Abstract
Epilithic periphyton communities were sampled at three sites on the Minnesota shoreline of Lake Superior from June 2004 to August 2005 to determine if fecal coliforms and Escherichia coli were present throughout the ice-free season. Fecal coliform densities increased up to 4 orders of magnitude in early summer, reached peaks of up to 1.4 × 105 CFU cm−2 by late July, and decreased during autumn. Horizontal, fluorophore-enhanced repetitive-PCR DNA fingerprint analyses indicated that the source for 2% to 44% of the E. coli bacteria isolated from these periphyton communities could be identified when compared with a library of E. coli fingerprints from animal hosts and sewage. Waterfowl were the major source (68 to 99%) of periphyton E. coli strains that could be identified. Several periphyton E. coli isolates were genotypically identical (≥92% similarity), repeatedly isolated over time, and unidentified when compared to the source library, suggesting that these strains were naturalized members of periphyton communities. If the unidentified E. coli strains from periphyton were added to the known source library, then 57% to 81% of E. coli strains from overlying waters could be identified, with waterfowl (15 to 67%), periphyton (6 to 28%), and sewage effluent (8 to 28%) being the major potential sources. Inoculated E. coli rapidly colonized natural periphyton in laboratory microcosms and persisted for several weeks, and some cells were released to the overlying water. Our results indicate that E. coli from periphyton released into waterways confounds the use of this bacterium as a reliable indicator of recent fecal pollution.
An ever-increasing number of studies completed during the past 40 years have provided evidence indicating that fecal coliforms and Escherichia coli can persist in secondary, nonhost habitats (7, 18, 20, 25, 31, 45). Prolonged survival of fecal coliforms and E. coli in freshwater has been studied for several decades (17, 25). In recent years, other studies indicate that E. coli can survive in sediments and soils over extended periods of time (1, 5, 28, 37, 43, 48, 52). More recently, growth or regrowth of fecal indicator bacteria in tropical and temperate soils has also been reported (6, 13, 23, 31, 50).
Other nonhost habitats need to be examined, not only to determine the survival and possible naturalization of fecal bacteria but also to estimate their potential contribution of fecal indicator bacteria to waterways. Only a few studies, however, have examined other potential habitats for fecal bacteria, primarily vegetation and algae (7, 56, 59), insects (18), zooplankton (49), turtles (24), and fish (12). Recently, Cladophora glomerata (L.) from several Lake Michigan beaches was shown to harbor not only high densities of E. coli and enterococci (57) but also potential human pathogens such as Salmonella and Campylobacter spp. (32). Thus, nonhost habitats can harbor and enhance the survival of pathogenic bacteria released into the environment from point and nonpoint sources (7, 32).
Microbial source tracking studies have revealed that although humans or sewage effluents can be sources of fecal indicator bacteria in water and at beaches, wildlife and waterfowl also make important contributions to fecal counts. Choi et al. (9) identified sewage, birds, marsh, sediments, and barn runoff as predominant sources of enterococci in seawater at Huntington Beach, CA, and Whitman and Nevers (57) reported that E. coli counts were correlated with the number of birds (gulls) in the morning and afternoon in the water at a Lake Michigan beach.
Periphyton is a biological community of diatoms, green and blue-green algae, bacteria, protozoa, and fungi, often found as biofilms (41). These biofilms are attached to most natural or artificial submerged surfaces (e.g., vegetation, rocks, sand, mud, steel, and concrete walls) and are therefore often abundant in rivers and lakes. Epilithic periphyton attached to rocks may provide habitat for populations of fecal coliforms and E. coli in nearshore aquatic environments.
Research on the persistence and growth of fecal coliforms or E. coli in sediments and soils has contributed greatly to our understanding of the survival of fecal bacteria in secondary habitats. While previous studies have shown that E. coli attaches to macroalgae (7, 32), we were interested in determining if fecal coliforms are also present, grow, and have adapted to periphyton communities in the nearshore zone of lakes and harbors. Consequently, the objectives of this study were to (i) determine if fecal coliforms and E. coli populations are present and persist in periphyton communities from a harbor and Lake Superior, (ii) identify the most probable sources of E. coli found in periphyton, (iii) use laboratory microcosms to examine colonization and survival of E. coli in natural periphyton communities, and (iv) estimate the contribution of periphyton-borne E. coli to overlying waters.
MATERIALS AND METHODS
Site descriptions.
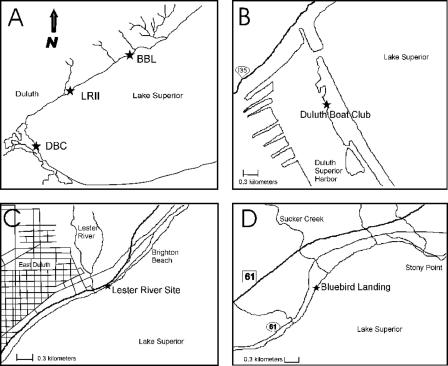
Field investigations were performed at three sampling sites (Fig. 1), the Duluth Boat Club (DBC), Lester River II (LRII), and Bluebird Landing (BBL), in the Duluth-Superior harbor and along the north shore of Lake Superior. The DBC site (Fig. 1B) is located inside the Duluth-Superior harbor (46°46.16′N, 92°05.40′W) in a rocky area between a sand beach and a dock for the U.S. Coast Guard. The LRII site (Fig. 1C) is located along the north shore of Lake Superior at the outer urban limits of Duluth, MN (46°50.23′N, 92°00.15′W), about 300 m northeast of the Lester River mouth. Residences in this part of Duluth are generally connected to sewer lines. The BBL site (Fig. 1D) is on the north shore of Lake Superior about 25 km from downtown Duluth (46°55.06′N, 91°51.15′W) and approximately 300 m southwest of the mouth of Sucker Creek. Residences in this area are not connected to the Duluth sewage treatment system.
FIG. 1.
Sampling sites in the western tip of Lake Superior and the Duluth-Superior harbor. (A) Locations of the DBC, LRII, and BBL sites. (B) DBC is located within the Duluth-Superior harbor (46°46.16′N, 092°05.40′W). (C) LRII is located along the north shore of Lake Superior at the outer urban limits of Duluth (46°50.23′N, 092°00.15′W). (D) BBL is located along the north shore of Lake Superior about 25 km from downtown Duluth (46°55.06′N, 091°51.15′W).
Sampling procedure.
Periphyton and water samples for most analyses were taken approximately monthly at each site from 19 July until 27 November 2004 and again from 1 April until 6 August 2005. The BBL site was sampled more intensively than the other sites between 25 June and 16 August 2004. Periphyton was collected at DBC in November 2004 but not at the other sites after repeated attempts, due to high waves. Ice prevented the taking of samples at all sites between November 2004 and the end of March 2005.
Periphyton at the DBC site was 2 to 6 mm thick and contained large amounts of sand and a few filamentous green algae, while periphyton at the Lake Superior sites was 1 to 3 mm thick and brownish olive in color. Microscopic analysis (46, 47) indicated that diatoms were a major component of the periphyton communities at all three sites.
Periphyton was sampled using a syringe brush sampler (54). The needle end of a 60-ml syringe was cut off, and brush fibers from a scrub brush were glued with epoxy to the end of the syringe plunger. Rocks with periphyton collected at each site were gently removed from the water. The brush sampler was then pressed against the rock and the plunger turned to scrub periphyton from a 5.2-cm2 area. The loose periphyton was rinsed into a sterile Whirl-Pak bag by using a plastic funnel pressed to the scrubbed area. Periphyton entangled in the brush was removed with a sterile wooden applicator stick and added to each sample.
Prior sampling indicated that the dry weights of periphyton and concentrations of fecal coliforms differ up to 10-fold on neighboring rocks (data not shown). Consequently, seven replicate periphyton samples per site were taken each time. Each replicate sample was a composite of eight subsamples (41.6 cm2 total) taken from individual rocks collected approximately 3 m from the shoreline in eight zones perpendicular to the shoreline. A single water sample was collected above the periphyton at each site, 3 m from shore, and subsampled (n = 4) to measure fecal coliforms and isolate E. coli. Water temperature was measured at the same depth as that at which the periphyton was collected. All periphyton and water samples were kept on ice in the dark while being transported to the lab and held at 4°C until processed; most analyses were done within 18 h of sampling.
Replicate periphyton samples were diluted to 200 ml with autoclaved and filtered (pore size, 0.2 μm) lake water and homogenized for 10 s using a commercial Hamilton Beach blender (19). Tween 80 (polyoxyethylene sorbitan monooleate) was added to the homogenized samples to a final concentration of 0.25% (33, 52), and each sample was mixed and sonicated for 3 min. This approach provided maximum cell recovery with minimal cell lysis (data not shown).
Analyses.
Several characteristics of the periphyton communities were measured for correlation with the abundance of periphyton-borne fecal coliforms at each site. The dry weight and ash-free dry weight of tripicate 10-ml aliquots of each replicate periphyton sample were determined after drying to constant weight at 60°C and subsequent combustion at 500°C for 2 h. Duplicate aliquots (10 to 40 ml) of each replicate were filtered onto Gelman A/E glass fiber filters and extracted with 10 ml of 90% acetone for 24 h at 4°C, and chlorophyll a concentrations were calculated using a spectrophotometric method (10, 51). Another aliquot of each replicate periphyton sample was fixed with formaldehyde (2% final concentration), and the abundances of total and dividing bacterial cells were estimated after 4′,6′-diamidino-2-phenylindole (DAPI) staining using epifluorescence microscopy (42, 44). Fecal coliform concentrations were measured in duplicate for all replicate periphyton samples by using the pour-plate technique. Ten milliliters of appropriate dilutions was used to inoculate m-Fecal Coliform Agar (m-FC agar) (Difco). After 24 h at 44.5°C, dark blue colonies on the surface and dark colonies in the agar were counted as fecal coliform bacteria. Fecal coliforms in water samples were quantified using the membrane filtration technique (0.45-μm-pore-size filters) and m-FC agar (10).
In 2005, gross primary production and bacterial protein production were measured in periphyton samples. Three replicate samples were chosen randomly from each site to estimate rates of gross primary production using the dissolved oxygen method (4). Rates of gross primary production were estimated from the differences between light and dark bottle incubations. Bacterial protein production was measured in each replicate periphyton sample as described previously (26, 35).
E. coli isolation and verification.
On several occasions, E. coli was isolated from periphyton and water samples for DNA fingerprinting. E. coli was isolated from periphyton at the DBC site on 19 July and 27 November 2004 and 23 May and 15 July 2005 and from water on 27 November 2004 and on both dates in 2005. At the LRII site, E. coli was isolated from periphyton on 19 July 2004 and 23 May and 15 July 2005 and from water only on 15 July 2005. At BBL, E. coli was isolated from periphyton on four occasions: 19 July and 27 November 2004 and 23 May and 15 July 2005. E. coli was isolated from water at this site on both dates in 2005.
All E. coli strains were isolated from m-FC agar plates and identified using a series of microbiological and biochemical tests (15, 31). Isolates confirmed as E. coli were cultured on plate count agar (Difco), transferred to 50% glycerol in cryovials with sterile swabs, and frozen at −80°C until DNA fingerprinting analyses could be done. A total of 996 E. coli strains were obtained from periphyton at the three sites (398, 270, and 328 strains from sites DBC, LRII, and BBL, respectively), and 207 strains were isolated from water overlying these periphyton communities (149, 37, and 21 strains from DBC, LRII, and BBL, respectively) during 2004 and 2005. The percentage of isolated colonies verified as E. coli was multiplied by the corresponding fecal coliform concentration to estimate E. coli densities in periphyton at each site.
HFERP DNA fingerprinting.
The DNA fingerprints of all isolated E. coli strains were obtained using the horizontal, fluorophore-enhanced repetitive-PCR (HFERP) method, imported into the BioNumerics software package (version 2.1; Applied Maths, Kortrijk, Belgium), and analyzed as previously described (31, 34). The most likely source of E. coli strains in periphyton and water samples was determined by comparing their HFERP fingerprints to fingerprints of E. coli strains from known animal and environmental sources in the Duluth-Superior harbor (27). The Duluth source library contained HFERP fingerprints of E. coli strains from deer (52 isolates), geese (64 isolates), gulls (127 isolates), terns (80 isolates), and beavers (38 isolates) and E. coli strains from the effluent of the Western Lake Superior Sanitary District sewage treatment plant (279 isolates). Jackknife analysis was used to determine the quality of the source library by determining the percentage of correct classifications of known source isolates. The fraction of E. coli isolates correctly assigned to their source group was expressed as a percentage of all strains correctly classified.
Dendrograms were constructed using the curve-based Pearson product-moment correlation coefficients and the unweighted pair group method with arithmetic means clustering (34). Multivariate analysis of variance was performed to cluster E. coli strains from each source group (15, 31). Identification bootstrap analysis (at P = 0.9) was performed using a BioNumerics script to identify the potential sources of environmental E. coli isolates from periphyton and water. Only source identifications with a P of ≥0.9 were accepted as correct. Since the number of E. coli strains isolated on each date was insufficient to make definitive conclusions about changes in E. coli sources over time, results from all dates were combined to estimate the annual contributions of E. coli sources in periphyton and water at each site. Periphyton-borne E. coli strains whose source could not be identified were later added to the Duluth source library (778 isolates) and used to identify which E. coli strains isolated from the overlying water might have originated from periphyton communities.
Microcosm experiment.
A microcosm experiment was conducted to examine the attachment of E. coli to periphyton-covered and bare rocks and to determine the contribution of detached E. coli to overlying water. Two rocks covered with naturally growing periphyton from the LRII site were placed in each of four replicate microcosms (38-liter aquaria). As a control treatment, rocks collected at the same time were meticulously scrubbed to remove attached periphyton, rinsed in Milli-Q water, autoclaved, and added to four replicate control microcosms. Each microcosm was filled with 16 liters of filtered (pore-size, 0.22 μm) lake water. Aquarium pumps provided constant water circulation (∼2 liters h−1). Water lost to evaporation was replaced every 10 days with fresh filtered lake water.
All microcosms were incubated at 13°C (average water temperature in the Duluth-Superior harbor during the ice-free season) in incubators lined with aluminum foil to reflect light. Gro-lux/Aquarium (Osram Sylvania, Versailles, KY) and Verilux Full Spectrum Instant Sun (Verilux, Stamford, CT) fluorescent bulbs (each 20 W) were mounted above each microcosm and programmed for a 16:8-h light-dark cycle. These wide-spectrum bulbs were selected because they have emission peaks in the photosynthetically important blue and red areas of the spectrum. Full illumination with these bulbs provided a photosynthetically active radiation irradiance of 16 to 26 μE m−2 s−1 reaching the periphyton.
Each microcosm was inoculated with a neomycin-, rifampin-, and nalidixic acid-resistant E. coli strain originally isolated from periphyton at site DBC. Resistance to neomycin and rifampin (>50 μg ml−1 each) was induced in this strain by exposure to UV light (16). A spontaneous mutant also resistant to nalidixic acid (>50 μg ml−1), designated E. coli strain NRR27, was grown in Minimal Broth Davies for 24 h (∼109 CFU ml−1), washed three times in sterile phosphate-buffered saline (pH 7.0), resuspended in phosphate-buffered saline, and used to inoculate all microcosms to a final concentration of about 2.7 × 105 CFU ml−1 microcosm water.
Two days after inoculation, the original microcosm water was removed and each microcosm was cleaned, rinsed, and refilled with 0.22-μm-filtered lake water (time zero). A composite periphyton sample, consisting of eight subsamples, and a water sample were taken from each replicate microcosm at 0, 2, 4, 6, 13, and 20 days after the water was exchanged. The abundance of E. coli NRR27 was determined using m-FC agar amended with neomycin, rifampin, and nalidixic acid (50 μg ml−1 each) after incubation for 24 h at 44.5°C. Concentrations of E. coli were also quantified on m-FC agar without added antibiotics to verify that a significant number of other E. coli strains were not present. Colony counts were never significantly different between these two media (P = 0.05). After the last sample (day 20), each microcosm was agitated by repeatedly lifting one side of the aquarium for 1 min, which created 10-cm waves to mimic conditions at the shore. Periphyton and water samples were taken again after this disturbance.
RESULTS
Annual variation in fecal coliform and E. coli concentrations in periphyton and overlying water.
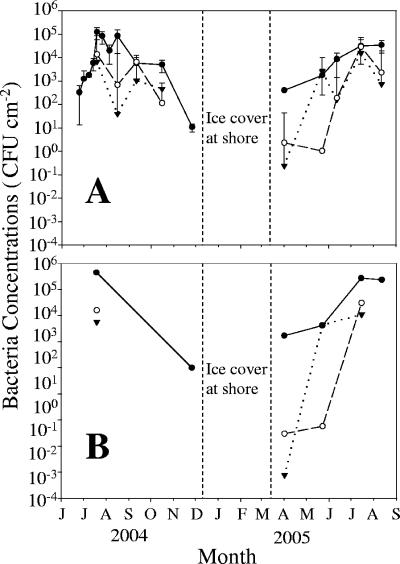
Fecal coliform concentrations in periphyton at the DBC site significantly increased (P < 0.05) in spring and the early summer, peaked in late summer, and subsequently decreased in both 2004 and 2005 (Fig. 2A). Fecal coliform concentrations at the other two sites, LRII and BBL, were more variable (Fig. 2A) but also generally increased from spring to summer and then decreased towards winter. During spring and early summer 2005, fecal coliform concentrations increased 100- to 10,000-fold in periphyton within 7 weeks at all sites but increased only fourfold during the same period in the overlying water (data not shown). E. coli concentrations in periphyton were correlated with fecal coliform concentrations (Pearson correlation, P < 0.05; Fig. 2B) and accounted for 41%, 46%, and 39% of the fecal coliforms isolated from periphyton at the DBC, LRII, and BBL sampling sites, respectively (Fig. 3A).
FIG. 2.
Seasonal changes in periphyton fecal coliforms (A) and estimated E. coli concentrations (B) at the DBC (•), LRII (○), and BBL (▾) sites. Error bars indicate standard deviations of measurements.
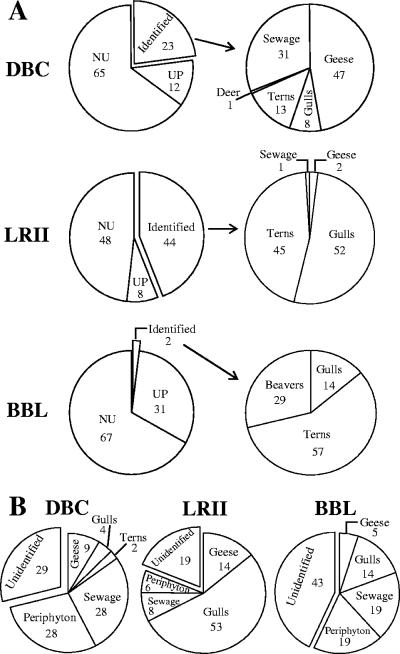
FIG. 3.
(A) Sources of E. coli in periphyton communities from the DBC, LRII, and BBL sites between July 2004 and July 2005. UP are unidentified E. coli strains that are unique to periphyton. NU are unidentified and nonunique periphyton E. coli strains. The distributions of source groups that could be identified are shown in the pie charts on the right, and the percentage in each source group is indicated. (B) Sources of E. coli in water overlying the periphyton communities at the three study sites during the same sample period. Numbers in each graph indicate percentages of each source group.
Fecal coliform and E. coli concentrations in periphyton at all sites were positively correlated (P < 0.01) with water temperature (r2 = 0.18 and 0.55, respectively) during the 13-month period. Dissolved nutrient concentrations (NH4-N, [NO2 + NO3]-N, and total P) measured in the overlying water were not correlated (P > 0.01) with periphyton fecal coliform concentrations (data not shown). When all sites were considered together, periphyton fecal coliforms were linearly related (P < 0.01) to periphyton ash-free dry weight (r2 = 0.72), chlorophyll a concentration (r2 = 0.34), and algal productivity (r2 = 0.77). However, there were no significant relationships (P > 0.01) between fecal coliforms and total bacterial abundance, dividing bacterial cell concentrations, or bacterial protein production in these periphyton communities (data not shown).
Sources of E. coli in periphyton communities.
We initially tried to identify the potential sources of E. coli isolated from each sampling site by combining the strains collected on all dates at each site and comparing their HFERP DNA fingerprints with similar DNA fingerprints of E. coli strains in the Duluth library that were isolated from known animal and environmental sources. The potential sources for the majority of the periphyton E. coli strains could not be identified from these comparisons (Fig. 3A). The percentages (and numbers) of E. coli isolates whose probable source could be identified at each site were 23% (n = 90), 44% (n = 118), and 2% (n = 7) at the DBC, LRII, and BBL sites, respectively (Fig. 3A). The major potential sources of periphyton E. coli at these sites were waterfowl (geese, terns, and gulls), sewage effluent, and beavers, but the percentages of E. coli attributed to each of these sources were different at each site (Fig. 3A). Two trends were observed. First, waterfowl were the largest potential source of E. coli in periphyton, for those strains whose source could be identified by comparison with the Duluth library of known strains. Together, E. coli strains from various waterfowl accounted for 68% (at DBC) to 99% (at LRII) of E. coli strains whose source could be identified. Second, the percentage of E. coli isolates in periphyton that originated from sewage effluent was smaller at the Lake Superior sites (BBL and LRII, 2 to 29%) than in the Duluth-Superior harbor (DBC, 47%).
Genetic relatedness of E. coli strains in periphyton.
E. coli isolates whose sources could not be identified using the Duluth library of known E. coli strains were examined further. The relative similarity values between HFERP DNA fingerprints of E. coli strains isolated from periphyton at all sites ranged from 5% to >99%. In general, HFERP fingerprints of E. coli strains from the same site clustered together. Fingerprints of E. coli strains isolated within a few months of each other were generally more similar than those of strains isolated within a year of each other.
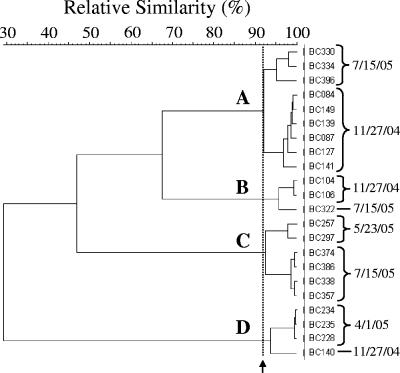
The HFERP fingerprints of some unidentified periphyton E. coli strains isolated from each of the three sites were very similar to one another (Fig. 4). For example, the fingerprints of several E. coli isolates from the DBC site obtained in November 2004, May 2005, and July 2005 were ≥92% similar. E. coli isolates whose HFERP fingerprints demonstrated ≥92% similarity were considered to be clones of the same strain (31, 34).
FIG. 4.
Partial dendrogram of unique E. coli strains isolated from epilithic periphyton at the DBC site. E. coli isolates with an HFERP fingerprint similarity of ≥92% were considered to be clones of the same strain. Four such E. coli strains, indicated by letters, were isolated from periphyton at the DBC site. The arrow indicates the 92% HFERP DNA fingerprint similarity value.
Representatives of several E. coli strains were also repeatedly isolated from each periphyton community over the course of a year, including over the winter months when air temperatures reached −40°C and the nearshore environment was frozen. For example, several isolates of one such E. coli strain were isolated from DBC periphyton in November 2004 and then again in May 2005 (cluster D in Fig. 4). Clones of two other strains were isolated in November 2004 and again in July 2005, while another DBC strain with a large number of clones (n = 48) was isolated in May and again in July 2005. Similarly, one E. coli strain was isolated from LRII periphyton in May 2005 and again in July 2005 (data not shown). At the BBL site, clones of a single strain with HFERP DNA fingerprints >96% similar (n = 99) were isolated in November 2004 and then again in May and July 2005 (data not shown).
E. coli strains from periphyton communities that were repeatedly isolated, whose HFERP fingerprints were ≥92% similar, and whose source could not be identified by comparison to the Duluth library of E. coli fingerprints were designated unique periphyton (UP) strains, indicating that they were most likely unique to the periphyton communities. The UP strains were a subset of all periphyton E. coli isolates whose source could not be identified and accounted for 8% (at LRII) to 31% (at BBL) of the E. coli strains isolated from the periphyton communities (Fig. 3A). The remaining unidentified periphyton strains were designated nonunique periphyton (NU) strains. Together, the UP and NU strains accounted for 56% (at LRII) to 98% (at BBL) of all E. coli strains that were isolated from the three periphyton communities.
Periphyton as a source of E. coli in overlying waters.
Unidentified E. coli strains isolated from periphyton were added to the Duluth library as a source group to identify the potential sources of E. coli in waters overlying periphyton communities. A jackknife analysis was performed using the UP (n = 169) and NU (n = 609) strains as individual source groups. The percentages of strains correctly classified to the UP and NU source groups were 100% and 94%, respectively, when their HFERP fingerprints were compared to those of other potential E. coli sources. When the UP and NU periphyton strains were combined, the correct classification for this larger unidentified periphyton strain group was 95%. Fingerprints of all unidentified periphyton strains were also compared to DNA fingerprints in the Duluth library using discriminant analysis and multivariate analysis of variance. Differences observed between all source groups, including the unidentified periphyton strains, were significant (P < 0.01). Low L values (0.032 and 0.133 for the first and second discriminants, respectively) also indicated that the unidentified periphyton strains came from a different population. Combined, these analyses supported addition of the unidentified periphyton strains to the Duluth source library and use of them as a separate potential source group for identifying the source of E. coli in waters overlying the periphyton communities.
Unlike those in periphyton, the majority of fecal coliforms isolated from water at two sites (70% at DBC, 86% at LRII) were E. coli, although E. coli strains accounted for only 33% of fecal coliforms isolated from water at the BBL site. When the HFERP fingerprints of these waterborne E. coli strains were compared to the Duluth fingerprint library containing the UP and NU strains, the percentages (and numbers) of E. coli isolates whose potential source could be identified were 71% (n = 106), 81% (n = 30), and 57% (n = 12) from the DBC, LRII, and BBL sites, respectively (Fig. 3B).
The major sources of E. coli in water at these sites were waterfowl, periphyton, and sewage effluent (Fig. 3B), although the distribution of these sources was different at each site. For example, while periphyton accounted for 28% of the E. coli strains isolated from water at the DBC site, sewage effluent accounted for 28%, and waterfowl (i.e., geese, gulls, and terns combined) contributed 15% of the E. coli strains found in the water. Unlike at the DBC site, waterfowl contributed most of the E. coli strains found in the water at the LRII site (67% of all E. coli strains isolated), with gulls being the largest source. Periphyton and sewage effluent were smaller sources of E. coli in water at the LRII site than at either the DBC or BBL site. At the BBL site, periphyton, sewage effluent, and waterfowl each accounted for about one-third of the E. coli strains isolated from water that could be identified.
Microcosm experiment.
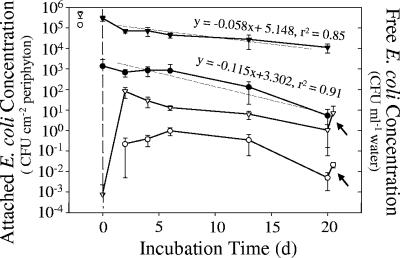
The antibiotic-resistant E. coli strain NRR27 used in the microcosm experiment attached faster to periphyton-covered rocks than to bare control rocks (Fig. 5). Two days after inoculation, densities of attached E. coli were 200-fold greater on periphyton-covered rocks than on the control rocks (2.94 × 105 versus 1.36 × 103 CFU cm−2). The abundance of attached E. coli cells decreased initially in both treatments after water in the microcosms was replaced and then stabilized briefly for 4 days before declining again. E. coli concentrations on the control rocks decreased almost twice as fast as did those of E. coli cells attached to periphyton-covered rocks (Fig. 5, P < 0.05).
FIG. 5.
Changes in E. coli NRR27 abundances in laboratory microcosms attached to periphyton-covered (▾) and bare control (•) rocks and in water overlying these rocks (periphyton-covered ▿ and bare control ○ rocks). The water in each microcosm was inoculated with E. coli NRR27 (open symbols to left of dashed line) 2 days before the experiment began and then replaced with 0.2-μm-filtered, sterile lake water at the start of the experiment (dashed line). Error bars represent standard deviations. Linear regressions indicated that the loss of E. coli from bare control rocks was almost twice as fast as that from periphyton-covered rocks. All microcosms were vigorously agitated at the end of the experiment (20 days) to simulate wave action, and E. coli concentrations in the overlying water were measured again (see arrows).
Concentrations of planktonic E. coli increased up to 5 orders of magnitude 2 days after exchange of the microcosm water. Subsequently, the maximum density of planktonic E. coli in the periphyton treatment (8.1 × 101 CFU ml−1 by day 2) slowly declined during the following 16 days of incubation. In contrast, planktonic E. coli in the bare rock control treatment reached a maximum concentration of 0.95 CFU ml−1 after 6 days before declining to about 5 × 10−3 CFU ml−1 by the end of the experiment. More than 99% of the E. coli cells remained attached to rocks in both microcosm treatments throughout the experiment. Briefly agitating the microcosms at the end of the experiment to mimic wave action released more E. coli cells from the periphyton. After the microcosms were agitated, E. coli concentrations in the overlying water increased (P < 0.01) to 5.8 CFU ml−1 (a 666% increase) in the periphyton-covered microcosms and to 2 × 10−2 CFU ml−1 (a 425% increase) in the bare control rock microcosms (indicated by arrows in Fig. 5).
DISCUSSION
Fecal coliforms and E. coli were found in natural periphyton communities from Lake Superior, and their populations grew during warm summer months and persisted through winter. While E. coli strains from waterfowl and sewage effluent were present, the original source for most E. coli strains isolated from these periphyton communities could not be identified. We contend that some E. coli strains have become naturalized members of these periphyton communities, and when detached, these bacteria contribute to fecal coliform loads detected in coastal waters near beaches.
Growth of fecal coliforms and E. coli in periphyton.
In our study, concentrations of fecal coliforms and E. coli increased several orders of magnitude in periphyton during spring and summer each year. These increases could be due to a large number of fecal coliforms in overlying waters that are available to colonize periphyton during summer when more waterfowl and animals are present; to the increased growth of active, self-sustaining fecal coliform populations in the periphyton; or to a combination of the two. The lower concentrations and smaller changes in concentrations of fecal coliform and E. coli in water than in periphyton at the field sites, however, make it unlikely that increased colonization was the only reason for the observed increase in abundances of E. coli in the periphyton communities that we studied.
Several previous studies have also demonstrated higher concentrations of fecal coliforms in water and sediments during summer (8, 29, 30, 53, 58), and growth of E. coli in nonhost environments has been reported previously (6, 11, 13, 23, 36, 50, 61). Whitman et al. (58) attributed a gradual increase of E. coli bacteria in water and sand at beaches during summer to higher survival and growth at warmer temperatures. Our data are in agreement with these observations; the abundances of fecal coliforms and E. coli in periphyton communities were positively correlated with water temperature, an important regulator of growth.
It has been previously demonstrated that enteric bacteria may persist longer in cold water than in warm water, with survival negatively related to temperature (17, 25), and that E. coli bacteria resist freezing better when previously adapted to the cold (3, 40). A downward temperature shift of ≥13°C normally induces a cold shock response in microorganisms (2). A shift from 37°C to 10°C provided optimal protection for E. coli later frozen in foods (3). Similar temperature shifts in natural environments may induce a cold shock response and may be one reason why fecal coliforms and E. coli persisted through the winter in the periphyton communities that we studied.
Sources of E. coli in periphyton communities.
Waterfowl can be an important source of fecal contamination in freshwater (56) and beach sand (57), and we demonstrated that this is also true for periphyton. Together, waterfowl (geese, terns, and gulls) contributed a large percentage of periphyton-borne E. coli strains that could be identified. Geese, gulls, and terns were frequently seen at the DBC site, and gulls were usually observed sitting on large boulders offshore at the LRII site. Although the source for the majority of periphyton E. coli isolates was not identified, and some of these strains may be unique to the periphyton communities that we examined, waterfowl were the largest source of E. coli strains from animals that could be identified.
Finding large percentages of unidentified bacterial isolates in environmental samples is not uncommon in microbial source tracking studies. Several investigators have postulated that the large fractions of unidentified isolates are due to source libraries with poor host animal representation or to limited strains from some hosts (34, 38, 39). In fact, some E. coli strains colonize very small niches within some hosts (14) and may not be well represented in fingerprint libraries like ours. E. coli strains that are better adapted to their host animal's intestinal tract may be more abundant and thus more likely to be represented in strain libraries when conventional sampling methods are used. However, other E. coli strains in host animals may be better adapted to nonhost environments like water, sediment, and periphyton. Whittam (60) found large changes in the clonal composition of E. coli populations during the transition from the host animal to an external environment. In some cases, little genetic similarity has been reported between E. coli populations in hosts and those in the environment where feces from those hosts accumulate (21, 22, 38).
Naturalization of E. coli in periphyton.
The differential survival of some fecal coliform and E. coli strains in freshwater and soils (1, 55) may lead to the development of unique environmental strains. McLellan (38) found that some E. coli strains isolated from river water were more closely related to isolates taken from different river sites or collected on other days than they were to E. coli strains from known sources of fecal contamination (e.g., gulls) at these sites. More recently, Ishii et al. (31) demonstrated that some E. coli strains from riparian soils in Lake Superior watersheds have unique HFERP fingerprints and have developed naturalized populations unique to specific soils and locations.
In this study, we found that several E. coli isolates (8% to 31% of all isolates) from each periphyton community had very similar HFERP fingerprints (≥92% relative similarity), and their origin could not be identified when their HFERP fingerprints were compared to E. coli strains isolated from known animal sources in the region. These E. coli strains were also repeatedly isolated from the periphyton communities over the course of a year, indicating that they likely persist over winter in the periphyton. Although we cannot rule out the possibility that these strains were from an animal source unknown to us, the available data strongly suggest that the strains are unique to these periphyton communities. Coupled with the knowledge that fecal coliforms and E. coli populations can grow in these periphyton communities during summer, these data imply that some E. coli strains become naturalized to periphyton communities and develop self-sustaining populations.
Periphyton as a source of E. coli in overlying waters.
E. coli rapidly colonized natural periphyton in microcosms in the laboratory and persisted in these communities for several weeks, and some of these cells were released to the overlying water (Fig. 5). Agitating the microcosms at the end of this experiment caused more E. coli cells to detach from periphyton-covered rocks and caused a subsequent large increase in planktonic E. coli abundance (>500 CFU 100 ml−1). If periphyton uniformly covered a flat bottom at our sampling sites and all periphyton E. coli cells were released simultaneously to the overlying water column when we measured the highest periphyton E. coli abundances, then we estimate that E. coli from periphyton could contribute 50 to 1,000 CFU 100 ml−1 of water at these field sites. Although it is unlikely that all E. coli cells would be released simultaneously from periphyton communities, this calculation and results from the microcosm study indicate that the number of E. coli cells potentially released from periphyton could be detected in water quality studies.
E. coli has been found to be associated with the surface of the aquatic alga Cladophora in Lake Michigan and in shoreline deposits of decaying vegetation, and E. coli attached to aquatic vegetation has been suggested to inoculate water in surrounding coastal areas (7, 32, 56, 57). When the unidentified E. coli strains that we isolated from periphyton (UP and NU strains combined) were added to our known source library, between 6% and 28% of all E. coli cells in water at the field sites that we examined may have originated from periphyton communities. Although some E. coli strains in periphyton and water could originate from a common but as-yet-unknown source, our microcosm and field data both indicate that E. coli attached to periphyton may be a relatively large source of fecal coliform bacteria detected in coastal waters.
In conclusion, although many E. coli strains isolated from periphyton may have originated from waterfowl and sewage effluent, other strains appeared to be unique to the periphyton that we studied and may have developed self-sustaining naturalized populations in these communities. E. coli cells attached to periphyton, whether they are unique to these periphyton communities or not, can detach and contribute to fecal coliform numbers measured in coastal waters. The presence, persistence, and possible naturalization of E. coli in periphyton communities further confound the use of fecal coliforms as a reliable indicator of recent fecal contamination of recreational waters. Future studies should consider periphyton and other nonhost habitats as potential sources of fecal coliform bacteria in aquatic environments.
Acknowledgments
We thank J. Schreiber, W. Hieb, M. Kading, P. Hoheisel, J. Bergin, D. Hansen, K. Alsharif, D. Gunasekera, J. Hanson, N. McCann, and J. Ferguson for technical assistance in the field and lab. We also thank R. Axler and L. Barker for reviewing our manuscript.
This work was supported, in part, by a grant from the Minnesota Sea Grant College Program, NOAA Office of Sea Grant, United States Department of Commerce, under grant no. NA03-OAR4170048 (to R.E.H. and M.J.S.).
This paper is journal reprint no. 537 of the Minnesota Sea Grant College Program.
Footnotes
Published ahead of print on 27 April 2007.
REFERENCES
- 1.Anderson, K. L., J. E. Whitlock, and V. J. Harwood. 2005. Persistence and differential survival of fecal indicator bacteria in subtropical waters and sediments. Appl. Environ. Microbiol. 71:3041-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berry, E. D., and P. M. Foegeding. 1997. Cold temperature adaptation and growth of microorganisms. J. Food Prot. 60:1583-1594. [DOI] [PubMed] [Google Scholar]
- 3.Bollman, J., A. Ismond, and G. Blank. 2001. Survival of Escherichia coli O157:H7 in frozen foods: impact of the cold shock response. Int. J. Food Microbiol. 64:127-138. [DOI] [PubMed] [Google Scholar]
- 4.Bott, T. L. 1996. Primary productivity and community respiration, p. 533-556. In F. R. Hauer and G. A. Lamberti (ed.), Methods in stream ecology. Academic Press, San Diego, CA.
- 5.Burton, G. A., D. Gunnison, and G. R. Lanza. 1987. Survival of pathogenic bacteria in various freshwater sediments. Appl. Environ. Microbiol. 53:633-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Byappanahalli, M. N., and R. S. Fujioka. 1998. Evidence that tropical soil environment can support the growth of Escherichia coli. Water Sci. Technol. 38:171-174. [Google Scholar]
- 7.Byappanahalli, M. N., D. A. Shively, M. B. Nevers, M. J. Sadowsky, and R. L. Whitman. 2003. Growth and survival of Escherichia coli and enterococci populations in the macro-alga Cladophora (Chlorophyta). FEMS Microbiol. Ecol. 1575:1-9. [DOI] [PubMed] [Google Scholar]
- 8.Byappanahalli, M. N., R. L. Whitman, D. A. Shively, M. J. Sadowsky, and S. Ishii. 2006. Population structure, persistence, and seasonality of autochthonous Escherichia coli in temperate, coastal forest soil from a Great Lakes watershed. Environ. Microbiol. 8:504-513. [DOI] [PubMed] [Google Scholar]
- 9.Choi, S., W. C. J. Brown, S. J. Becker, V. J. Harwood, and S. C. Jiang. 2003. Application of enterococci antibiotic resistance patterns for contamination source identification at Huntington Beach, California. Mar. Pollut. Bull. 46:748-755. [DOI] [PubMed] [Google Scholar]
- 10.Clesceri, L. S., A. E. Greenberg, and A. D. Eaton (ed.). 1998. Standard methods for the examination of water and wastewater, 20th ed. American Public Health Association, Washington, DC.
- 11.Davies, C. M., J. A. H. Long, M. Donald, and N. J. Ashbolt. 1995. Survival of fecal microorganisms in marine and freshwater sediments. Appl. Environ. Microbiol. 61:1888-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Del Rio-Rodriguez, R. E., V. Inglis, and S. D. Millar. 1997. Survival of Escherichia coli in the intestine of fish. Aquac. Res. 28:257-264. [Google Scholar]
- 13.Desmarais, T. R., H. M. Solo-Gabriele, and C. J. Palmer. 2002. Influence of soil on fecal indicator organisms in a tidally influenced subtropical environment. Appl. Environ. Microbiol. 68:1165-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dixit, S. M., D. M. Gordon, X.-Y. Wu, T. Chapman, K. Kailasapathy, and J. J.-C. Chin. 2004. Diversity analysis of commensal porcine Escherichia coli—associations between genotypes and habitat in the porcine gastrointestinal tract. Microbiology 150:1735-1740. [DOI] [PubMed] [Google Scholar]
- 15.Dombek, P. E., L. K. Johnson, S. T. Zimmerley, and M. J. Sadowsky. 2000. Use of repetitive DNA sequences and the PCR to differentiate Escherichia coli isolates from human and animal sources. Appl. Environ. Microbiol. 66:2572-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisenstadt, E., B. C. Carlton, and B. J. Brown. 1994. Gene mutation, p. 297-316. In P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.), Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC.
- 17.Flint, K. P. 1987. The long-term survival of Escherichia coli in river water. J. Appl. Bacteriol. 63:261-270. [DOI] [PubMed] [Google Scholar]
- 18.Geldreich, E. E., B. A. Kenner, and P. W. Kabler. 1964. Occurrence of coliforms, fecal coliforms, and streptococci on vegetation and insects. Appl. Microbiol. 12:63-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Genter, R., D. Cherry, E. P. Smith, and J. Cairns, Jr. 1987. Algal periphyton population and community changes from zinc stress in stream mesocosms. Hydrobiologia 153:261-275. [Google Scholar]
- 20.Gerba, C. P., and J. S. McLeod. 1976. Effect of sediment on the survival of Escherichia coli in marine waters. Appl. Environ. Microbiol. 32:114-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gordon, D. M. 2001. Geographical structure and host specificity in bacteria and the implications for tracing the source of coliform contamination. Microbiology 147:1079-1085. [DOI] [PubMed] [Google Scholar]
- 22.Gordon, D. M., S. Bauer, and J. R. Johnson. 2002. The genetic structure of Escherichia coli populations in primary and secondary habitats. Microbiology 148:1513-1522. [DOI] [PubMed] [Google Scholar]
- 23.Hardina, C. M., and R. S. Fujioka. 1991. Soil: the environmental source of Escherichia coli and enterococci in Hawaii's streams. Environ. Toxicol. Water Qual. 6:185-195. [Google Scholar]
- 24.Harwood, V. J., J. Butler, D. Parrisch, and V. Wagner. 1999. Isolation of fecal coliform bacteria from the diamondback terrapin (Malaclemys terrapin centrata). Appl. Environ. Microbiol. 65:865-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hendricks, C. W., and S. M. Morrison. 1967. Multiplication and growth of selected enteric bacteria in clear mountain stream water. Water Res. 1:567-576. [Google Scholar]
- 26.Hicks, R. E., P. Aas, and C. Jankovich. 2004. Annual and offshore changes in bacterioplankton communities in the western arm of Lake Superior during 1989 and 1990. J. Great Lakes Res. 30(Suppl. 1):196-213. [Google Scholar]
- 27.Hieb, W. S. 2005. Identifying the sources of fecal coliform bacteria in Lake Superior watersheds. M.S. thesis. University of Minnesota, St. Paul.
- 28.Hood, M. A., and G. E. Ness. 1982. Survival of Vibrio cholerae and Escherichia coli in estuarine waters and sediments. Appl. Environ. Microbiol. 43:578-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Howell, J. M., M. S. Coyne, and P. L. Cornelius. 1996. Effect of sediment particle size and temperature on fecal bacteria mortality rates and the fecal coliform/fecal streptococci ratio. J. Environ. Qual. 25:1216-1220. [Google Scholar]
- 30.Hyland, R., J. Byrne, B. Selinger, T. Graham, J. Thomas, I. Townshend, and V. Gannon. 2003. Spatial and temporal distribution of fecal indicator bacteria within the Oldman River basin of southern Alberta, Canada. Water Qual. Res. J. Can. 38:15-32. [Google Scholar]
- 31.Ishii, S., W. B. Ksoll, R. E. Hicks, and M. J. Sadowsky. 2006. Presence and growth of naturalized Escherichia coli in temperate soils from Lake Superior watersheds. Appl. Environ. Microbiol. 72:612-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ishii, S., T. Yan, D. A. Shively, M. N. Byappanahalli, R. L. Whitman, and M. J. Sadowsky. 2006. Cladophora (Chlorophyta) spp. harbor human bacterial pathogens in nearshore water of Lake Michigan. Appl. Environ. Microbiol. 72:4545-4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeng, D. K., L. I. Lin, and L. V. Hervey. 1990. Importance of ultrasonication conditions in recovery of microbial contamination from material surfaces. J. Appl. Bacteriol. 68:479-484. [Google Scholar]
- 34.Johnson, L.-A. K., M. B. Brown, E. A. Carruthers, J. A. Ferguson, P. E. Dombek, and M. J. Sadowsky. 2004. Sample size, library composition, and genotypic diversity among natural populations of Escherichia coli from different animals influence accuracy of determining sources of fecal pollution. Appl. Environ. Microbiol. 70:4478-4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kirchman, D., E. K'nees, and R. Hodson. 1985. Leucine incorporation and its potential as a measure of protein synthesis by bacteria in natural waters. Appl. Environ. Microbiol. 49:599-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.LaLiberte, P., and D. J. Grimes. 1982. Survival of Escherichia coli in lake bottom sediment. Appl. Environ. Microbiol. 43:623-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lopez-Torres, A. J., T. C. Hazen, and G. A. Toranzos. 1987. Distribution and in situ survival and activity of Klebsiella pneumoniae and Escherichia coli in a tropical rain forest. Curr. Microbiol. 15:213-218. [Google Scholar]
- 38.McLellan, S. L. 2004. Genetic diversity of Escherichia coli isolated from urban rivers and beach water. Appl. Environ. Microbiol. 70:4658-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McLellan, S. L., A. D. Daniels, and A. K. Salmore. 2003. Genetic characterization of Escherichia coli populations from host sources of fecal pollution using DNA fingerprinting. Appl. Environ. Microbiol. 69:2587-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mihoub, F., M. Y. Mistou, A. Guillot, J. Y. Leveau, A. Boubetra, and F. Billaux. 2003. Cold adaptation of Escherichia coli: microbiological and proteomic approaches. Int. J. Food Microbiol. 89:171-184. [DOI] [PubMed] [Google Scholar]
- 41.Moss, B. 1988. Ecology of fresh waters: man and medium, 2nd ed. Blackwell Scientific Publications, London, United Kingdom.
- 42.Newell, S. Y., and R. R. Christian. 1981. Frequency of dividing cells as an estimator of bacterial productivity. Appl. Environ. Microbiol. 42:23-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Obiri-Danso, K., and K. Jones. 2000. Intertidal sediments as reservoirs for hippurate negative campylobacters, salmonellae and faecal indicators in three EU recognized bathing waters in North West England. Water Res. 34:519-527. [Google Scholar]
- 44.Porter, K. G., and Y. S. Feig. 1980. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25:943-948. [Google Scholar]
- 45.Powers, M. L., J. Littlefield-Wyer, D. M. Gordon, D. A. Veal, and M. B. Slade. 2005. Phenotypic and genotypic characterization of encapsulated Escherichia coli isolated from blooms in two Australian lakes. Environ. Microbiol. 7:631-640. [DOI] [PubMed] [Google Scholar]
- 46.Prescott, G. W. 1973. How to know: the freshwater algae. W. C. Brown Company Publishers, Dubuque, IA.
- 47.Renberg, I. 1990. A procedure for preparing large sets of diatom slides from sediment cores. J. Paleolimnol. 4:87-90. [Google Scholar]
- 48.Sherer, B. B., J. R. Miner, J. A. Moore, and J. C. Buckhouse. 1992. Indicator bacterial survival in stream sediments. J. Environ. Qual. 21:591-595. [Google Scholar]
- 49.Signoretto, C., G. Burlacchini, M. del Mar Lleo, C. Pruzzo, M. Zampini, L. Pane, G. Franzini, and P. Canepari. 2004. Adhesion of Enterococcus faecalis in the nonculturable state to plankton is the main mechanism responsible for persistence of the bacterium in both lake and seawater. Appl. Environ. Microbiol. 70:6892-6896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Solo-Gabriele, H. M., M. A. Wolfert, T. R. Desmarais, and C. J. Palmer. 2000. Sources of Escherichia coli in a coastal subtropical environment. Appl. Environ. Microbiol. 66:230-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steinman, A. D., and G. A. Lamberti. 1996. Biomass and pigments of benthic algae, p. 295-313 In F. R. Hauer and G. A. Lamberti (ed.), Methods in stream ecology. Academic Press, San Diego, CA.
- 52.Stenstroem, T. A., and A. Carlander. 2001. Occurrence and die-off of indicator organisms in the sediment in two constructed wetlands. Water Sci. Technol. 44:223-230. [PubMed] [Google Scholar]
- 53.Stephenson, G. R., and L. V. Street. 1978. Bacterial variations in streams from a southwest Idaho rangeland watershed. J. Environ. Qual. 7:150-157. [Google Scholar]
- 54.Stockner, J. G., and F. A. J. Armstrong. 1971. Periphyton of the Experimental Lakes Area, northwestern Ontario. J. Fish Res. Board Can. 28:215-229. [Google Scholar]
- 55.Topp, E., M. Welsh, Y. C. Tien, A. Dang, G. Lazarovits, K. Conn, and H. Zhu. 2003. Strain-dependent variability in growth and survival of Escherichia coli in agricultural soil. FEMS Microbiol. Ecol. 44:303-308. [DOI] [PubMed] [Google Scholar]
- 56.Weiskel, P. K., B. L. Howes, and G. R. Heufelder. 1996. Coliform contamination of a coastal embayment: sources and transport pathways. Environ. Sci. Technol. 30:1872-1881. [Google Scholar]
- 57.Whitman, R. L., and M. B. Nevers. 2003. Foreshore sand as a source of Escherichia coli in nearshore water of a Lake Michigan beach. Appl. Environ. Microbiol. 69:5555-5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whitman, R. L., M. B. Nevers, and P. J. Gerovac. 1999. Interaction of ambient conditions and fecal coliform bacteria in southern Lake Michigan waters: monitoring program implications. Nat. Areas J. 19:166-171. [Google Scholar]
- 59.Whitman, R. L., D. A. Shively, H. Pawlik, M. B. Nevers, and M. N. Byappanahalli. 2003. Occurrence of Escherichia coli and enterococci in Cladophora (Chlorophyta) in nearshore water and beach sand of Lake Michigan. Appl. Environ. Microbiol. 69:4714-4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Whittam, T. S. 1989. Clonal dynamics of Escherichia coli in its natural habitat. Antonie Leeuwenhoek 55:23-32. [DOI] [PubMed] [Google Scholar]
- 61.Winfield, M. D., and E. A. Groisman. 2003. Role of nonhost environments in the lifestyles of Salmonella and Escherichia coli. Appl. Environ. Microbiol. 69:3687-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]