Abstract
Rhizobium leguminosarum bv. trifolii forms nitrogen-fixing root nodules on the pasture legume Trifolium repens, and T. repens seed is often coated with a compatible R. leguminosarum bv. trifolii strain prior to sowing. However, significant losses in bacterial viability occur during the seed-coating process and during storage of the coated seeds, most likely due to desiccation stress. The disaccharide trehalose is known to function as an osmoprotectant, and trehalose accumulation due to de novo biosynthesis is a common response to desiccation stress in bacteria. In this study we investigated the role of endogenous trehalose synthesis in desiccation tolerance in R. leguminosarum bv. trifolii strain NZP561. Strain NZP561 accumulated trehalose as it entered the stationary phase due to the combined actions of the TreYZ and OtsAB pathways. Mutants deficient in either pathway showed near-wild-type levels of trehalose accumulation, but double otsA treY mutants failed to accumulate any trehalose. The double mutants were more sensitive to the effects of drying, and their survival was impaired compared to that of the wild type when glass beads were coated with the organisms and stored at relative humidities of 5 and 32%. The otsA treY mutants were also less competitive for nodule occupancy. Gene expression studies showed that the otsA and treY genes were expressed constitutively and that expression was not influenced by the growth phase, suggesting that trehalose accumulation is controlled at the posttranscriptional level or by control of trehalose breakdown rates. Our results indicate that accumulated trehalose plays an important role in protecting R. leguminosarum bv. trifolii cells against desiccation stress and against stress encountered during nodulation.
Rhizobium leguminosarum bv. trifolii forms nitrogen-fixing root nodules on the pasture legume Trifolium repens (white clover). Inoculation of legumes is an efficient and convenient way of introducing rhizobia into soil and subsequently into the rhizosphere of legumes. The seeds of many clover species (including white clover) are coated with compatible R. leguminosarum bv. trifolii strains prior to sowing. One of the main reasons for the ineffectiveness of legume inoculants in the field is the rapid death of rhizobia due to desiccation (12, 55). Significant losses in inoculant viability occur during the seed-coating process, which involves a drying stage, during storage of the coated seeds and after introduction into the field environment. These losses in bacterial viability are likely to be due mainly to desiccation stress.
Bacteria accumulate osmoprotective compounds, referred to as compatible solutes or osmolytes, in response to osmotic or desiccation stress. Osmolytes can be obtained by uptake from the environment (exogenous) or through de novo biosynthesis (endogenous). De novo biosynthesis of trehalose, a nonreducing (α,α-1,1)-glucose disaccharide, is a common response to desiccation and osmotic stress in many bacteria (11, 35, 57). Trehalose has been shown to protect cell membranes and proteins from inactivation or denaturation caused by a variety of stress conditions, including osmotic shock, desiccation, cold, heat, and oxidation (for reviews, see references 3, 10, 15, 35, and 36).
Four pathways for trehalose biosynthesis in bacteria have been described to date (4, 15, 41). The common OtsAB pathway involves the condensation of glucose-6-phosphate with UDP-glucose by trehalose-6-phosphate synthase (OtsA) to form trehalose-6-phosphate, with subsequent dephosphorylation by trehalose-6-phosphate phosphatase (OtsB), yielding free trehalose. The TreYZ pathway involves the conversion of maltodextrins, such as glycogen, into trehalose. The terminal α-1,4-glycosylic bond at the reducing end of the glucan polymer is converted into an α-1,1-glycosidic bond via transglycosylation by maltooligosyltrehalose synthase (TreY). Free trehalose is subsequently released from the end of the polymer via hydrolysis by maltooligosyltrehalose trehalohydrolase (TreZ). The TreS pathway involves a reversible transglycosylation reaction in which trehalose synthase (TreS) converts the α-1,4 bond of maltose to the α-1,1 bond of trehalose. The recently discovered fourth pathway involves trehalose glycosyltransferring synthase (TreT), which catalyzes the reversible formation of trehalose from ADP-glucose and glucose. TreT has been characterized in hyperthermophilic bacteria (37, 39).
Many bacterial species possess a single pathway; for example, Escherichia coli (48) and Salmonella enterica (24) synthesize trehalose using the OtsAB pathway. In E. coli, the otsBA operon is induced by osmotic shock, desiccation, and entry into the stationary phase (48). In contrast, the OtsAB, TreYZ, and TreS pathways are all present in Mycobacterium bovis, Mycobacterium smegmatis (13), Corynebacterium glutamicum (53, 56), and Rhodobacter sphaeroides (29). In C. glutamicum and R. sphaeroides, the OtsAB and TreYZ pathways are important for biosynthesis, while the TreS pathway appears to be involved mainly in trehalose catabolism (29, 53, 56).
Trehalose is a common disaccharide in the root nodules of legumes and is present at high concentrations in bacteroids at the onset of nitrogen fixation (43). It is the major carbohydrate present in cultures of Bradyrhizobium strains and was also detected in cultures of other rhizobial species (Sinorhizobium meliloti, R. leguminosarum bv. trifolii, R. leguminosarum bv. viciae, and R. leguminosarum bv. phaseoli) (43). In R. leguminosarum bv. trifolii strain TA1, trehalose accumulates in the cells as an osmoprotectant in response to increasing osmotic pressure of the medium (6, 7). TreYZ enzyme activity was found in Bradyrhizobium japonicum, Bradyrhizobium elkanii, Rhizobium sp. strain NGR234, S. meliloti, R. leguminosarum bv. trifolii, and R. leguminosarum bv. viciae, suggesting that synthesis of trehalose using the TreYZ pathway is common in rhizobia (45). The accumulation of trehalose by B. japonicum cells, achieved by loading during growth, improved survival during desiccation (44). It has also been shown that trehalose acts as an osmoprotectant when it is exogenously supplied to S. meliloti, R. leguminosarum bv. trifolii, and R. leguminosarum bv. phaseoli, but in this case the mechanism does not involve accumulation; instead, trehalose and other disaccharides indirectly contribute to cell turgor by eliciting sharp increases in the levels of the osmolytes glutamate and N-acetylglutaminylglutamine amide (21).
It is likely that most rhizobia possess three trehalose biosynthesis pathways (all the pathways except the TreT pathway). Genes encoding putative enzymes belonging to the pathways have been annotated in the genome sequences of R. leguminosarum bv. viciae strain 3841 (58), Rhizobium etli strain CFN42 (20), S. meliloti strain 1021 (18), and B. japonicum strain USDA110 (28). Putative otsA, otsB, and treY genes have been identified in the partial genome sequence of Rhizobum sp. strain NGR234 (16, 47), but in Mesorhizobium loti strain MAFF303099 only genes for the OtsAB pathway were identified (27).
Recently, transcriptome profiling was used to investigate the responses of S. meliloti to a sudden increase in external osmolarity (14). A 200-kb region of the pSymB plasmid that contains a high density of osmoregulated genes was identified. Among these genes were treY (SMb20574), otsA (SMa0233), treS (SMb20099), and five other genes in the same putative operon as treS (SMb20095 to SMb20100).
Despite the interest in improving the desiccation tolerance of rhizobia used as agricultural inoculants, the roles of the various trehalose biosynthetic pathways and their contribution to the stress tolerance of the bacteria have not been characterized. In this study, we show that there are two pathways (otsA and treY) involved in trehalose biosynthesis in R. leguminosarum bv. trifolii strain NZP561 and that accumulated trehalose plays an important role in protecting R. leguminosarum bv. trifolii cells against desiccation stress.
MATERIALS AND METHODS
Strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| E. coli strains | ||
| HB101 | pro leu thi gal lacY recA str hsdD hsdM | 5 |
| S17-1 | RK2 tra regulon | 42 |
| R. leguminosarum bv. trifolii strains | ||
| NZP561 | Also known as CC275e; commercial inoculant strain; wild type | Our laboratory |
| HM1 | NZP561 otsA::EZTn5 (Neor) | This study |
| HM2 | NZP561 treY::EZTn5 (Neor) | This study |
| HM3 | NZP561 otsA::lacZ, pFUS2 IDM (Gmr) | This study |
| HM4 | NZP561 treY::lacZ, pFUS2 IDM (Gmr) | This study |
| HM5 | HM1 treY::lacZ, pFUS2 IDM (Neor Gmr) | This study |
| HM12 | NZP561 treY::lacZ, pFUS2 CMD (Gmr) | This study |
| TH1 | HM2 otsA::lacZ, pFUS2 IDM (Neor Gmr) | This study |
| TH3 | NZP561 otsA::lacZ, pFUS2 CMD (Gmr) | This study |
| Plasmids | ||
| pLAFR1 | Broad-host-range cosmid; Tcr | 17 |
| pFUS2 | oriVColE1oriTRK2lacZ transcriptional reporter; suicide vector, Gmr | 2 |
| pPH1J1 | Conjugative plasmid, IncP Gmr | 23 |
| pUC8 | Cloning vector, oriColE1lacZα Apr | 54 |
| pHM2 | pLAFR1 cosmid containing NZP561 DNA that hybridized to otsA probe | This study |
| pHM3 | pLAFR1 cosmid containing NZP561 DNA that hybridized to treY probe | This study |
| pHM4 | pUC8 containing 6.0-kb EcoRI fragment of pHM2 that contains the entire otsA gene | This study |
| pHM5 | pLAFR1 containing otsA::EZTn5 EcoRI fragment | This study |
| pHM6 | pHM3 treY::EZTn5 | This study |
Growth media and conditions.
R. leguminosarum bv. trifolii strains were grown at 28°C in TY or Rhizobium defined media (RDM) (49). Glucose and mannitol (0.4%, wt/vol) were routinely used as the carbon sources for RDM agar (GRDM) plates and RDM (MRDM) broth, respectively. Media were supplemented with antibiotics as required at the following concentrations: for E. coli, 15 μg tetracycline ml−1, 25 μg gentamicin ml−1, and 50 μg kanamycin ml−1; and for R. leguminosarum, 2 μg tetracycline ml−1, 30 and 15 μg gentamicin ml−1, and 50 and 25 μg neomycin ml−1 (the lower concentrations were used for double mutant strains).
For all growth phase experiments, a 1/50 dilution of cells from stationary-phase starter broth cultures (optical density at 600 nm adjusted to 0.5) was used to inoculate MRDM broth, which was then incubated at 28°C with shaking (160 rpm).
DNA manipulations.
Rhizobial DNA was prepared as described previously (49). Plasmid DNA extraction, agarose gel electrophoresis, cloning, and electroporation were carried out using established methods (40). A library of partially digested EcoRI fragments of NZP561 DNA in the cosmid vector pLAFR1 was constructed as described previously (17) and packaged into λ phage heads using the Packagene Lambda packaging system (Promega). Southern blotting was carried out by capillary transfer. DNA probes were labeled by random priming, and membranes were hybridized and washed as previously described (49). Oligonucleotide primers used for PCR and DNA sequencing are described in Table 2. DNA was amplified using the Expand High Fidelity and GC-rich PCR systems (Roche), using the conditions recommended by the manufacturer.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′ to 3′)a | Use |
|---|---|---|
| treYF | CACGACACCAAACGCGGTGA | treY probe |
| treYR | CGACGGTTGTCGGGATCAAC | treY probe |
| otsAF | TATGGATGGGCTGGTCGGGAAAGT | otsA probe |
| otsAR | TGCAGGCGGCCAGGAATGTG | otsA probe |
| treYIDMR | AAATTTGGATCCGTCGCCATAGGTGCGGTAGA | IDM |
| treYIDML | AAATTTAAGCTTGGCCGGACACCTATATCGTC | IDM |
| otsAIDMR | AAATTTGGATCCCGAATAATCGAGGCGGTCGA | IDM |
| otsAIDML | AAATTTAAGCTTCGGCTTCTTCCTGCACATTC | IDM |
| treYCMDR | AAATTTGGATCCATCGAAGGTCATGCCGTTGC | CMD |
| treYCMDL | AAATTTAAGCTTCTGTCACCGCTGATGCTGAA | CMD |
| otsACMDR | AAATTTGGATCCGGCAGCGGCGCTCCCATCTT | CMD |
| otsACMDL | AAATTTAAGCTTAGCGGTCGGCCTGGCACATC | CMD |
| lacZ | GCTATTACGCCAGCTGGCGA | Sequencing |
| KAN-FP1 | ACCTACAACAAAGCTCTCATCAACC | Sequencing |
| KAN-RP1 | GCAATGTAACATCAGAGATTTTGAG | Sequencing |
Restriction sites incorporated into primers are underlined.
DNA sequencing.
EcoRI restriction fragments of pHM2 and pHM3 were subcloned into pUC8, and plasmid templates were sequenced using the universal M13 forward and reverse primers and custom primers. A sequence was assembled using the Seqman package (DNASTAR). Databases at NCBI and Rhizobase (http://bacteria.kazusa.or.jp/rhizobase/) were searched using BLAST N, X, and P (1) for similar nucleotide and amino acid sequences.
Construction of R. leguminosarum bv. trifolii mutants.
An EZ::Tn5 <KAN-2> insertion kit (Epicenter Technologies) was used to create insertional mutations in pHM3 and pHM4. The EZ-Tn5 insertion reaction mixtures were electroporated into E. coli HB101 cells, and restriction digests and sequencing using Tn-specific primers KAN-FP1 and KAN-RP1 (Table 2) were used to precisely determine the sites of insertion in the mutated plasmids. The EcoRI fragment from a pHM4 derivative containing an EZ-Tn5 insertion within otsA was subcloned into pLAFR1, producing pHM5. A pHM6 cosmid from the pHM3::EZ-Tn5 library in which EZ-Tn5 had inserted within the treY gene was identified. Cosmids pHM5 and pHM6 were transferred to strain NZP561 by conjugation, and Tcr Neor transconjugants were selected. Marker exchange of the EZ-Tn5 insertions into the NZP561 genome was forced by plasmid incompatibility using pPH1J1 as described previously (25). To confirm that the expected recombination events had occurred, genomic DNA was extracted from Neor Gmr Tcs strains and analyzed by Southern hybridization using probes specific for the inserted DNA. Mutants that had lost pLAFR1 were selected after passage through nodules.
The suicide vector pFUS2 was used to create insertional duplication mutants (IDM) in which a pFUS2 insertion interrupted the gene and formed a lacZ transcriptional fusion. cis-Merodiploid transcriptional fusion (CMD) strains were also constructed using pFUS2. In this case insertion of pFUS2 fused the promoter region and 5′ end of the gene to the promoterless lacZ gene, and an intact copy of the gene and its promoter remained downstream of the integrated pFUS2 plasmid. Oligonucleotide primers pairs (Table 2) were designed to amplify 340- to 360-bp intragenic regions of treY and otsA for IDM pFUS2 constructs and 350-bp regions containing the promoter regions and 5′ ends of treY and otsA for the CMD constructs. The PCR products were cloned into pFUS2 adjacent to its promoterless lacZ gene. After confirmation by sequencing using a lacZ-specific primer, the pFUS2 constructs were transferred from E. coli S-17 into strain NZP561 by conjugation. Transconjugants were passaged four times on selective media before confirmation by Southern hybridization. Membranes were hybridized successively with probes derived from pFUS2 DNA and the PCR product used to construct the relevant pFUS2 construct. The pFUS2 IDM constructs were also used to create double trehalose biosynthesis mutant strains.
Preparation of trehalose extracts.
Cells were pelleted by centrifugation from 12 ml of MRDM broth and washed twice in an equal volume of 0.9% (wt/vol) NaCl. Two-milliliter aliquots were removed from each sample and stored at −70°C for protein determination assays. The cells in the remainder of each sample were pelleted, resuspended in 300 μl of water and 300 μl of 95% ethanol, and stored at −70°C. The internal standard (200 μl of a 25-μg ml−1 sucrose solution) and ethanol at a final concentration of 70% (vol/vol) were then added to each sample. Trehalose extraction was carried out at 80°C for 20 min. Cell debris was removed by centrifugation, and the supernatant was extracted with chloroform.
Derivatization of standards and trehalose extracts.
Two sets of calibration solutions were prepared. The first set contained 200 μl of a sucrose standard solution (25 μg ml−1, internal standard) and 20, 40, 80, and 160 μl of a 0.25-g liter−1 trehalose standard solution, and the second set contained 200 μl of the sucrose standard solution and 20, 40, 80, and 160 μl of a 1.25-g liter−1 trehalose standard solution. The total volumes were adjusted to 360 μl. Calibration solutions and the rhizobial extracts were dried for 13 h using a VirTis freezemobile 12SL. Aliquots (100 μl) of pyridine and N-methyl-N-trimethyl-silytrifluoroacetamide (Pierce Biotechnology) were then added to the freeze-dried samples. The solutions were heated to 70°C for 2 h and then left at room temperature for a minimum of 24 h. After derivatization, the samples were stored at 6 to 8°C.
Gas chromatography analysis of trehalose.
Aliquots of samples and sugar calibration solutions (150 to 200 μl) were transferred into 2-ml vials containing 0.2-ml glass inserts. Analyses of sugar trimethylsilyl derivatives were carried out with a gas chromatograph (Agilent Technologies 6890N). Samples (1 μl) were injected using an Agilent 7683 autosampler. The injections were carried out in split mode (30:1) at 250°C. A flame ionization detector at 320°C was used with the following gas flow rates: air, 300 ml min−1; hydrogen, 30 ml min−1; and nitrogen, 30 ml min−1. Separation was carried out on a df BPX5 capillary column (50 m by 0.32 mm [inside diameter] by 0.50 μm; SGE International). Hydrogen was used as the carrier gas at a constant flow rate of 2.5 ml min−1 (34 cm s−1 at 60°C). The gas chromatograph oven was programmed so that the temperature increased from 50 to 300°C at a rate of 10°C min−1 and then was kept at 300°C for 20 min. Data integration and computation were performed using HP Chemstation software (Hewlett Packard). The amounts of trehalose in the rhizobial extracts were determined based on the internal sucrose standard and were expressed as μg trehalose mg−1 protein.
Determination of protein.
Sigma Quanti Proassay (QP-BCA) and bicinchoninic acid (BCA-1) kits were used to the determine protein concentrations in cell suspensions.
Analysis of treY and otsA gene expression.
Samples were taken from 200-ml MRDM broth cultures of strains at specified time points corresponding to different phases in the growth cycle. The bacterial count (CFU ml−1), protein quantity, and β-galactosidase specific activity were determined for each sample. β-Galactosidase assays were performed as described previously (33).
Plant assays.
The symbiotic phenotypes of strain NZP561 and trehalose biosynthesis mutant strains were assessed using T. repens cv. Grasslands Huia seedlings grown aseptically on slopes of nitrogen-free agar. The plants were grown under controlled environmental conditions (70% humidity; day temperature, 22 to 25°C; night temperature, 14°C; cycle consisting of 16-h days and 8-h nights). The effectiveness of the symbiosis was determined by comparing the growth responses (wet weight of foliage of individual plants 6 weeks postinoculation) of the plants subjected to the different treatments with the growth responses of uninoculated and strain NZP561-inoculated control plants.
Coinoculation studies.
Cells from early-stationary-phase (42-h) MRDM broth cultures were pelleted and resuspended in water to an optical density at 600 nm of 0.3. Dilutions of the resulting suspensions were used for preparation of the various inocula. Individual plants were inoculated with 200 μl of a mixed suspension of strain NZP561 and the appropriate mutant strain. The exact proportions of the wild-type and mutant strains present in the inoculant suspensions were confirmed by viable plate counting. After 4 weeks, nodules were removed from the plant roots (49), and ex-nodule bacteria were streaked onto GRDM plates. To determine which strain(s) occupied the nodules, six to eight colonies from each plate were replica patched onto nonselective and selective (neomycin plus gentamicin) GRDM plates.
Drying and storage.
Cells from early-stationary-phase (42-h) MRDM cultures were harvested and resuspended in MilliQ water, 1 M trehalose, or 1 M lactose solutions. Aliquots (100 μl) of each treatment were transferred into 12-well culture plates (Falcon). The initial bacterial numbers (CFU ml−1) were determined for each treatment prior to drying. The culture plates were then air dried for 2.5 h (without lids) in a laminar flow hood, and replica plates were stored at 4 and 28°C. Plates were removed immediately after drying and sampled at regular intervals up to 14 days after drying. The samples were rehydrated by addition of 1 ml water, and the numbers of surviving bacteria were determined by plate counting. Viability was defined on the basis of the number of CFU of hydrated bacteria expressed as a percentage of the number of CFU of an undried control.
Desiccation experiments.
Cells from 1-liter early-stationary-phase MRDM broth cultures of strains NZP561, HM5, and TH1 were resuspended in 12 ml of a 1.5% (wt/vol) methylcellulose solution. For each strain, the cell suspension was mixed with 50 g sterile glass beads (3 mm) and shaken for 30 min to coat the beads, which were then air dried for 2 h. The number of viable rhizobia present on the surface of the glass beads prior to storage was determined by placing 10 beads in 2 ml water, shaking the preparation for 15 min with a wrist action shaker to remove the bacteria, and plating dilutions to obtain single colonies. The coated beads were then transferred to desiccators (1 liter), within which the atmospheres were maintained at different constant relative humidities (RHs). The RH was maintained using silica gel (5% RH), a saturated solution of potassium acetate (32% RH), or water (99% RH) at 28°C (32). At weekly intervals, bead samples were removed, and the numbers of viable bacteria on the beads were determined.
Nucleotide sequence accession numbers.
The DNA sequences of the otsA and treY gene regions of NZP561 have been deposited in the GenBank database under accession numbers EF444931 and EF444930, respectively.
RESULTS
R. leguminosarum bv. trifolii strain NZP561 accumulates trehalose upon entry into the stationary phase.
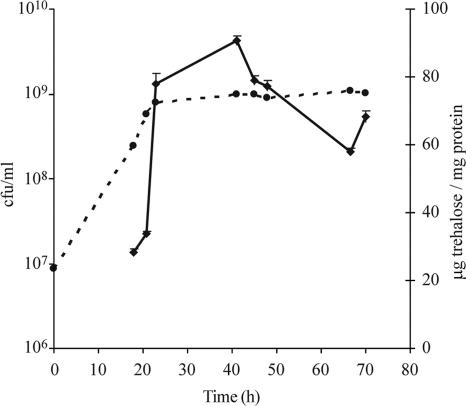
To determine whether trehalose may function as a protectant in strain R. leguminosarum bv. trifolii NZP561, the accumulation of trehalose during the growth cycle by cells grown in batch culture was measured by gas chromatography. Trehalose accumulation increased during the late exponential phase, reaching a peak during the early stationary phase (42 h), and the levels started to decline later in the stationary phase (Fig. 1). After 135 h, the level of trehalose (44 μg trehalose/mg protein) was approximately 50% of the level observed in the early stationary phase.
FIG. 1.
Trehalose accumulation by strain NZP561 during the growth cycle. The dashed line shows the growth curve (CFU ml−1) of the culture, and the solid line shows the mean trehalose concentration (μg mg protein−1); the error bars indicate standard errors. The values are the values for two independent samples, each with two replicates.
Identification of treY and otsA genes in R. leguminosarum bv. trifolii strain NZP561.
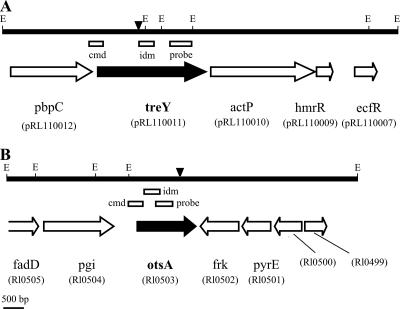
To determine the pathways used by NZP561 for trehalose biosynthesis, we decided to initially focus on the OtsA and TreY pathways as homologues of the treY and otsA genes that had been identified in other rhizobia. PCR primers (Table 2) treYF and treYR were designed from the treY gene of R. leguminosarum bv. viciae (accession number AF127795) (38), while primers otsAF and otsAR were designed from the otsA gene of R. leguminosarum bv. trifolii strain ICC105 (9). The latter gene was originally identified on the basis of its homology to otsA from pNGR234a (16). These primer pairs amplified products of appropriate sizes from NZP561 genomic DNA. The PCR products were then used to probe an NZP561 pLAFR1 cosmid library, leading to the identification of cosmids pHM3 and pHM2, respectively. The DNA sequence of the treY and otsA genes and adjacent regions was obtained by sequencing the ends of restriction fragments subcloned from the cosmids, primer walking, and sequencing out from the ends of transposon insertions (Fig. 2).
FIG. 2.
Maps of treY (A) and otsA (B) loci in R. leguminosarum bv. trifolii strain NZP561. The locations and orientations of open reading frames with homologs in databases are shown by arrows. The identification tags of corresponding genes in R. leguminosarum bv. viciae strain 3841 (58) are shown in parentheses. The positions of the transposon insertions in the EZTn5 mutants are indicated by inverted triangles. The locations of PCR products used for the construction of pFUS2 IDM and CMD strains or used as probes for library screening are shown by rectangles. E, EcoRI sites.
The treY gene from R. leguminosarum bv. trifolii strain NZP561 encoded a putative 869-amino-acid protein that shared 94% identity with a putative maltooligosyl trehalose synthase (pRL110011) located on plasmid pRL11 of R. leguminosarum bv. viciae strain 3841 (58). Hybridization of a Southern blot of an Eckhardt gel to the treY PCR product showed that treY was also on a plasmid in R. leguminosarum bv. trifolii strain NZP561 (data not shown). The arrangement of genes flanking treY in strain NZP561 (Fig. 2) was identical to the arrangement in corresponding regions in R. leguminosarum bv. viciae strains WR1-14 (38) and 3841 (58) and R. etli strain CFN42 (20). The flanking regions contained genes encoding a putative bifunctional penicillin-binding protein (PbpC), a copper-transporting P-type ATPase (ActP) involved in heavy metal stress tolerance, a copper export transcriptional regulator (HmrR), and an extracellular sigma factor (EcfR) (38).
The otsA gene from R. leguminosarum bv. trifolii strain NZP561 encoded a putative 471-amino-acid protein that shared 98% identity with a putative trehalose-6-phosphate synthase (RL0503) from R. leguminosarum bv. viciae strain 3841 (58). The genomic context of otsA was conserved in R. leguminosarum bv. trifolii strain NZP561, R. leguminosarum bv. viciae strain 3841, and R. etli strain CFN42. In all three strains, otsA was flanked by genes encoding putative proteins involved in carbohydrate, lipid, and nucleotide transport and metabolism (Fig. 2).
Trehalose synthesis in otsA, treY, and otsA treY mutants of R. leguminosarum bv. trifolii strain NZP561.
Mutations were introduced into both the otsA and treY genes using both the suicide vector pFUS2 and the EZ::Tn5 transposon, producing strains HM1, HM2, HM3, and HM4. Double otsA treY mutant strains (HM5 and TH1) were also constructed (Table 1). The amounts of trehalose accumulated by strain NZP561 and the trehalose biosynthesis mutant strains in the early stationary phase were then compared (Table 3). The highest level of trehalose accumulation was observed in strain NZP561, but the single trehalose biosynthesis mutants HM1 (otsA::EZTn5), HM2 (treY::EZTn5), and HM3 (otsA::lacZ, pFUS2) also accumulated high levels of trehalose. However, the double otsA treY mutants HM5 and TH1 failed to accumulate any trehalose (Table 3).
TABLE 3.
Trehalose accumulation by early-stationary-phase cultures of strain NZP561 and trehalose biosynthesis mutants
| Strain (genotype) | Trehalose concn (μg per mg protein)a
|
|
|---|---|---|
| 42 h | 46 h | |
| NZP561 (wild type) | 111.7 ± 3.5 (4) | 140.2 ± 4.4 (1) |
| HM1 (otsA) | 93.8 ± 2.6 (3) | 120.4 ± 1.9 (1) |
| HM2 (treY) | 101.8 ± 2.9 (2) | 134.2 ± 1.9 (1) |
| HM3 (otsA) | 91.17 ± 2.9 (3) | 130.4 ± 2.1 (1) |
| HM5 (otsA treY) | 0 (3) | 0 (1) |
| TH1 (otsA treY) | 0 (3) | 0 (1) |
The values are means ± standard errors. The number of independent assays for each strain is shown in parentheses. For each assay, two samples were analyzed in triplicate.
Growth and survival phenotypes of mutant strains.
Studies were carried out to determine if mutations in the trehalose biosynthesis genes conferred any type of growth or viability defect. The doubling times of strain NZP561 and the otsA treY mutants HM5 and TH1 in MRDM broth cultures were identical, and there was no difference in the survival rates of these strains during long-term (83 days) stationary-phase culture at 28°C (data not shown).
To investigate the effect of drying and storage on the survival of the wild-type and otsA treY mutant strains, early-stationary-phase cells of strains NZP561 and HM5 were resuspended in water, 1 M trehalose, or 1 M lactose (a nonosmoprotectant sugar) and then air dried and stored at 28°C (Table 4). This concentration of trehalose has been used previously for anhydrobiotic engineering of gram-negative bacteria (19). The survival of strains was greatest when the cells were dried in a 1 M trehalose solution. Drying had an immediate impact on cell viability, and after 1 day of storage the survival of wild-type cells dried in both water and trehalose was significantly higher than the survival of the mutant cells. This trend continued, and after 5 days of storage only strain NZP561 cells dried in trehalose remained viable. Similar but less marked trends were seen when the cells were stored at 4°C (data not shown).
TABLE 4.
Survival of early-stationary-phase cells of strain NZP561 and otsA treY mutant strain HM5 after 2.5 h of air drying and subsequent storage at 28°Ca
| Sample | % Survival of cells dried in:
|
|||||
|---|---|---|---|---|---|---|
| Water
|
1 M trehalose
|
1 M lactose
|
||||
| NZP561 | HM5 | NZP561 | HM5 | NZP561 | HM5 | |
| Initial | 100 | 100 | 100 | 100 | 100 | 100 |
| After drying | 70.2 | 62.9 | 57.6 | 53.3 | 22.8 | 24.1 |
| After storage for: | ||||||
| 1 day | 38.7 | <0.001 | 65.6 | 9.5 | <0.001 | <0.001 |
| 2 days | 8.5 | <0.001 | 17.3 | 1.0 | <0.001 | <0.001 |
| 5 days | 0 | 0 | 0.04 | <0.001 | 0 | 0 |
| 7 days | 0 | 0 | 0 | 0 | 0 | 0 |
Results from a single representative experiment are shown.
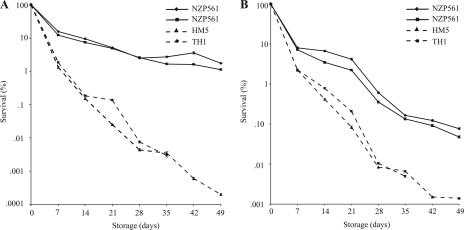
The abilities of strain NZP561 and double trehalose biosynthesis mutants to tolerate storage at different RHs were examined. Glass beads coated with 1.5% methylcellulose solutions containing strain NZP561, HM5, or TH1 were stored at RHs of 5, 32, and 99% at 28°C, and bacterial viability over time was determined (Fig. 3). No viable bacteria were detected for any strain after 7 days of storage at 99% RH. Strain NZP561 displayed significantly better survival after storage at both 32 and 5% RH than the double trehalose biosynthesis mutant strains. After 7 days at 5% RH, approximately 85 and 98% reductions in cell numbers were observed for wild-type strain NZP561 and the otsA treY mutant strains (HM5 and TH1), respectively. Thereafter, the viability of the mutant strains continued to decline at a relatively constant rate, in contrast to the viability of strain NZP561, which lost viability at a much lower rate. After storage at 5% RH for 6 to 7 weeks, there was a >1,000-fold difference in survival between strain NZP561 (>1%) and the otsA treY mutants (<0.001%). For coated beads stored at 32% RH, a similar initial reduction in cell numbers was observed, with 7 and 2% survival of the wild type and the otsA treY mutant strains, respectively, after 7 days of storage. After an additional 5 to 6 weeks of storage, strain NZP561 exhibited approximately 100-fold-higher survival than the otsA treY mutant strains.
FIG. 3.
Survival of strain NZP561 and double trehalose biosynthesis mutant strains HM5 and TH1 on glass beads after storage at RHs of 5% (A) and 32% (B) at 28°C. For each strain, the values represent the mean numbers of CFU bead−1 ± standard errors for five samples, each containing 10 beads, divided by the initial mean number of CFU bead−1 prior to storage, expressed as a percentage. Note that the y axis scales of panels A and B are different.
Expression of otsA and treY genes in strain NZP561.
To examine the expression of the otsA and treY genes, CMD strains TH3 (otsA::lacZ) and HM12 (treY::lacZ) were assayed to determine their β-galactosidase activities (Table 5). Both genes were expressed throughout the growth cycle, and their levels of expression were not affected by the growth phase. As the single trehalose biosynthesis mutants accumulated levels of trehalose comparable to the levels accumulated by wild-type strain NZP561, it was hypothesized that when one trehalose biosynthesis pathway was inactivated, expression of the remaining pathway might be increased. The treY::lacZ CMD and otsA::lacZ CMD fusions were introduced into strains HM1 (otsA::EZTn5) and HM2 (treY::EZTn5), respectively. However, β-galactosidase assays showed no increase in otsA or treY expression in the single mutant strain backgrounds compared to the corresponding CMD strains which contained intact otsA and treY genes (data not shown).
TABLE 5.
β-Galactosidase specific activities of strain NZP561 and the CMD strains HM12 (treY::lacZ) and TH3 (otsA::lacZ) throughout the growth cycle
| Growth cycle
|
β-Galactosidase sp acta
|
|||
|---|---|---|---|---|
| Time (h) | Phase | NZP561 | HM12 (treY::lacZ) | TH3 (otsA::lacZ) |
| 18 | Exponential | 38.0 ± 5.5 (4) | 164.1 ± 11.1 (4) | 94.5 ± 7.3 (3) |
| 24 | Exponential | 21.2 ± 0.8 (4) | 161.2 ± 10.4 (4) | 97.0 ± 4.6 (6) |
| 42 | Early stationary | 24.7 ± 2.6 (3) | 163.1 ± 11.2 (3) | 102.8 ± 3.2 (4) |
| 47 | Early stationary | 20.9 ± 1.6 (3) | 159.1 ± 10.3 (3) | 92.4 ± 1.2 (4) |
| 69 | Stationary | 25.1 ± 1.6 (6) | 155.3 ± 9.0 (5) | 77.6 ± 3.7 (6) |
| 90 | Stationary | 14.0 ± 2.9 (3) | 110.1 ± 72 (3) | 55.7 ± 4.9 (4) |
Expressed in nanomoles of O-nitrophenol released per minute per milligram of protein. Each sample was assayed in triplicate. The number of independent samples assayed for each strain is shown in parentheses.
Symbiotic properties of trehalose biosynthesis mutant strains.
All the trehalose biosynthesis mutant strains formed nitrogen-fixing nodules like those formed by strain NZP561 when they were inoculated onto T. repens plants (data not shown). However, coinoculation experiments which assessed the levels of nodule occupancy of strain NZ561 and the otsA treY mutant strains showed that mutant strains HM5 and TH1 occupied a lower-than-expected proportion of the nodules formed at the inoculum ratios tested (Table 6).
TABLE 6.
Competition for T. repens nodule occupancy between strain NZP561 and double trehalose biosynthesis mutants at saturating inoculum density
| Strain mixture | Inoculum ratio (NZP561/mutant) | Total no. of nodules screeneda | Expected no. of mutant nodules (%) | Observed no. of mutant nodules (%) | P valueb |
|---|---|---|---|---|---|
| NZP561 + HM5 (106 cells/plant) | 1:1 | 67 | 50 | 28.3 | 0.0002 |
| 1:3 | 57 | 75 | 43.8 | 0 | |
| 3:1 | 65 | 25 | 9.2 | 0.002 | |
| NZP561 + TH1 (106 cells/plant) | 1:1 | 66 | 50 | 33.3 | 0.009 |
| 1:3 | 25 | 75 | 52 | 0.018 | |
| NZP561 + TH1 (2 × 106 cells/plant) | 1:1 | 71 | 50 | 15.5 | 0 |
| 1:3 | 20 | 75 | 50 | 0.02 |
Nodules that contained both strains were excluded from the analysis.
A two-sided bionomial test was used to compare the observed outcomes with the expected outcomes. P values indicate the probability that the observed values differ from the expected outcomes by chance.
DISCUSSION
We have shown that R. leguminosarum bv. trifolii strain NZP561 accumulates trehalose as it enters the stationary phase due to the combined actions of the TreYZ and OtsAB pathways. A failure to accumulate trehalose rendered the strain more sensitive to the effects of drying and reduced the ability of the strains to survive storage at reduced RHs. This is the first demonstration of a direct link between endogenous synthesis of trehalose and protection against desiccation stress in rhizobia and opens up the possibility of improving desiccation tolerance of inoculant strains by manipulating intracellular trehalose levels. Gene expression studies showed that the otsA and treY genes were expressed constitutively and that expression was not influenced by the growth phase, suggesting that trehalose accumulation is controlled at the posttranscriptional level or by control of trehalose breakdown rates. Our studies also indicated that trehalose levels declined later during the stationary phase, suggesting that the cells may metabolize the accumulated trehalose at the expense of their desiccation tolerance.
The TreYZ, OtsAB, and TreS pathways were detected by an enzyme assay in B. japonicum USDA110 and B. elkanii (46). In contrast, only the OtsAB pathway was detected in S. meliloti cultures, and only the TreYZ pathway was detected in R. leguminosarum bv. viciae (46). However, osmotic stress induces genes involved in all three trehalose biosynthesis pathways in S. meliloti (14), and our studies show that two pathways are active and apparently functionally redundant in R. leguminosarum bv. trifolii strain NZP561. Putative genes for more than one trehalose biosynthesis pathway have been identified in the genome sequences of many other rhizobial species. Taken together, these observations suggest that the presence of multiple pathways for trehalose biosynthesis is widespread in rhizobia.
The OtsAB and TreYZ pathways both involve a second step, catalyzed by the OtsB and TreZ proteins, both of which belong to a large family of alpha-amylases (51). In many bacteria the otsA and otsB genes form a single operon, but neither the otsB nor treZ gene was identified from cosmid sequencing of strain NZP561. However, putative otsB and treZ genes are present in the genome sequences of all the other rhizobial species examined. In R. leguminosarum bv. viciae strain 3841, otsA is chromosomally encoded and otsB is found on plasmid pRL10, while treY and treZ are located on plasmids pRL11 and pRL12, respectively (58). Therefore, it seems highly likely that strain NZP561 has both otsB and treZ genes located elsewhere on the genome. It is also likely that R. leguminosarum bv. trifolii NZP561 has the TreS pathway as well, given that treS homologues are present in all rhizobial strains (with the possible exception of M. loti MAFF303099) for which complete genome sequences are available. This raises the question of why the otsA treY double mutants failed to accumulate trehalose. It is possible that TreS functions only in the reverse direction to degrade trehalose (see below).
Investigation of the timing of trehalose accumulation in strain NZP561 during the growth cycle (Fig. 2) revealed that the highest levels of accumulation occurred during the late exponential and early stationary phases. This result is consistent with trehalose accumulation kinetics observed for other bacteria, such as E. coli (22, 48), C. glutamicum (53), S. meliloti (50), and Pseudomonas putida (19). The timing of trehalose accumulation is also consistent with the predicted role of trehalose as a stress metabolite involved in the transition to stationary-phase survival. In E. coli, otsA expression is under the control of the stationary-phase sigma factor, RpoS (22). However, an rpoS gene has not been annotated in R. leguminosarum bv. viciae (58), and our studies indicate that otsA and treY in strain NZP561 are not subject to growth phase regulation at the transcriptional level.
R. sphaeroides and C. glutamicum have genes from all three trehalose biosynthesis pathways, and the roles of the different pathways have been characterized through analysis of mutant strains defective in individual biosynthetic routes. In R. sphaeroides, trehalose synthesis is mediated mainly by the OtsAB pathway and to a lesser extent by TreYZ (29, 53, 56). Osmoregulated trehalose synthesis in C. glutamicum is mainly dependent on TreYZ and only marginally dependent on OtsAB (56). However, under certain growth conditions, C. glutamicum mutant strains lacking a single pathway show a decrease in trehalose synthesis, but none display a total lack of trehalose production (53). In contrast, double treY otsA mutant strains of both C. glutamicum and R. sphaeroides fail to accumulate any trehalose, and their ability to recover from osmotic shock is severely impaired (53). C. glutamicum and R. sphaeroides also contain treS, but ΔtreS mutant strains show an increase in intracellular trehalose levels. In these bacteria, it appears that TreS compensates for the absence of a classical trehalase by degrading internal trehalose to maltose, thereby facilitating recycling of trehalose as a carbon source if the stress is alleviated (29, 53, 56). It may be that treS plays a similar role in strain NZP561. TreS is the dominant trehalose enzyme detected in B. japonicum bacteroids, but the concentration of maltose, the substrate for TreS, is very low in nodules (46), providing further support for the suggestion that TreS is involved in trehalose degradation.
R. leguminosarum bv. trifolii strain NZP561 was more resistant to drying and subsequent storage than a trehalose biosynthesis (otsA treY) mutant (Table 4). The enhanced survival of cells dried from trehalose solutions compared to cells dried from water is consistent with other studies that have shown that maximum protection against desiccation stress is achieved when trehalose is present on both sides of the bacterial cell membrane. Osmotic induction of intracellular trehalose synthesis combined with drying cells from trehalose solutions enhanced the desiccation tolerance and long-term viability of E. coli, P. putida, and S. enterica serovar Typhimurium (8, 19, 30, 52). The survival of S. meliloti strain 1021 during desiccation is enhanced when cells are dried in the stationary phase (55), and it was suggested that the increase in desiccation tolerance is due to the accumulation of trehalose by S. meliloti during stationary-phase growth (50). B. japonicum is unable to utilize trehalose as a carbon source, and the accumulation of intracellular trehalose by B. japonicum is enhanced by supplying trehalose during growth. The survival of trehalose-loaded cells was significantly better than that of nonloaded cells when soybean seeds were coated with cells and subjected to desiccation (44). These studies demonstrate that both intra- and extracellular trehalose are required for successful anhydrobiotic engineering of gram-negative bacteria (19).
To investigate the effectiveness of trehalose in protecting against desiccation stress under conditions more relevant to inoculant production and storage, glass beads were coated with bacteria, air dried, and stored at different RHs (Fig. 3). Strain NZP561 survived considerably better during storage at both 5 and 32% RH than the otsA treY mutant strains. This indicated that accumulated trehalose plays an important role in protecting R. leguminosarum bv. trifolii cells from desiccation stress. For strain NZP561, cells stored under the driest conditions (5% RH) displayed better survival than cells stored at a higher RH (32%). This finding, although unexpected, is not without precedent, as many conditions that affect the survival of rhizobia during desiccation have been identified (55). It has previously been shown that the driest environment (<7% RH) leads to the highest survival rates of rhizobia entrapped in polysaccharide gels (34), while storage in the presence of protecting agents (skim milk and sucrose) at a low RH (<22%) enhanced the stability of B. japonicum inocula (31). Exopolysaccharides have also been shown to protect rhizobia stored at 3% RH, but they were detrimental for survival at higher RHs (32).
Trehalose accumulation by R. leguminosarum bv. trifolii strain NZP561 was not required for nodulation and nitrogen fixation per se. However, the double trehalose biosynthesis mutant strains were less competitive for white clover nodule occupancy than strain NZP561 (Table 4). These findings are consistent with a previous study that investigated the role of trehalose in S. meliloti-alfalfa interactions (26). In S. meliloti, thuA and thuB are thought to encode a major pathway for trehalose catabolism. Mutants defective in thuA or thuB were not impaired in nodulation or nitrogen fixation of alfalfa. However, when the mutant strains were applied at a high level (106 bacteria/root), they outcompeted the wild-type S. meliloti strain 1021 when they were infecting alfalfa roots and forming nodules (26). Jensen et al. proposed that incidental accumulation of trehalose in the thu mutants due to a lack of trehalose breakdown improved the tolerance of the strains to infection-related stresses. Our results are also consistent with the conclusion that trehalose accumulation plays a role in the stress tolerance of rhizobia during nodulation.
In summary, we have shown that accumulated trehalose plays an important role in protecting R. leguminosarum bv. trifolii cells against desiccation stress and identified the pathways involved. Our objective now is to enhance desiccation tolerance and hence inoculant survival, through optimizing trehalose accumulation in stationary-phase bacteria. This may be achieved by developing strains that overproduce trehalose and/or display a reduced rate of trehalose catabolism.
Acknowledgments
We thank Michelle Leus for assistance with gas chromatography, Sheila Williams for statistical advice, and John Sullivan for advice and comments on the manuscript.
This work was supported by contract C10X0301 from the New Economy Research Fund administered by the Foundation for Research, Science and Technology, New Zealand.
Footnotes
Published ahead of print on 20 April 2007.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antoine, R., S. Alonso, D. Raze, L. Coutte, S. Lesjean, E. Willery, C. Locht, and F. Jacob-Dubuisson. 2000. New virulence-activated and virulence-repressed genes identified by systematic gene inactivation and generation of transcriptional fusions in Bordetella pertussis. J. Bacteriol. 182:5902-5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arguelles, J. C. 2000. Physiological roles of trehalose in bacteria and yeasts: a comparative analysis. Arch. Microbiol. 174:217-224. [DOI] [PubMed] [Google Scholar]
- 4.Avonce, N., A. Mendoza-Vargas, E. Morett, and G. Iturriaga. 2006. Insights on the evolution of trehalose biosynthesis. BMC Evol. Biol. 6:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyer, H. W., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 6.Breedveld, M. W., C. Dijkema, L. Zevenhuizen, and A. J. B. Zehnder. 1993. Response of intracellular carbohydrates to a NaCl shock in Rhizobium leguminosarum biovar trifolii TA1 and Rhizobium meliloti SU47. J. Gen. Microbiol. 139:3157-3163. [Google Scholar]
- 7.Breedveld, M. W., L. Zevenhuizen, and A. J. B. Zehnder. 1991. Osmotically-regulated trehalose accumulation and cyclic beta-(1,2)-glucan excretion by Rhizobium leguminosarum biovar trifolii TA1. Arch. Microbiol. 156:501-506. [Google Scholar]
- 8.Bullifent, H. L., K. Dhaliwal, A. M. Howells, K. Goan, K. Griffin, C. D. Lindsay, A. Tunnacliffe, and R. W. Titball. 2000. Stabilisation of Salmonella vaccine vectors by the induction of trehalose biosynthesis. Vaccine 19:1239-1245. [DOI] [PubMed] [Google Scholar]
- 9.Challis, B. C. 1998. Survival of Rhizobium leguminosarum. Ph.D. thesis. University of Otago, Dunedin, New Zealand.
- 10.Crowe, J. H. 2007. Trehalose as a “chemical chaperone”: fact and fantasy. Adv. Exp. Med. Biol. 594:143-158. [DOI] [PubMed] [Google Scholar]
- 11.Csonka, L. N. 1989. Physiological and genetic responses of bacteria to osmotic stress. Microbiol. Rev. 53:121-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deaker, R., R. J. Roughley, and I. R. Kennedy. 2004. Legume seed inoculation technology: a review. Soil Biol. Biochem. 36:1275-1288. [Google Scholar]
- 13.De Smet, K. A., A. Weston, I. N. Brown, D. B. Young, and B. D. Robertson. 2000. Three pathways for trehalose biosynthesis in mycobacteria. Microbiology 146:199-208. [DOI] [PubMed] [Google Scholar]
- 14.Dominguez-Ferreras, A., R. Perez-Arnedo, A. Becker, J. Olivares, M. J. Soto, and J. Sanjuan. 2006. Transcriptome profiling reveals the importance of plasmid pSymB for osmoadaptation of Sinorhizobium meliloti. J. Bacteriol. 188:7617-7625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elbein, A. D., Y. T. Pan, I. Pastuszak, and D. Carroll. 2003. New insights on trehalose: a multifunctional molecule. Glycobiology 13:17R-27R. [DOI] [PubMed] [Google Scholar]
- 16.Freiberg, C., R. Fellay, A. Bairoch, W. J. Broughton, A. Rosenthal, and X. Perret. 1997. Molecular basis of symbiosis between Rhizobium and legumes. Nature 387:394-401. [DOI] [PubMed] [Google Scholar]
- 17.Friedman, A. M., S. R. Long, S. E. Brown, W. J. Buikema, and F. M. Ausubel. 1982. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene 18:289-296. [DOI] [PubMed] [Google Scholar]
- 18.Galibert, F., T. M. Finan, S. R. Long, A. Puhler, P. Abola, F. Ampe, F. Barloy-Hubler, M. J. Barnett, A. Becker, P. Boistard, G. Bothe, M. Boutry, L. Bowser, J. Buhrmester, E. Cadieu, D. Capela, P. Chain, A. Cowie, R. W. Davis, S. Dreano, N. A. Federspiel, R. F. Fisher, S. Gloux, T. Godrie, A. Goffeau, B. Golding, J. Gouzy, M. Gurjal, I. Hernandez-Lucas, A. Hong, L. Huizar, R. W. Hyman, T. Jones, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, E. Kiss, C. Komp, V. Lelaure, D. Masuy, C. Palm, M. C. Peck, T. M. Pohl, D. Portetelle, B. Purnelle, U. Ramsperger, R. Surzycki, P. Thebault, M. Vandenbol, F. J. Vorholter, S. Weidner, D. H. Wells, K. Wong, K. C. Yeh, and J. Batut. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668-672. [DOI] [PubMed] [Google Scholar]
- 19.Garcia de Castro, A., H. Bredholt, A. R. Strom, and A. Tunnacliffe. 2000. Anhydrobiotic engineering of gram-negative bacteria. Appl. Environ. Microbiol. 66:4142-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonzalez, V., R. I. Santamaria, P. Bustos, I. Hernandez-Gonzalez, A. Medrano-Soto, G. Moreno-Hagelsieb, S. C. Janga, M. A. Ramirez, V. Jimenez-Jacinto, J. Collado-Vides, and G. Davila. 2006. The partitioned Rhizobium etli genome: genetic and metabolic redundancy in seven interacting replicons. Proc. Natl. Acad. Sci. USA 103:3834-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gouffi, K., N. Pica, V. Pichereau, and C. Blanco. 1999. Disaccharides as a new class of nonaccumulated osmoprotectants for Sinorhizobium meliloti. Appl. Environ. Microbiol. 65:1491-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hengge-Aronis, R., W. Klein, R. Lange, M. Rimmele, and W. Boos. 1991. Trehalose synthesis genes are controlled by the putative sigma factor encoded by rpoS and are involved in stationary-phase thermotolerance in Escherichia coli. J. Bacteriol. 173:7918-7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirsch, P. R., and J. E. Beringer. 1984. A physical map of pPH1J1 and pJB4J1. Plasmid 12:139-141. [DOI] [PubMed] [Google Scholar]
- 24.Howells, A. M., H. L. Bullifent, K. Dhaliwal, K. Griffin, A. Garcia de Castro, G. Frith, A. Tunnacliffe, and R. W. Titball. 2002. Role of trehalose biosynthesis in environmental survival and virulence of Salmonella enterica serovar Typhimurium. Res. Microbiol. 153:281-287. [DOI] [PubMed] [Google Scholar]
- 25.Hubber, A., A. C. Vergunst, J. T. Sullivan, P. J. J. Hooykaas, and C. W. Ronson. 2004. Symbiotic phenotypes and translocated effector proteins of the Mesorhizobium loti strain R7A VirB/D4 type IV secretion system. Mol. Microbiol. 54:561-574. [DOI] [PubMed] [Google Scholar]
- 26.Jensen, J. B., O. Y. Ampomah, R. Darrah, N. K. Peters, and T. V. Bhuvaneswari. 2005. Role of trehalose transport and utilization in Sinorhizobium meliloti-alfalfa interactions. Mol. Plant-Microbe Interact. 18:694-702. [DOI] [PubMed] [Google Scholar]
- 27.Kaneko, T., Y. Nakamura, S. Sato, E. Asamizu, T. Kato, S. Sasamoto, A. Watanabe, K. Idesawa, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, C. Kiyokawa, M. Kohara, M. Matsumoto, A. Matsuno, Y. Mochizuki, S. Nakayama, N. Nakazaki, S. Shimpo, M. Sugimoto, C. Takeuchi, M. Yamada, and S. Tabata. 2000. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 7:331-338. [DOI] [PubMed] [Google Scholar]
- 28.Kaneko, T., Y. Nakamura, S. Sato, K. Minamisawa, T. Uchiumi, S. Sasamoto, A. Watanabe, K. Idesawa, M. Iriguchi, K. Kawashima, M. Kohara, M. Matsumoto, S. Shimpo, H. Tsuruoka, T. Wada, M. Yamada, and S. Tabata. 2002. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 9:189-197. [DOI] [PubMed] [Google Scholar]
- 29.Makihara, F., M. Tsuzuki, K. Sato, S. Masuda, K. V. Nagashima, M. Abo, and A. Okubo. 2005. Role of trehalose synthesis pathways in salt tolerance mechanism of Rhodobacter sphaeroides f. sp. denitrificans IL106. Arch. Microbiol. 184:56-65. [DOI] [PubMed] [Google Scholar]
- 30.Manzanera, M., S. Vilchez, and A. Tunnacliffe. 2004. Plastic encapsulation of stabilized Escherichia coli and Pseudomonas putida. Appl. Environ. Microbiol. 70:3143-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mary, P., N. Moschetto, and R. Tailliez. 1993. Production and survival during storage of spray-dried Bradyrhizobium japonicum cell concentrates. J. Appl. Bacteriol. 74:340-344. [Google Scholar]
- 32.Mary, P., D. Ochin, and R. Tailliez. 1986. Growth status of rhizobia in relation to their tolerance to low water activities and desiccation stresses. Soil Biol. Biochem. 18:179-184. [Google Scholar]
- 33.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 34.Mugnier, J., and G. Jung. 1985. Survival of bacteria and fungi in relation to water activity and the solvent properties of water in biopolymer gels. Appl. Environ. Microbiol. 50:108-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pichereau, V., A. Hartke, and Y. Auffray. 2000. Starvation and osmotic stress induced multiresistances. Influence of extracellular compounds. Int. J. Food Microbiol. 55:19-25. [DOI] [PubMed] [Google Scholar]
- 36.Potts, M. 2001. Desiccation tolerance: a simple process? Trends Microbiol. 9:553-559. [DOI] [PubMed] [Google Scholar]
- 37.Qu, Q., S. J. Lee, and W. Boos. 2004. TreT, a novel trehalose glycosyltransferring synthase of the hyperthermophilic archaeon Thermococcus litoralis. J. Biol. Chem. 279:47890-47897. [DOI] [PubMed] [Google Scholar]
- 38.Reeve, W. G., R. P. Tiwari, N. B. Kale, M. J. Dilworth, and A. R. Glenn. 2002. ActP controls copper homeostasis in Rhizobium leguminosarum bv. viciae and Sinorhizobium meliloti preventing low pH-induced copper toxicity. Mol. Microbiol. 43:981-991. [DOI] [PubMed] [Google Scholar]
- 39.Ryu, S. I., C. S. Park, J. Cha, E. J. Woo, and S. B. Lee. 2005. A novel trehalose-synthesizing glycosyltransferase from Pyrococcus horikoshii: molecular cloning and characterization. Biochem. Biophys. Res. Commun. 329:429-436. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 41.Schiraldi, C., I. Di Lernia, and M. De Rosa. 2002. Trehalose production: exploiting novel approaches. Trends Biotechnol. 20:420-425. [DOI] [PubMed] [Google Scholar]
- 42.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilisation system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 43.Streeter, J. G. 1985. Accumulation of alpha, alpha-trehalose by Rhizobium bacteria and bacteroids. J. Bacteriol. 164:78-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Streeter, J. G. 2003. Effect of trehalose on survival of Bradyrhizobium japonicum during desiccation. J. Appl. Microbiol. 95:484-491. [DOI] [PubMed] [Google Scholar]
- 45.Streeter, J. G., and A. Bhagwat. 1999. Biosynthesis of trehalose from maltooligosaccharides in rhizobia. Can. J. Microbiol. 45:716-721. [DOI] [PubMed] [Google Scholar]
- 46.Streeter, J. G., and M. L. Gomez. 2006. Three enzymes for trehalose synthesis in Bradyrhizobium cultured bacteria and in bacteroids from soybean nodules. Appl. Environ. Microbiol. 72:4250-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Streit, W. R., R. A. Schmitz, X. Perret, C. Staehelin, W. J. Deakin, C. Raasch, H. Liesegang, and W. J. Broughton. 2004. An evolutionary hot spot: the pNGR234b replicon of Rhizobium sp. strain NGR234. J. Bacteriol. 186:535-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strøm, A. R., and I. Kaasen. 1993. Trehalose metabolism in Escherichia coli: stress protection and stress regulation of gene expression. Mol. Microbiol. 8:205-210. [DOI] [PubMed] [Google Scholar]
- 49.Sullivan, J. T., H. N. Patrick, W. L. Lowther, D. B. Scott, and C. W. Ronson. 1995. Nodulating strains of Rhizobium loti arise through chromosomal symbiotic gene transfer in the environment. Proc. Natl. Acad. Sci. USA 92:8985-8989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Talibart, R., M. Jebbar, K. Gouffi, V. Pichereau, G. Gouesbet, C. Blanco, T. Bernard, and J. Pocard. 1997. Transient accumulation of glycine betaine and dynamics of endogenous osmolytes in salt-stressed cultures of Sinorhizobium meliloti. Appl. Environ. Microbiol. 63:4657-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Timmins, J., H. K. Leiros, G. Leonard, I. Leiros, and S. McSweeney. 2005. Crystal structure of maltooligosyltrehalose trehalohydrolase from Deinococcus radiodurans in complex with disaccharides. J. Mol. Biol. 347:949-963. [DOI] [PubMed] [Google Scholar]
- 52.Tunnacliffe, A., A. Garcia de Castro, and M. Manzanera. 2001. Anhydrobiotic engineering of bacterial and mammalian cells: is intracellular trehalose sufficient? Cryobiology 43:124-132. [DOI] [PubMed] [Google Scholar]
- 53.Tzvetkov, M., C. Klopprogge, O. Zelder, and W. Liebl. 2003. Genetic dissection of trehalose biosynthesis in Corynebacterium glutamicum: inactivation of trehalose production leads to impaired growth and an altered cell wall lipid composition. Microbiology 149:1659-1673. [DOI] [PubMed] [Google Scholar]
- 54.Vieira, J., and J. Messing. 1982. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19:259-268. [DOI] [PubMed] [Google Scholar]
- 55.Vriezen, J. A., F. J. de Bruijn, and K. Nusslein. 2006. Desiccation responses and survival of Sinorhizobium meliloti USDA 1021 in relation to growth phase, temperature, chloride and sulfate availability. Lett. Appl. Microbiol. 42:172-178. [DOI] [PubMed] [Google Scholar]
- 56.Wolf, A., R. Kramer, and S. Morbach. 2003. Three pathways for trehalose metabolism in Corynebacterium glutamicum ATCC13032 and their significance in response to osmotic stress. Mol. Microbiol. 49:1119-1134. [DOI] [PubMed] [Google Scholar]
- 57.Wood, J. M., E. Bremer, L. N. Csonka, R. Kraemer, B. Poolman, T. van der Heide, and L. T. Smith. 2001. Osmosensing and osmoregulatory compatible solute accumulation by bacteria. Comp. Biochem. Physiol. Part A 130:437-460. [DOI] [PubMed] [Google Scholar]
- 58.Young, J. P. W., L. C. Crossman, A. W. B. Johnston, N. R. Thomson, Z. F. Ghazoui, K. H. Hull, M. Wexler, A. R. J. Curson, J. D. Todd, P. S. Poole, T. H. Mauchline, A. K. East, M. A. Quail, C. Churcher, C. Arrowsmith, I. Cherevach, T. Chillingworth, K. Clarke, A. Cronin, P. Davis, A. Fraser, Z. Hance, H. Hauser, K. Jagels, S. Moule, K. Mungall, H. Norbertczak, E. Rabbinowitsch, M. Sanders, M. Simmonds, S. Whitehead, and J. Parkhill. 2006. The genome of Rhizobium leguminosarum has recognizable core and accessory components. Genome Biol. 7:R34. [DOI] [PMC free article] [PubMed] [Google Scholar]