Abstract
Phytophthora ramorum is the causal agent of sudden oak death. The pathogen also affects a wide range of tree, shrub, and herbaceous species in natural and landscaped environments as well as plants in the nursery industry. A TaqMan real-time PCR method for the detection of this pathogen in the field has been described previously; this paper describes the development of a number of assays based on this method which have various advantages for use in the field. A scorpion real-time PCR assay that is twice as fast as TaqMan was developed, allowing the detection of P. ramorum in less than 30 min. Also designed was a loop-mediated isothermal amplification (LAMP) assay, which allowed sensitive and specific detection of P. ramorum in 45 min using only a heated block. A positive reaction was identified by the detection of the LAMP product by color change visible to the naked eye.
Phytophthora ramorum is a recently described pathogen (41) that causes extensive mortality of tanoak (Lithocarpus densiflorus [Hook. & Arn.] Rehd.) and Quercus spp. (by sudden oak death) in forests on the west coast of the United States (27). This pathogen also causes dieback and leaf blight in a wide range of woodland trees, shrubs, and herbaceous plant species (5). Suitable detection methods are needed in order to facilitate effective measures for the control and eradication of this pathogen. In particular, rapid methods that can be carried out in the field, minimizing the delay between sampling and diagnosis, could help reduce the spread of this pathogen.
The use of nucleic acid-based methods, especially PCR, for the identification and detection of plant pathogens is becoming increasingly widespread (17, 21, 37). In particular, real-time PCR methods have the advantages of speed, accuracy, and sensitivity over other methods (7, 28, 30, 43) and can be closed-tube systems involving no postamplification steps. TaqMan (11, 16) is the most widely used real-time PCR system, and assays using this detection chemistry have been described for a range of plant pathogens (3, 4, 14, 39, 38, 40), including assays for the detection of Phytophthora ramorum both in the laboratory (8, 12, 34) and in the field (33). A single round of real-time PCR has been found to be sufficiently sensitive to achieve the detection of 10 to 100 fg P. ramorum DNA in plant material (8, 33). However, nested PCR has been used to improve the sensitivity of detection in some matrices (8). These assays may have a number of limitations which could restrict their use in the field. For instance, many TaqMan assays use standard thermal cycling conditions, but even a rapid cycling real-time PCR instrument such as the Smart Cycler II (Cepheid, Sunnyvale, CA) takes over an hour to complete 40 cycles of standard TaqMan thermal cycling (33). This amount of time may be prohibitive when carrying out testing in the field. The use of rapid cycling instruments to perform real-time PCR in very short periods of time has been described previously (1), but increasing the speed of thermal cycling has been reported to have a detrimental effect on the sensitivity and reproducibility of some real-time PCR assays (10). Despite this effect, a reduction in the absolute sensitivity of an assay may be an acceptable compromise in circumstances where a faster result would be particularly advantageous, provided that the material to be tested is likely to contain levels of pathogen significantly in excess of the assay's limit of detection. Other real-time PCR systems commonly in use are reported to have potential advantages over TaqMan (43). In particular, scorpion primers (42), which act by a unimolecular mechanism, can be used with fast thermal cycling conditions to allow a result to be obtained in a much shorter period of time (32).
Another potential limitation of existing assays for P. ramorum is specificity. Previously described assays for P. ramorum designed within the internal transcribed spacer 1 (ITS 1) region of the nuclear ribosomal DNA have shown cross-reactions with high concentrations of DNA extracted from Phytophthora lateralis (9, 33), which differs from P. ramorum by only 11 bp in the ITS region (9). Cross-reactivity has been observed at only high DNA concentrations (more than 1 ng P. lateralis DNA in the PCR), and material being tested for P. ramorum is very unlikely to contain P. lateralis (a root pathogen). Improved specificity would allow undiluted extracts from cultures to be tested without the risk of false-positive results. One approach to improving the discrimination of similar species is to design a new assay for a different region of the genome (2, 13, 29). However, modifications to existing assays can also be made to improve specificity without the need to completely redesign the primers and/or probe. For example, the introduction of 3′ locked nucleic acid (LNA) residues into PCR primers has been shown to improve specificity in allele-specific PCRs (15). Alternatively, the use of a different real-time PCR detection chemistry could result in greater specificity. It has been reported that molecular beacons (35) can offer improved specificity over systems such as TaqMan, in which the probe does not contain a hairpin structure (36).
Despite the numerous advantages of real-time PCR over conventional PCR and other nucleic acid-based detection methods, such techniques may not be ideal for use in the field due to the complexity (and expense) of the equipment required. Loop-mediated isothermal amplification (LAMP) is a method which uses a set of four or six primers and the strand displacement activity of Bst DNA polymerase to amplify DNA with high specificity under isothermal conditions in less than 1 h (22, 23, 24). LAMP products can be visualized by gel electrophoresis, by measuring the increased turbidity (due to the production of large amounts of magnesium pyrophosphate) either in real time or at the end of the reaction (19, 20), or by using fluorescent intercalating dyes (6, 25). The simplicity of methods such as LAMP, which do not require thermal cycling, makes them well suited to field testing and potentially valuable to laboratories without real-time PCR facilities.
This paper describes modifications to a previously described TaqMan assay for P. ramorum and the development of alternative assays (scorpion, molecular beacon, and LAMP) designed using the same ITS target sequence in order to increase the feasibility of nucleic acid-based testing in the field. The relative advantages and disadvantages of each assay are considered.
MATERIALS AND METHODS
DNA extraction.
DNA was extracted from 0.5-cm2 plugs taken from cultures of P. ramorum and other Phytophthora species grown on semiselective P5ARP-(H) agar or carrot piece agar. A NucleoSpin plant kit (Machery-Nagel, Düren, Germany) was used following the manufacturer's protocol for fungi.
Real-time PCR primers and probes.
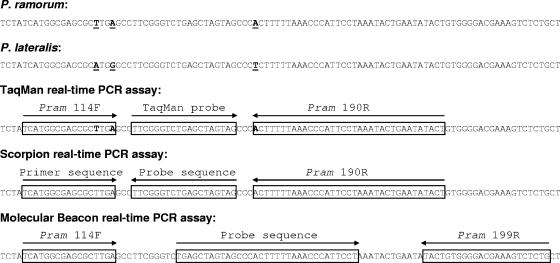
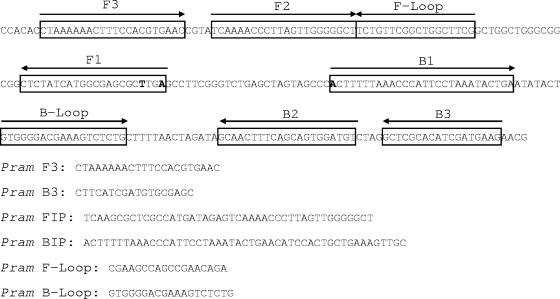
P. ramorum-specific primers (Pram-114F and Pram-190R) and a TaqMan probe (Pram probe), designed based on the ITS sequence of a range of Phytophthora spp. as previously described (12), were used as the basis for the design of a duplex scorpion primer, molecular beacon, and alternative reverse primer (Pram-199R) (Fig. 1). Primer and probe sequences, modifications, and reporter/quencher dyes are shown in Table 1. The duplex scorpion consists of two oligonucleotides: the scorpion primer itself, composed of a primer sequence attached to the reverse complement of a probe sequence (42), and a separate quencher oligonucleotide (31) which is complementary to the probe segment (Fig. 1 and Table 1). The molecular beacon is composed of a probe sequence flanked by complementary arm sequences, giving a stem-loop structure. A second reverse primer was designed to accommodate the molecular beacon sequence (Fig. 1). Six LAMP primers (external primers F3 and B3, internal primers FIP and BIP, and loop primers F-Loop and B-Loop) were designed in the same region (Fig. 2) (for details of the principle of the LAMP method, see the work of Notomi et al. [24]). TaqMan primers and probe and LAMP primers were synthesized by MWG Biotech (Ebersberg, Germany), and the duplex scorpion primer and molecular beacon were synthesized by Eurogentec (Seraing, Belgium). TaqMan primers containing LNA bases were synthesized by Sigma Proligo (Boulder, CO).
FIG. 1.
Primer and probe design for TaqMan, scorpion, and molecular beacon assays. Sequence differences between P. ramorum and P. lateralis are underlined. The positions of the LNA bases in the primers used for the LNA TaqMan are shown in bold. Sequence accession numbers are AY785958 for P. ramorum and AY785952 for P. lateralis.
TABLE 1.
Characteristics of primers and probes used for real-time PCRa
| Primer/probe | Sequence (5′-3′) | Reporter (5′) | Quencher (3′) | Final concn (μM) |
|---|---|---|---|---|
| Pram-114F | TCATGGCGAGCGCTTGA | 0.3 | ||
| Pram-190R | AGTATATTCAGTATTTAGGAATGGGTTTAAAAAGT | 0.3, 0.7e | ||
| Pram-199R | CAGAGACTTTCGTCCCCACAGTA | 0.3 | ||
| Pram-114F-LNA3 | AGTATATTCAGTATTTAGGAATGGGTTTAAAAAGTb | 0.3 | ||
| Pram-190R-LNA1 | TCATGGCGAGCGCTTGAb | 0.3 | ||
| Pram TaqMan probe | TTCGGGTCTGAGCTAGTAG | FAM | BHQ1 | 0.1 |
| Pram scorpion | CTACTAGCTCAGACCCGAA[HEG]TCATGGCGAGCGCTTGAc | FAM | 0.4 | |
| Pram quencher | TTCGGGTCTGAGCTAGTAG | DDQ | 1.6 | |
| Pram molecular beacon | CGCGGCGTGAGCTAGTAGCCCACTTTTTAAACCCATTCCTCGCCGCGd | FAM | Dabcyl | 0.3 |
FAM, 6-carboxyfluorescein; BHQ1, black hole quencher 1 (Biosearch Technologies, Novato, CA); DDQ, deep dark quencher 1 (Eurogentec, Seraing, Belgium); Dabcyl, 4-(4′-dimethylaminophenylazo)benzoic acid.
LNA bases are shown in bold.
The Scorpion primer consists of a probe sequence (complementary to the target sequence) (shown in bold) plus a primer sequence separated by a hexaethylene glycol (HEG) PCR blocker.
Molecular Beacon arm sequences are shown in bold.
The final concentration was 0.3 with the TaqMan assay and 0.7 with the scorpion assay.
FIG. 2.
Primer design for LAMP assay. Sequence differences between P. ramorum and P. lateralis are shown in bold. Internal primer FIP is composed of the complementary sequence to F1 plus the sequence F2; internal primer BIP is composed of the sequence B1 plus the complementary sequence to B2. The P. ramorum sequence accession number is AY785958.
Real-time PCR.
Real-time PCR was carried out on a Smart Cycler II (Cepheid, Sunnyvale, CA). In all cases, 1 μl of DNA extract was added to 24 μl of master mix, and negative controls containing nuclease-free water instead of DNA were included in each run. DNA extracts were tested in duplicate. TaqMan real-time PCR was carried out using a master mix consisting of 0.025 U/μl Hot Taq DNA polymerase (BioGene, Kimbolton, United Kingdom), 1× PCR buffer, 0.2 mM each deoxynucleoside triphosphate (dNTP), 5.5 mM MgCl2, 5% trehalose (wt/vol), 300 nM Pram-114F (or Pram-114F-LNA3), 300 nM Pram-190R (or Pram 190R-LNA1), and 100 nM Pram probe. Cycling conditions were 10 min at 95°C, followed by 40 two-step cycles of 15 s at 95°C and 1 min at 60°C, except where indicated otherwise. Scorpion real-time PCR was carried out using a master mix consisting of 0.08 U/μl ThermoPrime Plus DNA polymerase (ABgene, Epsom, United Kingdom), 1× buffer IV, 0.2 mM each dNTP, 5 mM MgCl2, 250 ng/μl bovine serum albumin, 400 nM Pram scorpion, 1.6 μM Pram quencher, and 700 nM Pram-190R. Cycling conditions were 15 s at 95°C, followed by 40 two-step cycles of 5 s at 95°C and 20 s at 63°C. Molecular beacon real-time PCR was carried out using a master mix consisting of 0.05 U/μl Hot Taq DNA polymerase (BioGene, Kimbolton, United Kingdom), 1× PCR buffer, 0.2 mM each dNTP, 5 mM MgCl2, 300 nM Pram-114F, 300 nM Pram-199R, and 300 nM Pram molecular beacon. Cycling conditions were 10 min at 95°C, followed by 40 three-step cycles of 30 s at 95°C, 30 s at 60°C, and 30 s at 72°C.
Real-time PCR results were analyzed in terms of threshold cycle (CT) value (the PCR cycle at which background-corrected fluorescence exceeds the threshold value). The default threshold setting (30 fluorescent units) was used throughout.
Loop-mediated isothermal amplification.
DNA extract (1 μl) was added to 24 μl of master mix, and negative controls containing nuclease-free water instead of DNA were included in each run. LAMP master mix consisted of 0.32 U/μl Bst DNA polymerase (New England Biolabs, Ipswich, MA), 1× ThermoPol buffer, 1.4 mM each dNTP, 6 mM MgCl2 (including 2 mM in Thermopol buffer), 1.2 M betaine, 200 nM each external primer (F3 and B3), 2 μM each internal primer (FIP and BIP), and 1 μM each loop primer (F-Loop and B-Loop). Reactions were incubated at 65°C for 40 min and then at 80°C for 5 min to inactivate the Bst polymerase. Amplified products were visualized by gel electrophoresis or by adding 2 μl Quant-iT PicoGreen double-stranded DNA (dsDNA) reagent (Invitrogen, Carlsbad, CA) and observing the color change (from orange to yellow). For real-time monitoring of LAMP reactions, EvaGreen dye (Biotium, Hayward, CA) (20× concentration in phosphate-buffered saline) was added to give a final concentration of 0.5×, and the reaction mixtures were held at 65°C for 40 min on the Smart Cycler, with a fluorescence reading every 60 s. Real-time LAMP results were analyzed in terms of Tp (time to a positive result) values using the default threshold setting as for real-time PCR.
RESULTS
Characterization of TaqMan assay.
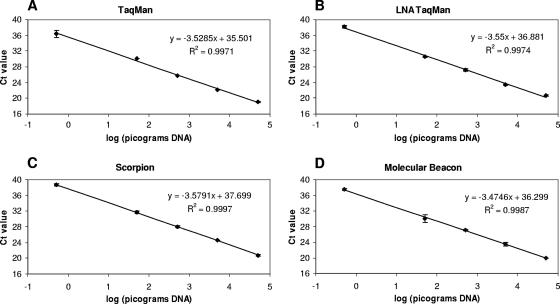
A dilution series of DNA extracted from P. ramorum culture was tested using the P. ramorum TaqMan assay in order to characterize its sensitivity and efficiency (Fig. 3A). The response of the assay was linear down to 500 fg, and the estimated efficiency of amplification [calculated from the slope of the standard curve as equal to 10(−1/slope) − 1] was 0.92. Below 500 fg, the CT values were more variable, but the lowest amount of P. ramorum DNA reliably amplified was 100 fg. DNA extracts from cultures of 28 other species of Phytophthora (Table 2) were also tested (approximately 50 to 100 ng DNA per reaction). The only species to be amplified by the TaqMan assay was P. lateralis (approximately 70 ng per reaction), which had a mean CT value of 36.39. A dilution series of DNA extracted from P. lateralis culture was also tested using the TaqMan assay, and the lowest amount of DNA detected was 7 ng (mean CT value of 39.53).
FIG. 3.
Standard curves of CT values for serial dilutions of P. ramorum DNA. CT values are mean values for duplicate reactions; error bars show standard deviations. (A) TaqMan real-time PCR. (B) TaqMan real-time PCR with LNA reverse primer. (C) Scorpion real-time PCR. (D) Molecular beacon real-time PCR.
TABLE 2.
Phytophthora sp. isolates used for specificity testing
| Phytophthora sp. | Isolate |
|---|---|
| P. boehmeriae | P 6950a |
| P. botryosa | P 6945a |
| P. cactorum | CSL 2151c |
| P. cambivora | CBS 376.61b |
| P. cinnamomi | SCRI CIN5d |
| P. citricola | SCRI CIT1d |
| P. citrophthora | IMI 132217e |
| P. cryptogea | SCRI P521d |
| P. erythroseptica | P 7889a |
| P. europaea | CBS 109053b |
| P. fragariae var. rubi | FR-163f |
| P. gonopodyides | P 10337a |
| P. heveae | CBS 958.87b |
| P. hibernalis | P 3822a |
| P. ilicis | P 3939a |
| P. insolita | P 6195a |
| P. kernoviae | CSL 2169c |
| P. lateralis | P 1728a |
| P. macrochlamydospora | P 10263a |
| P. megasperma | CBS 320.49b |
| P. nemorosa | P 10288a |
| P. nicotianae | CBS 411.87b |
| P. palmivora | SCRI P488d |
| P. pseudosyringae | P 10444a |
| P. quercina | P 10334a |
| P. richardiae | P 7788a |
| P. syringae | 4N0247-6f |
| P. uliginosa | CBS 109055b |
Provided by Michael Coffey, University of California.
Provided by Centraalbureau voor Schimmelcultures, Utrech, The Netherlands.
Provided by Central Science Laboratory, York, United Kingdom.
Provided by David Cooke, Scottish Crop Research Institute Invergowrie, Dundee, United Kingdom.
Provided by CABI Bioscience, Egham, United Kingdom.
Provided by Laboratoire National de la Protection des Vegataux, Nancy, France.
Effect of LNA substitution in PCR primers.
The effect of incorporating an LNA residue at the 3′ end of the forward and reverse primers of the TaqMan assay was investigated in terms of CT values obtained for P. ramorum and high concentrations of P. lateralis DNA. The incorporation of an LNA residue at the 3′ end of the forward primer Pram-114F (Pram-114F-LNA1) resulted in an increase in the P. ramorum CT value of approximately six cycles (data not shown). However, the incorporation of an LNA residue at the 3′ end of the reverse primer Pram-190R (Pram-190R-LNA3) resulted in a much smaller increase in CT value (fewer than two cycles). No decrease in assay efficiency was observed when a dilution series of P. ramorum DNA was tested and a standard curve was constructed (Fig. 3B). The lowest amount of P. ramorum DNA amplified was 250 fg, and 70 ng P. lateralis DNA was not amplified within 40 cycles. None of the 27 other Phytophthora species tested were amplified by this assay.
P. ramorum scorpion assay.
Amplification of P. ramorum DNA by scorpion real-time PCR was initially carried out using a range of concentrations of MgCl2 (3.5 to 5.5 mM), dNTPs (0.1 to 0.4 mM), Pram scorpion (300 to 600 nM), Pram-190R reverse primer (400 to 800 nM), ThermoPrime Plus DNA polymerase (0.02 to 0.08 U/μl), and bovine serum albumin (0 to 500 ng/μl), plus different scorpion-to-quencher ratios (1:1 to 1:10) and annealing/fluorescence monitoring temperatures (50 to 65°C, with separate or combined annealing and fluorescence-monitoring steps), in order to optimize the reaction conditions. Optimal conditions were selected on the basis of the lowest CT value and/or the highest fluorescence signal. A dilution series of DNA extracted from P. ramorum culture was tested using the optimized scorpion assay in order to characterize its sensitivity and efficiency (Fig. 3C). The response of the assay was linear down to 500 fg, and the estimated efficiency of amplification was 0.90. The lowest amount of P. ramorum DNA reliably amplified within 40 cycles was 500 fg. None of the other Phytophthora species tested, including P. lateralis (70 ng DNA), were amplified by the scorpion assay within 40 cycles.
P. ramorum molecular beacon assay.
Amplification of P. ramorum DNA by molecular beacon real-time PCR was initially carried out using a range of concentrations of MgCl2 (4 to 6 mM), dNTPs (0.1 to 0.4 mM), Pram molecular beacon (50 to 300 nM), Pram-114F forward primer (50 to 900 nM), Pram-199R reverse primer (50 to 900 nM), and Hot Taq DNA polymerase (0.025 to 0.1 U/μl), plus a range of annealing temperatures (45 to 60°C) and different thermal cycling conditions (with or without a separate extension step at 72°C) in order to optimize the reaction conditions. Optimal conditions were selected on the basis of the lowest CT value and/or highest fluorescence signal. A dilution series of DNA extracted from P. ramorum culture was tested using the molecular beacon assay in order to characterize its sensitivity and efficiency (Fig. 3D). The response of the assay was linear down to 500 fg, and the efficiency of amplification was 0.94. Of the other 28 species of Phytophthora tested, P. lateralis was amplified with a mean CT value of 36.24, and Phytophthora cactorum was amplified with a CT value of 39.30. A mean CT value of 38.83 was obtained for a second isolate of P. cactorum (isolate reference MUCL9638; Belgian Co-ordinated Collections of Micro-organisms). A dilution series of DNA extracted from P. lateralis culture was also tested using the molecular beacon assay, and the lowest amount of DNA detected was 7 ng (mean CT value of 39.83).
P. ramorum LAMP assay.
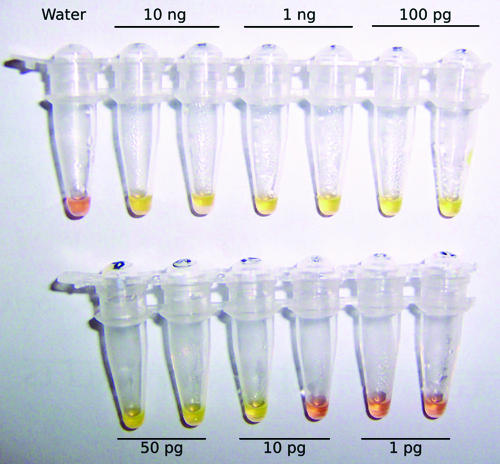
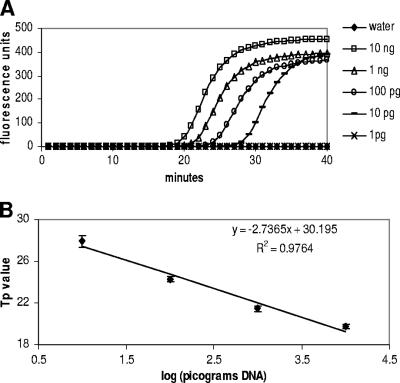
Optimization of the P. ramorum LAMP assay was carried out using gel electrophoresis to visualize the LAMP products. A range of concentrations of MgSO4 (2 to 8 mM), dNTPs (0.2 to 2 mM), primers (0.2 to 2 μM), betaine (0.8 to 1.6 M), and Bst DNA polymerase (0.32 to 0.64 U/μl), plus different incubation times (10 to 90 min), were used in order to optimize the reaction conditions. Optimal conditions were selected on the basis of the amount of product as assessed by gel electrophoresis (suboptimal conditions often resulted in no amplification) and also on the basis of cross-reactivity with P. lateralis DNA. A dilution series of P. ramorum DNA was tested using an incubation period of 40 min (Fig. 4A). Fifty picograms of P. ramorum DNA was consistently amplified, and 10 pg was amplified in one out of two replicate reactions, indicating that 10 pg is close to the limit of detection for this assay. PicoGreen dsDNA reagent also was added to the LAMP reaction tubes, and the color change from orange to yellow was observed to indicate a positive reaction. The PicoGreen results were consistent with the gel electrophoresis results (Fig. 5). In order to characterize the LAMP assay further, LAMP reactions were also run on the Smart Cycler with a fluorescent intercalating dye (EvaGreen) added to the reaction mix to allow amplification to be monitored in real time (Fig. 6A). A linear relationship between DNA concentration and Tp value was observed (Fig. 6B). In the real-time assay, there was an unambiguous distinction between positive reactions (amplification plots for 10 ng to 10 pg DNA) and negative reactions (1 pg DNA, water) after 40 min.
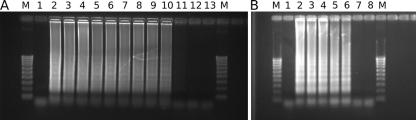
FIG. 4.
LAMP. (A) A dilution series of P. ramorum DNA was amplified by LAMP, and the product was visualized by gel electrophoresis. M, marker; lane 1, negative control (water); lanes 2 and 3, 10 ng P. ramorum DNA; lanes 4 and 5, 1 ng P. ramorum DNA; lanes 6 and 7, 100 pg P. ramorum DNA; lanes 8 and 9, 50 pg P. ramorum DNA; lanes 10 and 11, 10 pg P. ramorum DNA; lanes 12 and 13, 1 pg P. ramorum DNA. (B) Cross-reactivity of LAMP assay with P. lateralis DNA. M, marker; lane 1, negative control (water); lane 2, positive control (10 ng, P. ramorum DNA); lanes 3 and 4, 70 ng P. lateralis DNA; lanes 5 and 6, 7 ng P. lateralis DNA; lanes 7 and 8, 700 pg P. lateralis DNA.
FIG. 5.
LAMP products visualized by adding 2 μl PicoGreen dsDNA reagent at the end of the reaction. The presence of a large amount of LAMP product in positive reaction mixtures causes a color change from orange to yellow. The results shown are for the same reaction mixtures as in Fig. 4A. The amount of Phytophthora ramorum DNA added to each reaction mixture is indicated for each tube.
FIG. 6.
Real-time monitoring of LAMP reactions. (A) Amplification plots for LAMP amplification of a dilution series of P. ramorum DNA. (B) Standard curve of mean Tp values. Tp values are mean values for duplicate reactions; error bars show standard deviations.
The 28 species of Phytophthora listed in Table 2 were also tested with the LAMP assay, and the only other species to be amplified was P. lateralis, as determined by color change with PicoGreen and by gel electrophoresis. The lowest amount of P. lateralis DNA to be consistently amplified was 7 ng per reaction (Fig. 4B), while 700 pg occasionally gave a very weak smear visible with ethidium bromide staining in some replicates (data not shown). However, this was not enough product to produce a visible color change with PicoGreen.
Speed of assays.
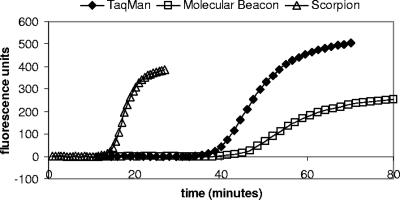
Since the Smart Cycler software displays results in real time, it is possible to obtain a positive result before the run is completed. TaqMan real-time PCR was carried out using modified cycling conditions in order to determine how much the time taken to detect P. ramorum DNA (approximately 10 ng) could be reduced without making any other modifications: mean CT values are shown in Table 3. The time taken to achieve a positive result was reduced from 39 min using standard cycling conditions to 30 min using a denaturation time of 5 s and an annealing/extension time of 30 s. This reduction in time was accompanied by an increase in CT value from 19.37 to 24.03. Further reductions of the annealing/extension period caused a greater increase in CT value, with the effect that the time taken to obtain a positive result was not reduced below 30 min (Table 3). A dilution series of P. ramorum DNA was tested using a denaturation time of 5 s and an annealing/extension time of 30 s, and a standard curve was constructed (R2 = 0.9865). The efficiency of amplification was reduced from 0.92 using standard cycling conditions to 0.65 (data not shown). The run times and time to result for the different real-time PCR assays were also compared. The Smart Cycler run times for the optimized TaqMan, molecular beacon, and scorpion assays were 70 min, 80 min, and 27 min, respectively. The time taken to detect 10 ng P. ramorum DNA was approximately 39 min for the TaqMan assay, 45 min for the molecular beacon assay, and 14 min for the scorpion assay (Fig. 7). A DNA extract from a rhododendron leaf sample containing 0.1% symptomatic P. ramorum-inoculated leaf by weight (containing approximately 10 pg P. ramorum DNA per μl) was also tested using each real-time PCR assay. The time taken to obtain a positive result was approximately 57 min for the TaqMan assay (mean CT value of 31.27), 65 min for the molecular beacon assay (mean CT value of 31.68), and 22 min for the scorpion assay (mean CT value of 32.54). A positive LAMP result for this extract was obtained in 45 min using PicoGreen detection.
TABLE 3.
Effect of changing TaqMan real-time PCR cycling conditions on CT values for the detection of P. ramorum DNAa
| Cycling conditions | Approximate run time for 40 cycles (min) | Mean CT value | Approximate time to positive result (min) |
|---|---|---|---|
| 15 s denature, 60 s anneal | 70 | 19.37 | 39 |
| 5 s denature, 60 s anneal | 63 | 19.93 | 37 |
| 5 s denature, 30 s anneal | 43 | 24.03 | 30 |
| 5 s denature, 25 s anneal | 40 | 26.98 | 30 |
| 5 s denature, 20 s anneal | 37 | 29.38 | 30 |
Approximately 10 ng DNA per reaction mixture.
FIG. 7.
Speed of real-time PCR assays. Amplification plots showing fluorescence versus time in minutes for the detection of 10 ng P. ramorum DNA using TaqMan, molecular beacon, and scorpion real-time PCR.
DISCUSSION
The aim of the work described in this paper was to develop improved methods for P. ramorum detection more specifically suited for use in the field. Some aspects of our existing TaqMan method (33), such as the run time and the complexity of the equipment required, are not ideally suited to on-site testing. The assay on which this work is based was itself adapted from a laboratory TaqMan assay (12) which is better suited to large-scale use in a routine diagnostic setting. The original assay uses “universal” thermal cycling conditions which are often used for TaqMan real-time PCR (many TaqMan primers and probes are designed to have melting temperatures which allow universal cycling conditions to be used). The use of universal cycling conditions allows different assays to be run in the same 96- or 384-well plate or two or more assays to be multiplexed in the same tube, allowing greater flexibility and efficiency for high-throughput testing. However, the run time of 90 to 120 min (depending on the real-time PCR machine used) for this assay, while acceptable in a laboratory, is not ideal for testing in the field. This is particularly true in situations in which speed is critical, for example, where material is held subject to quarantine controls. In previous direct comparisons, scorpion PCR has been found to perform better under fast cycling conditions than either TaqMan or molecular beacons (32). We also found that using faster thermal cycling conditions caused a reduction in the efficiency of the TaqMan assay, while scorpion PCR could achieve a similar level of sensitivity and efficiency in less than half the time of the other assays. Phytophthora ramorum was detected in a DNA extract from a leaf sample containing a low level of pathogen in less than 25 min using the scorpion assay, compared to nearly an hour using the previously described on-site TaqMan assay. Such time savings could clearly influence whether testing in the field is genuinely practical but also could increase throughput substantially in a routine testing laboratory.
Other potential obstacles to the use of the P. ramorum TaqMan assay in the field are the expense and complexity of the portable equipment required to perform simultaneous thermal cycling and fluorescence monitoring. However, alternative approaches, such as serological or morphological methods, can have significant disadvantages of speed, sensitivity, and specificity which limit their use in the field. Isothermal amplification of a specific DNA sequence by LAMP combined sensitivity and specificity (approaching the performance of real-time PCR methods) with requirements for only basic equipment (a water bath or heated block) and endpoint detection using a color-change reaction visible with the naked eye. The LAMP assay was found to be somewhat less sensitive than the real-time PCR assays tested but was still able to detect P. ramorum in plant material containing a low level of the pathogen. LAMP has the potential to be used in the field by nonspecialists (for example, to carry out surveillance at ports of entry or in the nursery industry) and also in small or regional laboratories in which nucleic acid-based testing is not currently performed and equipment is limited. Methods have been described for monitoring LAMP reactions in real-time using the increase in turbidity which accompanies amplification (19, 20) or using fluorescent intercalating dyes (6, 25). While the use of real-time monitoring may be very useful for characterizing assays during development and optimization, the main advantage of LAMP over real-time PCR is the ability to perform reactions without the need for any specialized equipment. The use of a simple color change detection method is particularly well suited for use in the field, and the extremely high product yield and amplification efficiency of the LAMP reaction ensure that results obtained with this method of detection are unambiguous. However, the very high yield of the LAMP reaction could also be a significant disadvantage of this method. PicoGreen dsDNA reagent completely inhibits the LAMP reaction at the concentration needed to produce a color change visible with the naked eye. However, opening the tube after the reaction has finished in order to add the PicoGreen reagent makes the method extremely vulnerable to carryover contamination due to the very large amount of product generated. Possible methods for closed-tube endpoint detection have been suggested (18). The results of some initial experiments using either a compartmentalized reaction tube (26) or a reaction tube with PicoGreen dried onto the lid (to keep the PicoGreen separate from the reaction mix until after amplification) have been promising (data not shown).
The assays described in this paper are all designed within the same target sequence in the ITS 1 region of the nuclear ribosomal DNA; however, it has been reported that assays based on other genes, including β-tubulin and elicitin (2), or on mitochondrial genes (cox I and cox II) (34) may have greater sensitivities and/or specificities than ITS-based assays. In particular, it has been reported that assays designed in regions other than ITS may distinguish between P. ramorum and P. lateralis more reliably. We have found that the specificity of the P. ramorum TaqMan assay can be improved by the introduction of an LNA base in one of the primers, without the need to design new primers and probe in a different region and without any significant adverse effect on sensitivity or efficiency. This modification has the major advantage that only one reagent needs to be replaced, reducing the cost of implementing this change and avoiding the need for reoptimization. The scorpion PCR assay also failed to amplify levels of P. lateralis DNA that were amplified by standard TaqMan PCR. The ability of the assays described in this paper to discriminate P. lateralis from P. ramorum was determined using DNA concentrations in excess of the levels likely to be encountered in the field and also somewhat higher than the extracts used to characterize the specificities of real-time PCR assays for P. ramorum in the past (8, 34). It has been reported that greater specificity can be exhibited from molecular beacons than from other probe types which do not have hairpin structures (36). While our molecular beacon assay had a sensitivity level similar to that of the original TaqMan assay, we did not observe any improvement in specificity; in fact, the molecular beacon assay was the only assay tested that amplified P. cactorum DNA.
A particular advantage of the original TaqMan assay is that it has been optimized for use in multiplex with an internal control assay for the detection of DNA from the host plant. The development of suitable internal control assays to be used with the assays described here will be extremely important for the interpretation of results, as will thorough validation of the methods in comparison with established methods. It also should be emphasized that in order to perform nucleic acid-based detection methods in the field it is crucial to have suitable nucleic acid extraction methods which can be performed with minimal equipment in the shortest possible amount of time. The DNA extraction method described previously for P. ramorum detection in the field (33) can be completed in approximately 40 to 50 min and requires only a heated block and a PickPen hand-held magnetic separation device (Bio-Nobile, Turku, Finland). This methodology could be used in conjunction with, for example, the scorpion real-time PCR assay to obtain results within 1 h of sample collection or with the LAMP assay to allow detection using only extremely basic equipment. However, this extraction method has a number of disadvantages, such as the small sample size that can be processed and the relatively low efficiency of extraction compared to those of established laboratory protocols. The development of more versatile DNA extraction methods which are even more rapid and simple to perform will be the next step in realizing the potential for performing nucleic acid-based testing for plant pathogens in the field.
Acknowledgments
This work was funded by Plant Health Division, Defra, project PH0305, and by the European Union under the 6th framework program, project EU-SSPE-RTD-502348, for the development of generic “on site” molecular diagnostics for EU quarantine pests and pathogens (PortCheck).
Footnotes
Published ahead of print on 20 April 2007.
REFERENCES
- 1.Belgrader, P., W. Benett, D. Hadley, J. Richards, P. Stratton, R. Mariella, and F. Milanovich. 1999. PCR detection of bacteria in seven minutes. Science 284:449-450. [DOI] [PubMed] [Google Scholar]
- 2.Bilodeau, G. J., C. A. Lévesque, A. W. A. M. De Cock, C. Duchaine, G. Kristjansson, and R. C. Hamelin. 2005. Molecular detection of Phytophthora ramorum by real-time PCR using TaqMan, SYBR green and molecular beacons with three genes, p. 139-140. In S. J. Frankel, P. J. Shea, and M. I. Haverty (ed.), Proceedings of the Sudden Oak Death Second Science Symposium: the state of our knowledge. Gen. Tech. Rep. PSW-GTR-196. Pacific Southwest Research Station, Forest Service, U.S. Department of Agriculture, Albany, CA.
- 3.Bonants, P. J. M., M. P. E. van Gent-Pelzer, R. Hooftman, D. E. L. Cooke, D. C. Guy, and J. M. Duncan. 2004. A combination of baiting and different PCR formats, including measurement of real-time quantitative fluorescence, for the detection of Phytophthora fragariae in strawberry plants. Eur. J. Plant Pathol. 110:689-702. [Google Scholar]
- 4.Boonham, N., L. G. Pérez, M. S. Mendez, E. L. Peralta, A. Blockley, K. Walsh, I. Barker, and R. A. Mumford. 2004. Development of a real-time RT-PCR assay for the detection of potato spindle tuber viroid. J. Virol. Methods 116:139-146. [DOI] [PubMed] [Google Scholar]
- 5.Davidson, J. M., S. Werres, M. Garbelotto, E. Hansen, and D. M. Rizzo. 7 July 2003, posting date. Sudden oak death and associated diseases caused by Phytophthora ramorum. Plant Health Prog. doi: 10.1094/PHP-2003-0707-01-DG. [DOI]
- 6.Dukes, J. P., D. P. King, and S. Alexandersen. 2006. Novel reverse transcription loop-mediated isothermal amplification for rapid detection of foot-and-mouth disease virus. Arch. Virol. 151:1093-1106. [DOI] [PubMed] [Google Scholar]
- 7.Gachon, C., A. Mingam, and B. Charrier. 2004. Real-time PCR: what relevance to plant studies? J. Exp. Bot. 55:1445-1454. [DOI] [PubMed] [Google Scholar]
- 8.Hayden, K., K. Ivors, C. Wilkinson, and M. Garbelotto. 2006. TaqMan chemistry for Phytophthora ramorum detection and quantification, with a comparison of diagnostic methods. Phytopathology 96:846-854. [DOI] [PubMed] [Google Scholar]
- 9.Hayden, K. J., D. Rizzo, J. Tse, and M. Garbelotto. 2004. Detection and quantification of Phytophthora ramorum from Californian forests using a real-time polymerase chain reaction assay. Phytopathology 94:1075-1083. [DOI] [PubMed] [Google Scholar]
- 10.Hilscher, C., W. Vahrson, and D. P. Dittmer. 2005. Faster quantitative real-time PCR protocols may lose sensitivity and show increased variability. Nucleic Acids Res. 33:e182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holland, P. M., R. D. Abramson, R. Watson, and D. H. Gelfand. 1991. Detection of specific polymerase chain reaction product by utilizing the 5′ to 3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. USA 88:7276-7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hughes, K. J. D., J. A. Tomlinson, R. L. Griffin, N. Boonham, A. J. Inman, and C. R. Lane. 2006. Development of a one-step real-time polymerase chain reaction assay for diagnosis of Phytophthora ramorum. Phytopathology 96:975-981. [DOI] [PubMed] [Google Scholar]
- 13.Ioos, R., L. Laugustin, N. Schenck, S. Rose, C. Husson, and P. Frey. 2006. Usefulness of single copy genes containing introns in Phytophthora for the development of detection tools for the regulated species P. ramorum and P. fragariae. Eur. J. Plant Pathol. 116:171-176. [Google Scholar]
- 14.Ippolito, A., L. Schena, F. Nigro, V. S. Ligorio, and T. Yaseen. 2004. Real-time detection of Phytophthora nicotianae and P. citrophthora in citrus roots and soil. Eur. J. Plant Pathol. 110:833-843. [Google Scholar]
- 15.Latorra, D., K. Arar, and J. M. Hurley. 2003. Design considerations and effects of LNA in PCR primers. Mol. Cell. Probes 17:253-259. [DOI] [PubMed] [Google Scholar]
- 16.Lee, L. G., C. R. Connell, and W. Bloch. 1993. Allelic discrimination by nick-translation PCR with fluorogenic probes. Nucleic Acids Res. 21:3761-3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin, R. R., D. James, and C. A. Lévesque. 2000. Impacts of molecular diagnostic technologies on plant disease management. Annu. Rev. Phytopathol. 38:207-239. [DOI] [PubMed] [Google Scholar]
- 18.Mori, Y., T. Hirano, and T. Notomi. 2006. Sequence specific visual detection of LAMP reactions by addition of cationic polymers. BMC Biotech. 6:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mori, Y., M. Kitao, N. Tomita, and T. Notomi. 2004. Real-time turbidimetry of LAMP reaction for quantifying template DNA. J. Biochem. Biophys. Methods 59:145-157. [DOI] [PubMed] [Google Scholar]
- 20.Mori, Y., K. Nagamine, N. Tomita, and T. Notomi. 2001. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 289:150-154. [DOI] [PubMed] [Google Scholar]
- 21.Mumford, R. A., N. Boonham, J. Tomlinson, and I. Barker. 2006. Advances in molecular phytodiagnostics—new solutions for old problems. Eur. J. Plant Pathol. 116:1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagamine, K., T. Hase, and T. Notomi. 2002. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes 16:223-229. [DOI] [PubMed] [Google Scholar]
- 23.Nagamine, K., K. Watanabe, K. Ohtsuka, T. Hase, and T. Notomi. 2001. Loop-mediated isothermal amplification reaction using a nondenatured template. Clin. Chem. 47:1742-1743. [PubMed] [Google Scholar]
- 24.Notomi, T., H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino, and T. Hase. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohtsuka, K., K. Yanagawa, K. Takatori, and Y. Hara-Kudo. 2005. Detection of Salmonella enterica in naturally contaminated liquid eggs by loop-mediated isothermal amplification, and characterization of Salmonella isolates. Appl. Environ. Microbiol. 71:6730-6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olmos, A., M. Cambra, O. Esteban, M. T. Gorris, and E. Terrada. 1999. New device and method for capture, reverse transcription and nested PCR in a single closed tube. Nucleic Acids Res. 27:1564-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rizzo, D. M., M. Garbelotto, J. M. Davidson, G. W. Slaughter, and S. T. Koike. 2002. Phytophthora ramorum as the cause of extensive mortality of Quercus spp. and Lithocarpus densiflorus in California. Plant Dis. 86:205-214. [DOI] [PubMed] [Google Scholar]
- 28.Schaad, N. W., and R. D. Frederick. 2002. Real-time PCR and its application for rapid plant disease diagnostics. Can. J. Plant Pathol. 24:250-258. [Google Scholar]
- 29.Schena, L., and D. E. L. Cooke. 2006. Assessing the potential of regions of the nuclear and mitochondrial genome to develop a “molecular toolbox” for the detection and characterization of Phytophthora species. J. Microbiol. Methods 67:70-85. [DOI] [PubMed] [Google Scholar]
- 30.Schena, L., F. Nigro, A. Ippolito, and D. Gallitelli. 2004. Real-time quantitative PCR: a new technology to detect and study phytopathogenic and antagonistic fungi. Eur. J. Plant Pathol. 110:893-908. [Google Scholar]
- 31.Solinas, A., L. J. Brown, C. McKeen, J. M. Mellor, J. T. G. Nicol, N. Thelwell, and T. Brown. 2001. Duplex scorpion primers in SNP analysis and FRET applications. Nucleic Acids Res. 29:e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thelwell, N., S. Millington, A. Solinas, J. Booth, and T. Brown. 2000. Mode of action and application of scorpion primers to mutation detection. Nucleic Acids Res. 28:3752-3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomlinson, J. A., N. Boonham, K. J. D. Hughes, R. L. Griffin, and I. Barker. 2005. On-site DNA extraction and real-time PCR for detection of Phytophthora ramorum in the field. Appl. Environ. Microbiol. 71:6702-6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tooley, P. W., F. N. Martin, M. M. Carras, and R. D. Frederick. 2006. Real-time fluorescent polymerase chain reaction detection of Phytophthora ramorum and Phytophthora pseudosyringae using mitochondrial gene regions. Phytopathology 96:336-345. [DOI] [PubMed] [Google Scholar]
- 35.Tyagi, S., and F. R. Kramer. 1996. Molecular beacons: probes that fluoresce upon hybridisation. Nat. Biotechnol. 14:303-308. [DOI] [PubMed] [Google Scholar]
- 36.Tyagi, S., D. Bratu, and F. R. Kramer. 1998. Multicolor molecular beacons for allele discrimination. Nat. Biotechnol. 16:49-53. [DOI] [PubMed] [Google Scholar]
- 37.Ward, E., S. J. Foster, B. A. Fraaije, and H. A. McCartney. 2004. Plant pathogen diagnostics: immunological and nucleic acid-based approaches. Ann. Appl. Biol. 145:1-16. [Google Scholar]
- 38.Ward, L., M. Fenn, and C. Henry. 2004. A rapid method for detection of Polymyxa DNA in soil. Plant Pathol. 53:485-490. [Google Scholar]
- 39.Ward, L. I., P. A. Beales, A. V. Barnes, and C. R. Lane. 2004. A real-time PCR assay based method for routine diagnosis of Spongospora subterranea on potato tubers. J. Phytopathol. 152:633-638. [Google Scholar]
- 40.Weller, S. A., J. G. Elphinstone, N. C. Smith, N. Boonham, and D. E. Stead. 2000. Detection of Ralstonia solanacearum strains with a quantitative, multiplex, real-time, fluorogenic PCR (TaqMan) assay. Appl. Environ. Microbiol. 66:2853-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Werres, S., R. Marwitz, W. A. Man In't Veld, A. W. A. M. De Cock, P. J. M. Bonants, M. De Weerdt, K. Themann, E. Ilieva, and R. P. Baayen. 2001. Phytophthora ramorum sp. nov., a new pathogen on Rhododendron and Viburnum. Mycol. Res. 105:1155-1165. [Google Scholar]
- 42.Whitcombe, D., J. Theaker, S. P. Guy, T. Brown, and S. Little. 1999. Detection of PCR products using self-probing amplicons and fluorescence. Nat. Biotechnol. 17:804-807. [DOI] [PubMed] [Google Scholar]
- 43.Wong, M. L., and J. F. Medrano. 2005. Real-time PCR for mRNA quantitation. BioTechniques 39:75-85. [DOI] [PubMed] [Google Scholar]