergilbungskrankheit) MLO in grapevine, alternative hosts and a vector by a specific PCR procedure. Eur. J. Plant Pathol. 101:241-250. [Google Scholar]
- 40.Maixner, M., R. C. Pearson, E. Boudon-Padieu, and A. Caudwell. 1993. Scaphoideus titanus, a possible vector of grapevine yellows in New York. Plant Dis. 77:408-413. [Google Scholar]
- 41.Maixner, M., and W. Reinert. 1999. Oncopsis alni (Schrank) (Auchenorrhyncha: Cicadellidae) as a vector of the alder yellows phytoplasma of Alnus glutinosa (L.) Gaertn. Eur. J. Plant Pathol. 105:87-94. [Google Scholar]
- 42.Maixner, M., W. Reinert, and H. Darimont. 2000. Transmission of grapevine yellows by Oncopsis alni (Schrank) (Auchenorrhyncha: Macropsinae). Vitis 39:83-84. [Google Scholar]
- 43.Maixner, M., M. Rudel, X. Daire, and E. Boudon-Padieu. 1995. Diversity of grapevine yellows in Germany. Vitis 34:235-236. [Google Scholar]
- 44.Marcone, C., A. Ragozzino, B. Schneider, U. Lauer, C. D. Smart, and E. Seemüller. 1996. Genetic characterization and classification of two phytoplasmas associated with spartium witches'-broom disease. Plant Dis. 80:365-371. [Google Scholar]
- 45.Marcone, C., A. Ragozzino, and E. Seemüller. 1996. Detection of an elm yellows-related phytoplasma in eucalyptus trees affected by little-leaf disease in Italy. Plant Dis. 80:669-673. [Google Scholar]
- 46.Marcone, C., A. Ragozzino, and E. Seemüller. 1997. Identification and characterization of the phytoplasma associated with elm yellows in southern Italy and its relatedness to other phytoplasmas of the elm yellows group. Eur. J. For. Pathol. 27:45-54. [Google Scholar]
- 47.Martini, M., S. Botti, C. Marcone, C. Marzachi, P. Casati, P. A. Bianco, R. Benedetti, and A. Bertaccini. 2002. Genetic variability among flavescence dorée phytoplasmas from different origins in Italy and France. Mol. Cell Probes 16:197-208. [DOI] [PubMed] [Google Scholar]
- 48.Martini, M., E. Murari, N. Mori, and A. Bertaccini. 1999. Identification and epidemic distribution of two flavescence dorée-related phytoplasmas in Veneto (Italy). Plant Dis. 83:925-930. [DOI] [PubMed] [Google Scholar]
- 49.Marzachi, C., S. Palermo, A. Boarino, F. Veratti, M. D'Aquilio, A. Loria, and G. Boccardo. 2001. Optimization of a one-step PCR assay for the diagnosis of Flavescence dorée-related phytoplasmas in field-grown grapevines and vector populations. Vitis 40:213-217. [Google Scholar]
- 50.Maurer, R., and E. Seemüller. 1995. Nature and genetic relatedness of the mycoplasma-like organism causing rubus stunt in Europe. Plant Pathol. 44:244-249. [Google Scholar]
- 51.Maurer, R., E. Seemüller, and W. A. Sinclair. 1993. Genetic relatedness of mycoplasmalike organisms affecting elm, alder, and ash in Europe and North America. Phytopathology 83:971-976. [Google Scholar]
- 52.McCoy, R. E., A. Caudwell, C. J. Chang, T. A. Chen, L. N. Chiykowski, M. T. Cousin, J. E. Dale, G. T. N. de Leeuw, D. A. Golino, K. J. Hackett, B. C. Kirkpatrick, R. Marwitz, H. Petzold, R. C. Sinha, M. Sugiura, R. F. Whitcomb, I. L. Yang, B. M. Zhu, and E. Seemüller. 1989. Plant diseases associated with mycoplasma-like organisms, p. 545-640. In R. F. Whitcomb and J. G. Tully (ed.), The mycoplasmas, vol. V. Spiroplasmas, acholeplasmas and mycoplasmas of plants and arthropods. Academic Press Inc., New York, NY. [Google Scholar]
- 53.Mori, N., A. Bressan, M. Martini, M. Guadagnini, V. Girolami, and A. Bertaccini. 2002. Experimental transmission by Scaphoideus titanus Ball of two Flavescence dorée-type phytoplasmas. Vitis 41:99-102. [Google Scholar]
- 54.Murray, R. G. E., and K. H. Schleifer. 1994. Taxonomic notes: a proposal for recording the properties of putative taxa of procaryotes. Int. J. Syst. Bacteriol. 44:174-176. [DOI] [PubMed] [Google Scholar]
- 55.Oshima, K., S. Kakizawa, H. Nishigawa, H. Y. Jung, W. Wei, S. Suzuki, R. Arashida, D. Nakata, S. Miyata, M. Ugaki, and S. Namba. 2004. Reductive evolution suggested from the complete genome sequence of a plant-pathogenic phytoplasma. Nat. Genet. 36:27-29. [DOI] [PubMed] [Google Scholar]
- 56.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 57.Schneider, B., E. Seemüller, C. D. Smart, and B. C. Kirkpatrick. 1995. Phylogenetic classification of plant pathogenic mycoplasmalike organisms or phytoplasmas, p. 369-380. In S. Razin and J. G. Tully (ed.), Molecular and diagnostic procedures in mycoplasmology, vol. 1. Academic Press, San Diego, CA. [Google Scholar]
- 58.Schvester, D., P. Carle, and G. Moutous. 1961. Sur la transmission de la flavescence dorée des vignes par une cicadelle. C. R. Acad. Sci. 18:1021-1024. [Google Scholar]
- 59.Seemüller, E., C. Marcone, U. Lauer, A. Ragozzino, and M. Göschl. 1998. Current status of molecular classification of the phytoplasmas. J. Plant Pathol. 80:3-26. [Google Scholar]
- 60.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 61.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Valiunas, D., A. Alminaite, J. Staniulis, R. Jomantiene, and R. E. Davis. 2001. First report of alder yellows phytoplasma in the eastern Baltic region. Plant Dis. 85:1120. [DOI] [PubMed] [Google Scholar]
- 63.Weintraub, P. G., and L. Beanland. 2006. Insect vectors of phytoplasmas. Annu. Rev. Entomol. 51:91-111. [DOI] [PubMed] [Google Scholar]