Abstract
Noroviruses have received increased attention in recent years because their role as etiologic agents in acute gastroenteritis outbreaks is now clearly established. Our inability to grow them in cell culture and the lack of an animal model hinder the characterization of these viruses. More recently, molecular approaches have been used to study the genetic relationships that exist among them. In the present study, environmental samples from seawater, estuarine water, and effluents of sewage treatment plants were analyzed in order to evaluate the role of environmental surface contamination as a possible vehicle for transmission of norovirus genogroups I and II. Novel broad-range reverse transcription-PCR/nested assays targeting the region coding for the RNA-dependent RNA polymerase were developed, amplifying fragments of 516 bp and 687 bp in the nested reactions for genogroups II and I, respectively. The assays were evaluated and compared against widely used published assays. The newly designed assays provide long regions for high-confidence BLAST searches in public databases and therefore are useful diagnostic tools for molecular diagnosis and typing of human noroviruses in clinical and environmental samples, as well as for the study of molecular epidemiology and the evolution of these viruses.
Noroviruses (NoVs) cause the majority of acute viral gastroenteritis cases worldwide (2, 9, 24). These viruses belong to the genus Norovirus within the family Caliciviridae and are divided into five separate genogroups (GI, GII, GIII, GIV, and GV) further divided into genetic clusters or antigenic types, of which GI, GII, and GIV have been detected in humans (59). NoV infections cause symptoms of severe vomiting, watery diarrhea, nausea, abdominal cramps, fever, and general malaise (17, 58). Primary infection results from the ingestion of fecally contaminated food or water, while secondary infection results from person-to-person contact, aerosolized vomitus, fomites, and infected food handlers (6, 23, 25, 28, 33, 46). Low-level transmission can occur via contaminated drinking supplies when surface water or groundwater supplies are contaminated (4, 16, 26, 31, 47). Fresh and marine waters are subjected to fecal contamination because of intense human activities which lead to continuous inputs in specific areas. Unintentional ingestion of contaminated recreational waters while swimming can then lead to gastrointestinal illness, even in nonoutbreak settings (41). Moreover, filter feeders, such as oysters, that live in contaminated waters become contaminated food sources (44, 48, 54).
NoVs have traditionally been detected using molecular methods since they cannot be cultured in cells; among these methods, reverse transcription-PCR (RT-PCR) is currently the most sensitive diagnostic assay for the detection of NoVs in stool and environmental samples. Coupled with sequence-based approaches, RT-PCR has begun to unravel the complex epidemiology of NoV infections. The application of these newer assays has significantly increased the recognition of the importance of human NoV as the cause of sporadic and outbreak-associated gastroenteritis.
Italy, unlike other countries, has no specific surveillance system for viral gastroenteritis, and laboratory diagnosis is rarely performed; therefore, few epidemiological data are available regarding NoV gastroenteritis (34, 39, 43).
Moreover, even if the role of water as a viral dissemination vehicle is widely known and reported for NoVs, to our knowledge no documentation exists regarding water environment diffusion of NoVs in Italy.
In this work we investigated the presence of human NoV GI and GII in seawaters, estuarine waters, and effluents of sewage treatment plants in Rome in order to evaluate environmental surface contamination. Finding enteric viruses in the water contributes to the understanding of the mechanisms of viral transmission and to the determination of the possible role played by water as a vehicle for transmission.
Further, in order to study the similarity of viruses found in water to those causing clinical problems in the community, we analyzed clinical samples collected from children with symptoms of acute sporadic gastroenteritis hospitalized at the Policlinico Umberto I in Rome.
Sequencing and phylogenetic analysis of a highly conserved region of the RNA-dependent RNA polymerase (RdRp) region were used to detect and characterize NoVs in clinical and environmental samples.
Molecular characterization of NoV isolates was first performed using previously published seminested RT-PCR assays. Newly designed nested RT-PCR tests able to amplify longer PCR products were designed for GI and GII, in order to increase the detection rate of NoVs and to further display the high genetic polymorphism of different strains.
MATERIALS AND METHODS
Environmental samples. (i) River, estuarine, and seawater samples.
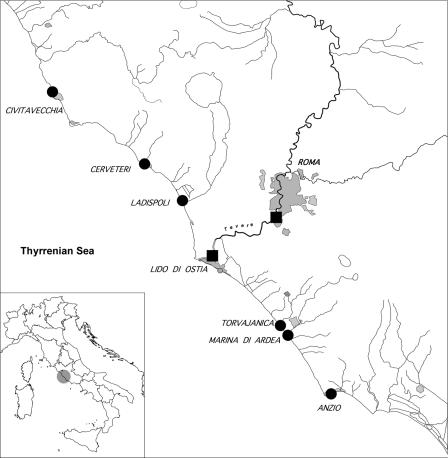
Fourteen river, estuarine, and seawater samples were collected in June 2004 at the major and minor branches of the Tiber River (Table 1, samples 323 to 336) with the help of the Coast Guard of Roma-Fiumicino, who kindly provided a suitable boat and specialized personnel. Nineteen seawater samples were collected in swimming areas during 2005 by the Arpalazio Environmental Agency (Table 1, samples 632 to 651) along 120 km of the Lazio coast (Fig. 1). Ten liters of each sample was collected with a tank and stored at +4°C upon arrival in the laboratory; each sample was assigned a code in our PostgreSQL database based on its location and history. Samples were usually processed within 1 day in order to concentrate viral particles.
TABLE 1.
Environmental samples
| Sample ID | Water source | Sampling site | Date of sampling
|
PCR result | Genogroup(s) | |
|---|---|---|---|---|---|---|
| Yr | Mo | |||||
| 323 | River | Fiumicino | 2004 | June | − | |
| 324 | Estuary | Ostia | 2004 | June | − | |
| 325 | Seawater | Fiumicino | 2004 | June | − | |
| 326 | River | Fiumicino | 2004 | June | − | |
| 327 | Estuary | Fiumicino | 2004 | June | − | |
| 328 | Seawater | Fiumicino | 2004 | June | − | |
| 329 | Estuary | Ostia | 2004 | June | − | |
| 330 | Seawater | Ostia | 2004 | June | − | |
| 331 | Seawater | Fiumicino | 2004 | June | − | |
| 332 | Seawater | Fiumicino | 2004 | June | − | |
| 333 | Estuary | Fiumicino | 2004 | June | − | |
| 334 | Estuary | Fiumicino | 2004 | June | − | |
| 335 | Seawater | Ostia | 2004 | June | − | |
| 336 | Seawater | Ostia | 2004 | June | − | |
| 632 | Seawater | Anzio | 2005 | July | − | |
| 633 | Seawater | S. Marinella | 2005 | August | − | |
| 634 | Seawater | Fiumicino | 2005 | August | − | |
| 635 | Seawater | Nettuno | 2005 | August | − | |
| 636 | Seawater | Cerveteri | 2005 | August | − | |
| 637 | Seawater | Ardea | 2005 | August | − | |
| 638 | Seawater | Cerveteri | 2005 | August | − | |
| 639 | Seawater | Fiumicino | 2005 | August | − | |
| 640 | Seawater | Ladispoli | 2005 | August | − | |
| 641 | Seawater | Pomezia | 2005 | August | − | |
| 642 | Seawater | Roma | 2005 | September | − | |
| 644 | Seawater | Fiumicino | 2005 | September | − | |
| 645 | Seawater | Fiumicino | 2005 | September | + | GII |
| 646 | Seawater | Anzio | 2005 | September | − | |
| 647 | Seawater | Ardea | 2005 | September | − | |
| 648 | Seawater | Pomezia | 2005 | September | − | |
| 649 | Seawater | Cerveteri | 2005 | September | + | GII |
| 650 | Seawater | Civitavecchia | 2005 | September | + | GII |
| 651 | Seawater | Fiumicino | 2005 | September | − | |
| 671 | Sewage | Roma Sud | 2006 | January | + | GII + GI |
| 672 | Sewage | Roma Sud | 2006 | January | + | GII |
| 677 | Sewage | Roma Sud | 2006 | February | + | GII + GI |
| 678 | Sewage | Roma Sud | 2006 | February | + | GII + GI |
| 679 | Sewage | Roma Sud | 2006 | March | + | GII + GI |
| 680 | Sewage | Roma Sud | 2006 | March | + | GII + GI |
| 699 | Sewage | Ostia | 2006 | April | + | GII |
| 700 | Sewage | Ostia | 2006 | April | + | GII |
| 703 | Sewage | Roma Sud | 2006 | May | + | GII |
| 704 | Sewage | Roma Sud | 2006 | May | + | GII + GI |
FIG. 1.
Sample collection sites along 120 km of the Latium coast. Symbols: circles, seawater samples; squares, sewage samples.
(ii) Sewage samples.
Ten sewage samples (raw influent at the wastewater treatment plant) of 50 ml (samples 671 to 704) were collected, two every 15 days, during 2006 and stored at 4°C upon arrival in the laboratory (Table 1, samples 671 to 704). They were immediately centrifuged at 3,000 × g to remove debris and then ultracentrifuged at 200,000 × g for 2 h to pellet viruses. Pellets were resuspended in 1 ml of phosphate-buffered saline (PBS) and stored at −80°C for future use.
(iii) Concentration of river and seawater samples by PrepScale tangential flow filtration.
A PrepScale tangential flow filtration membrane cartridge (type PTHK) of polysulfone, 0.23 m2 in size and with a 10-kDa nominal molecular size limit, equipped with a peristaltic pump, adapters, and a holder tool kit, was used (Millipore Corporation, Bedford, MA).
Briefly, a sandwich of two prefilters of 80 and 25 μm was set up before the cartridge and the whole system was cleaned with 20 liters of water and then pretreated with 250 ml of 3% Todd-Hewitt broth at pH 7.4. The cleaned prefiltered sample was collected in a cleaned tank. Concentration of the samples proceeded until 100 ml of retentate was obtained. Elution of the concentrated suspension was carried out by washing the suspension with 250 ml of 3% Todd-Hewitt broth at pH 9 for 35 min. A total of 100 ml of the eluate was collected and added to the concentrates. A postconcentration step was carried out by precipitation with 10% polyethylene glycol 6000 (Promega Corporation, Madison, WI) overnight at 4°C. Pellets were recovered at 12,000 × g for 30 min at 10°C in a fixed-angle rotor and recovered in 10 ml PBS. The sanitation step for the cartridge was carried out with 20 liters of distilled water and 5 liters of 0.1 N NaOH at 50°C for 40 min as recommended by the manufacturer. Viruses were recovered by ultracentrifugation at 200,000 × g for 2 h at 4°C, and the pellet was resuspended in 2 ml of PBS.
Clinical samples.
Nineteen stool samples were collected during 2006 from children with symptoms of acute sporadic gastroenteritis hospitalized at the Policlinico Umberto I of Rome. Upon arrival, each sample was assigned a code (identification [ID]) in the PostgreSQL database based on its location and history. A convenient web interface allows internet connection to this database, available to registered users at https://cosmos.bio.uniroma1.it. The repository keeps track of all primers, PCRs, and samples used. All specimens were prepared in a 10% suspension in phosphate buffer (PBS), clarified by low-speed centrifugation, and stored at −80°C until processing.
Four positive GII samples (already characterized as NoV GII) were kindly provided from the National Institute for Infectious Diseases Lazzaro Spallanzani (INMI), Rome (samples 372 and 374), and from the Institute of Microbiology, University of Parma (samples 616 and 617). Moreover, a positive control (653) was provided from the Health Protection Agency (HPA; United Kingdom). Table 2 shows the complete list of clinical samples used in this study.
TABLE 2.
Clinical samples collected from the Policlinico Umberto I, Rome, and elsewhere
| Sample | Collection date (yr, mo) | Origin | Symptoms | PCR result | Genogroup |
|---|---|---|---|---|---|
| 668 | 2006, January | Policlinico Umberto I | Vomiting and diarrhea | + | GII |
| 669 | Vomiting, diarrhea, fever | + | GII | ||
| 670 | 2006, February | Vomiting and diarrhea | + | GII | |
| 673 | Vomiting, fever, abdominal pains | − | |||
| 675 | Vomiting and diarrhea | − | |||
| 676 | Vomiting and diarrhea | − | |||
| 681 | Vomiting, diarrhea, abdominal pains | − | |||
| 682 | Vomiting, diarrhea, fever | − | |||
| 683 | Vomiting, diarrhea, fever | − | |||
| 684 | Vomiting, diarrhea, fever | − | |||
| 685 | 2006, March | Vomiting, diarrhea, fever | − | ||
| 686 | Vomiting, diarrhea, fever | − | |||
| 691 | Vomiting, diarrhea, abdominal pains | + | GII | ||
| 692 | Vomiting and diarrhea | + | GII | ||
| 693 | Vomiting and diarrhea | − | |||
| 694 | Vomiting and diarrhea | − | |||
| 695 | Vomiting, diarrhea, and fever | − | |||
| 928 | 2006, August | Vomiting, diarrhea, and fever | − | ||
| 929 | Vomiting, diarrhea, abdominal pains | − | |||
| 372 | 2002, October | INMI | + | GII | |
| 374 | 2002, February | + | GII | ||
| 616 | 2005, May | University of Parma | + | GII | |
| 617 | + | GII | |||
| 653 | 2005, September | HPA | + | GII |
Extraction of nucleic acid.
Nucleic acids were extracted from 300 μl of PBS-resuspended pellet for environmental samples by using the EXTRAgen kit (Amplimedical, Torino, Italy) according to the manufacturer's instructions. RNA from clinical samples was extracted from 140 μl of stool suspension by using the QIAamp viral RNA kit (QIAGEN, Hilden, Germany), according to the manufacturer's instructions. The NoV GII sample provided from the HPA was included in each round of extraction, to control for the quality of the extraction. Extracted RNA was diluted in 60 μl of sterile water and used directly in RT-PCR assays or stored at −80°C for future use.
RT-PCR assays.
Clinical and environmental extracts were tested with a published seminested RT-PCR assay, based on the method of Vennema et al. (56). It consists of an amplification of both GI and GII of NoV in the RdRp gene region by RT-PCR (RT-PCR 443; see Table 3), followed by specific seminested PCR assays for each genogroup (PCRs 444 and 446 for GII and GI, respectively). The target amplicon sizes are 327 bp in the RT-PCR (for both genogroups), 188 bp in the seminested PCR for GI, and 237 bp in the seminested PCR for GII.
TABLE 3.
PCRs used in this work for PCR and cloning purposes
| PCR ID | Primer ID | Sequence (5′→3′) | Product length (bp) | Primer position, 5′ to 3′a |
|---|---|---|---|---|
| 443 | 1421-f | ATACCACTATGATGCAGAYTA | 327 | 4279-4299 |
| 1422-r | TCATCATCACCATAGAAIGAG | 4605-4585 | ||
| 444 | 1421-f | ATACCACTATGATGCAGAYTA | 237 | 4279-4299 |
| 1424-r | AGCCAGTGGGCGATGGAATTC | 4515-4495 | ||
| 446 | 1423-f | TCNGAAATGGATGTTGG | 188 | 4691-4707 |
| 1422-r | TCATCATCACCATAGAAIGAG | 4878-4858 | ||
| 437 | 1356-f | AGCCNTNGAAATNATGGT | 1,048 | 4342-4359 |
| 1367-r | CGATTTCATCATCACCATA | 5389-5367 | ||
| 438 | 1364-f | YTCYTTCTATGGYGATGATGA | 516 | 4585-4605 |
| 1319-r | TCGACGCCATCTTCATTCACA | 5100-5080 | ||
| 475 | 1449-f | GGGACTCAACACAAAATAGAC | 1,013 | 4581-4604 |
| 1448-r | ACATCACCGGGGGTATTRTTT | 5593-5571 | ||
| 476 | 1423-f | TCTGAGATGGATGTAGG | 687 | 4691-4707 |
| 1316-r | TCCTTAGACGCCATCATCAT | 5377-5358 |
To discriminate true-negative results from false-negative results due to PCR failure, a NoV internal amplification control (IAC) RNA (NOROIAC; Yorkshire Bioscience Ltd., Biocentre, York, United Kingdom), which contains the target region for primers used in each round, was included. The IAC amplicon sizes are 369 bp in the first cycle, 228 bp in the GI seminested PCR, and 277 bp in the GII seminested reaction. A positive reaction control and a negative reaction control (ultrapure water) were included with each series of tests. All standard precautions were followed and strict laboratory practices were adhered to in order to prevent any PCR contamination. The pre-PCR manipulations (RNA isolation and PCR setup) were performed in a clean room that was physically isolated from the post-PCR processing area; dedicated pipettes and reagents were used for each location and washed daily with Microsol 3+ laboratory decontaminant (Anachem, Luton, United Kingdom).
In the first cycle (RT-PCR 443), 10 μl of the extracted RNA and 22 pmol of each primer (1421 and 1422) were used in a final mixture of 50 μl, for reverse transcription amplifications using the GeneAmp RNA PCR kit (Applied Biosystems) according to the manufacturer's instructions. RT-PCRs were carried out in a GeneAmp PCR System 9600 Thermocycler (Applied Biosystems) under the following conditions: reverse transcription at 50°C for 30 min; one cycle of template denaturation at 94°C for 5 min; and 40 cycles of 94°C for 30 s, 45°C for 30 s, and 72°C for 1 min, followed by one cycle of elongation for 10 min at 72°C. At the end of the amplification 2 μl of product amplicon was used for each seminested reaction for GI and GII (PCR 446 and PCR 444, respectively) in 35 cycles of amplification at the same temperature as that of the first cycle.
All clinical and environmental extracts were then also amplified by newly designed nested RT-PCR assays for GI and GII in order to verify the typing results obtained by the method of Vennema et al. and to further explore genetic diversity within the two genogroups by phylogenetic analysis. Two different strategies were used to design the new assay for the two genogroups in order to amplify roughly the same target region: for detection of GII NoVs, in the first cycle of amplification (RT-PCR 437 [see Table 3]) a combination of published primers (1356 and 1367) in the RdRp gene was used (15, 19), modified by us, in order to obtain a fragment of 1,048 bp. To increase the sensitivity of the assay, a nested PCR (PCR 438) was performed with a combination of published primers (1364 and 1319) (18, 19) to obtain an amplification product of 516 bp.
For detection of GI NoV, newly designed primers were engineered on the basis of the multialignment of GI sequences in the database. An amplification of 1,013 bp of NoV GI was obtained (RT-PCR 475) by using primers 1449 and 1448; in the nested PCR (PCR 476) primers 1423 and 1316 were used to amplify a fragment of 687 bp. Table 3 shows the list of PCRs and primers used in this study with ID codes and references.
For the new PCR assays, the annealing temperature was increased to 50°C in both the initial and nested cycles. Visualization of amplification products was carried out by horizontal electrophoresis in a 1.5% agarose gel stained with ethidium bromide. A marker with a known molecular size (1-kb DNA ladder; Invitrogen) and a negative control (diethyl pyrocarbonate-treated water) were run on the same electrophoresis gel to confirm the validity of results. Gel images were visualized under a UV transilluminator and digitally recorded. The PCR products were purified prior to sequencing by using the Wizard SV Gel and PCR Clean-Up System (Promega Corporation, Madison, WI), following the manufacturer's instructions.
Gene sequencing and data analysis.
Amplicons were sequenced from both directions with the ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems, Foster City, CA) following the manufacturer's instructions, by using the same primers as those for the PCRs. After cycle sequencing, unincorporated dyes were removed with the Montage SEQ96 Sequencing Reaction Cleanup Kit (Millipore Corporation, Billerica, MA). Sequencing analysis was performed in a capillary automatic sequencer (ABI PRISM 310 Genetic Analyzer; Applied Biosystems) as previously described (35).
The consensus sequences were constructed by comparing forward and reverse electropherograms using the AutoAssembler sequence assembly software, version 2.1.1 (Applied Biosystems, Foster City, CA), and exported in GCG format to a Sun Blade 2000 workstation (Sun Microsystems, Palo Alto, CA) implemented with Wisconsin GCG version 10.3 (University of Wisconsin Genetics Computer Group, Madison, WI). Database searches were run using the BLAST (Basic Local Alignment Search Tool) service provided by the National Center for Biotechnology Information (NCBI, Bethesda, MD) web server.
Multiple alignment was carried out using the GCG PILEUP program, and the subsequent phylogenetic analyses were performed using the Clustal W program, version 1.8 (51), with the neighbor-joining method based on a matrix of distances. The significance of constructed phylogenies was estimated by bootstrap analysis with 100 pseudoreplicate data sets, with values above 70% being considered significant.
The trees were displayed with the Njplot program (40), and the postscript files were imported in the CorelDraw (version 10) program for adjustments.
Cloning PCR product.
Since environmental samples may contain multiple virus strains (30), positive PCR products were cloned into the PCR4-TOPO vector (Invitrogen) by the TA cloning strategy, according to the manufacturer's protocol, in order to obtain a cDNA library of NoV sequences. The amplified products ligated into the TA cloning vector were plated onto Luria-Bertani medium-ampicillin agar plates containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal).
At least five colonies that appeared to be white when screened for the appropriate-size insert by PCR were sequenced using vector-specific primers (T3 and T7).
Nucleotide sequence accession numbers.
The nucleotide sequence data obtained in this study have been submitted to GenBank and assigned accession numbers AM712389 to AM712434.
RESULTS
An amplicon of 237 bp for GII was detected by using RT-PCR 443, followed by PCR 444 for 10 samples (five reference samples and five clinical samples from the Policlinico Umberto I of Rome [Table 2]); no GI signals were detected. A fragment of 277 bp for NOROIAC in the negative samples was amplified, excluding the presence of inhibitors in amplification reactions or other causes of false-negative results.
Concordant results were obtained with the newly designed assays for GI and GII: an amplicon of 516 bp was obtained by using the newly designed assay for NoV GII (RT-PCR 437 followed by the nested PCR 438) from the 10 GII samples (five reference samples and five clinical samples); no GI signals were detected.
Genotyping of the five clinical NoV strains detected in our study showed that three strains (668, 669, and 692) belonged to the GII.4 cluster, one strain (691) belonged to the GII.3 cluster, and one strain (670) belonged to the GIIb cluster. Of the five samples used as reference strains, four belonged to the GII.4 group (653, 616, 372, and 374) and one (617) belonged to the GIIb cluster.
Environmental samples were first tested with the method of Vennema et al. (56). None of the 15 river, estuarine, and seawater samples collected during 2004 were positive for GI or GII NoV. Of 19 seawater samples collected during 2005, three (samples 645, 649, and 650) turned out to be positive for NoV GII (Table 1). All 10 sewage samples collected during 2006 were positive for NoV GII; moreover, in six samples (671, 677, 678, 679, 680, and 704) both GI and GII were detected. All samples yielding negative PCR results for GI and GII produced the expected band corresponding to the internal standard control.
When environmental samples were tested with the newly designed assays for GI and GII, concordant results were obtained in comparison with the reference method: a fragment of 516 bp for NoV GII was obtained from the three seawater samples collected during 2005 and from all sewage samples collected during 2006. In 6 out of the 10 sewage samples (Table 1), in addition to the signal for GII, a fragment of 687 bp for GI was amplified, indicating a simultaneous contamination by the two genogroups. Moreover, two sewage samples (703 and 704) were contaminated besides strains of different genogroups even with different strains of GII.
Genotyping of GII strains showed that the three seawater samples belonged to the GII.4 group while the sewage samples contained, in addition to GII.4 strains (6 out of 10), three GIIb strains and one GII.3 strain (Table 1).
Sequences obtained from both environmental and clinical samples from the different tests used in this study were used to construct different phylogenetic trees, in order to characterize NoV strains and to study their genetic relationships.
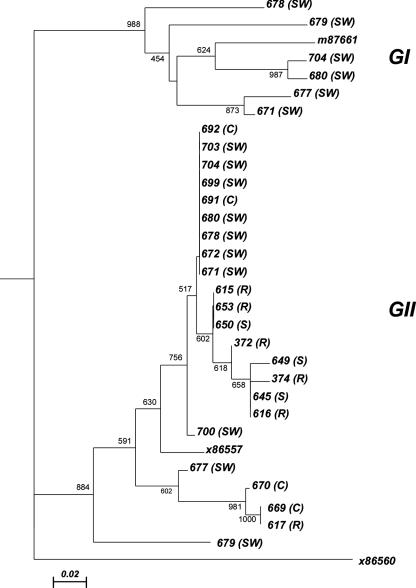
Figure 2 shows the tree constructed on the basis of multialignment of GI and GII sequences obtained with the Vennema method. In the tree, prototype sequences from GI and GII obtained from GenBank (accession numbers M87661 and X86557, respectively) and an outgroup sequence from a Sapporo virus (Manchester; X86560) were included.
FIG. 2.
Phylogenetic tree constructed in the RdRp region (56). The type of sample from which the sequence was obtained is listed in parentheses (C, clinical samples; S, seawater, SW, sewage; R, reference).
The tree shows two separated clusters for GI and GII samples, supported by high bootstrap values; moreover, a within-cluster variability is evident for both genogroups. In particular, GII samples all differ from each other; GII samples showed a certain degree of genetic diversity but no clearly distinct clusters supported by significant bootstrap values. A group of samples were indistinguishable because of 100% identical nucleotide sequences. Figure 3 shows the tree constructed with GI and GII sequences obtained from the newly designed nested assays. Besides the Sapporo virus Manchester (X86560) as outgroup, prototypes from GenBank were included for each GII cluster (AY502023, ab089871, and X86557 for GII.4; AY773210 for GIIb; and AF414415 for GII.3) and for each GI cluster (af414404 for GI.4, M87661 for GI.1, and ay694413 for GI.2). The tree shows two clearly separated clusters for GI and GII samples, supported by high bootstrap values; moreover, the tree shows a greater genetic diversity within GII samples. In particular, tree clusters referring to GII.4, GIIb, and GII.3 are clearly separated (bootstrap value, >98.3%). For samples 703 and 704, in which two different sequences were simultaneously detected by direct sequencing and by cloning of PCR products, the different strains are indicated as 703.2 and 704.2. In particular, of 26 GII sequences, 19 belonged to the GII.4 cluster, 5 to the GIIb cluster, and 2 to the GII.3 cluster; of six GI strains, two belonged to the GI.4 group, one to the GI.1 cluster, and three to the GI.2 cluster.
FIG. 3.
Phylogenetic tree constructed in the RdRp region for NoV (this study). The abbreviations in parentheses are the same as those listed in the legend to Fig. 2.
DISCUSSION
Human NoVs are important pathogens associated with food- and waterborne outbreaks of acute gastroenteritis in children and adults. These viruses, transmitted by the fecal-oral route, are estimated to be responsible for almost all nonbacterial gastroenteritis worldwide. Contaminated water poses an especially serious health risk, and waterborne outbreaks have been caused by contaminated surface water, groundwater, drinking water, and mineral water (4, 26, 46, 53).
A large European project called VIROBATHE (Methods for the Detection of Adenoviruses and Noroviruses in European Bathing Waters with Reference to the Revision of the Bathing Water Directive 76/160/EEC) is under way to acquire data about “emerging waterborne pathogens” such as NoVs and adenoviruses, in order to standardize reliable techniques for the detection and quantification of such viruses in water samples for water quality assessment.
While the occurrence of these viruses in oysters or other seafood has been widely reported (3, 15, 20), the fate of viruses in seawater is unknown and reports of detection of NoVs from seawater are limited. Nevertheless, the viral contamination of seawater is one of the important epidemiological issues.
In the present study, environmental samples from seawater, estuarine water, and effluents of sewage treatment plants were analyzed in order to evaluate the role of environmental surface contamination as a possible vector for transmission of NoVs from GI and GII. Moreover, in order to study the similarity of viruses found in water to those causing clinical problems in the community, we analyzed clinical samples collected from children with symptoms of acute sporadic gastroenteritis hospitalized at the Policlinico Umberto I of Rome.
Sequencing and phylogenetic analysis in the highly conserved region of the RdRp region were used to detect and characterize NoVs in environmental and clinical samples collected from 2004 to 2006.
Firstly we used a published assay (56) able to amplify a fragment of 237 bp for GII and 188 bp for GI in seminested reactions. The use of double-round PCR is needed when analyzing water samples due to the low concentration of viruses. Because NoV detection in water samples requires concentration of viruses from large volumes of water, a process that can coconcentrate inhibitors which may test falsely negative by conventional RT-PCR methods (42, 53), an IAC was used in the PCR assays to discriminate a true-negative result from a false-negative result due to PCR failure.
Nucleotide sequences obtained from GI and GII clinical and environmental samples were used to construct a phylogenetic tree. In the tree, GI and GII strains are clearly separated into different clusters and genetic diversity within each genogroup is shown. Still, two groups of sequences in the GII cluster showed 100% nucleotide sequence identity. Because of the short length of the analyzed region and the conserved genomic region targeted by the primers used, we cannot confirm if those viruses are identical or if we are unable to discriminate between them.
To address the problem, in addition to published primers, we have also designed a new broadly reactive RT-PCR/nested assay for GI and GII NoV within the RNA polymerase region which gives longer amplicons (516 bp for GII and 687 bp for GI), in order to better display the high genetic polymorphism of NoVs and to understand the relatedness and phylogeny of different strains.
The Vennema method is a useful tool for rapid detection and identification of NoVs belonging to GI and GII; nevertheless, because of the short length of PCR products, it yields limited phylogenetic information of the sort useful for revealing variability within the genogroups. In fact, it is well known that sequence data generated from small regions should be interpreted with caution. To our knowledge, to date, published nested assays (required when analyzing environmental samples) produce fragments of no more than 158 bp (15, 21, 37, 38, 43, 50, 57). The newly designed assays for GI and GII provided more extensive regions of the polymerase gene, which allows high-confidence BLAST searches of the public database. The new assays are therefore a useful tool for molecular typing of NoVs, able to display the high genetic polymorphism of these viruses and to help us understand the relatedness and phylogeny of different strains.
In order to discriminate a true-negative result from a false-negative result due to PCR failure we are preparing an IAC for the new primer sets, for detection of inhibitory effects.
In conclusion, the two novel assays, coupled with phylogeny-based clustering in the RdRp region, should be of use not only for the rapid diagnosis and typing of human NoVs in environmental and clinical samples but also for global epidemiological study of these viruses. Although this work shows only preliminary epidemiological data based on a relatively low number of samples, results appear to be promising. Further studies with more field isolates are needed in order to evaluate the sensitivity of the new primers to detect a wide variety of NoV genetic types, to confirm the value of this approach.
Of 19 clinical samples analyzed 26% were infected with NoV, all belonging to GII; negative samples showed infection with enteric viruses other than NoV (data not shown). Despite the small number of clinical samples analyzed in this work, these data confirm NoVs as important causative agents of pediatric gastroenteritis and underline the importance of NoV GII strains as the main cause of the majority of sporadic cases of infantile gastroenteritis. Moreover, this result is in agreement with previous reports of a higher prevalence of GII than of GI strains in outbreaks, as well as studies of sporadic gastroenteritis (5, 11, 12, 13, 22, 32, 36, 49, 52).
Regarding environmental contamination by NoVs, different types of water were contaminated by viruses differently. River, estuarine, and seawater samples were scarcely contaminated: 3 out 33 (0.9%) samples were positive for GII; sewage samples were 100% positive for NoV, of which 60% presented multiple NoV contaminations (simultaneous presence of both GI and GII). Moreover, mixed infections with different strains of GII were detected in two samples (703 and 704) by direct sequencing of the amplified products or by sequencing of the cloned fragments by using vector-specific primers. While directly sequencing PCR products without first cloning the fragment provides distinct advantages in terms of time and cost, the drawback is that where “single bands” are really two or more superimposed products—not a particularly rare event if amplifying related but nonidentical sequences—only the much more representative product is visible. Since environmental samples may contain multiple virus strains, cloning before sequencing of RT-PCR products is important to highlight genetic variability.
It is interesting that despite the broad diffusion of both GI and GII NoVs in sewage samples, human diseases are mainly caused by GII strains (5, 12, 13, 22, 32, 36, 49, 52). The reason for this is unknown, although differences in biological properties such as virulence, routes of transmission, or stability of the virus in the environment are possible explanations (7). This discrepancy, already described by different authors, suggests the need for further investigation of possible differences in the pathogenic potential of different genogroups, different degrees of immunity, and water environmental stability.
Even though the clinical and environmental sample numbers are small, we noted correlations between sewage samples (SW) and clinical samples (C) collected during 2006 (Fig. 3). Particularly, the 670 (C) and 679 (SW) samples, which are identical and belong to genotype GII.b, were collected in February-March; samples 691 (C) and 699 (SW), which were collected in March-April, clustered together in the GII.3 group; sample 692 (C) clustered with sample 680 (SW) in the GII.4 group, both collected in March 2006.
The wide circulation of NoVs and a widespread dissemination of NoV variants in raw and treated sewage waters, with a predominance of GII strains, are already known (55), and waterborne outbreaks of NoV in community settings caused by sewage contamination of wells and recreational water have been described elsewhere (6, 18, 27). In fact, sewage treatment processes, when present, are only partially effective at viral removal and discharges constantly release human viruses into the marine environment. Once in the environment, viruses can survive for weeks to months either in the water or by attaching themselves to particulate matter and accumulating in sediments. While viral presence in seawater samples decreases with distance from the pollution source, even slightly contaminated waters are potentially a risk because of NoV characteristics: low infectious doses (8, 29), prolonged asymptomatic shedding (14), great strain diversity (1, 10), and environmental stability (45).
Moreover, the coexistence of several GI and GII NoVs and different genotypes of each genogroup as detected in this study can lead to exposure of human or animal hosts to multiple NoV strains with a potential risk of recombination: emerging new strains which may be more virulent and pathogenic than strains currently circulating in the population can better adapt to infect susceptible individuals or better survive in the environment, with new potential risks for the community (55).
To complement qualitative PCR assays, we are performing quantitative PCR assays to detect NoV concentrations in environmental samples by broadly reactive one-step TaqMan RT-PCR assays (unpublished data). Quantitative data on the concentrations of NoVs present in recreational water are indispensable for assessment of the public health risks caused by NoV infections.
Few studies have investigated the prevalence and epidemiology of NoVs in Italy, and, to our knowledge, this work is the first to demonstrate the diffusion of NoV GI and GII in water environments in Italy. Finding enteric viruses in water helps us to understand the mechanisms of viral transmission and to determine the possible role played by water as a vector for transmission. As virally contaminated waters can serve as vehicles for NoV transmission and can be expected to cause further outbreaks, continued surveillance of NoVs and typing in clinical samples as well as in surface water are warranted, and in our opinion, legislative measures for viral monitoring as part of the microbial risk assessment in seawater should be seriously considered.
Acknowledgments
We are grateful to C. Moretti, F. Midulla, and M. Battaglia of the Policlinico Umberto I of Rome for providing clinical samples and to M. C. Medici of Università degli Studi di Parma, M. R. Capobianchi of INMI-Spallanzani of Rome, and J. Sellwood for UK-HPA for providing reference samples. We thank Paolo Pastori for his contribution to the implementation of the PostgreSQL database.
Footnotes
Published ahead of print on 4 May 2007.
REFERENCES
- 1.Ando, T., J. S. Noel, and R. L. Fankhauser. 2000. Genetic classification of Norwalk-like viruses. J. Infect. Dis. 181(Suppl. 2):S336-S348. [DOI] [PubMed] [Google Scholar]
- 2.Atmar, R. L., and M. K. Estes. 2006. The epidemiologic and clinical importance of norovirus infection. Gastroenterol. Clin. N. Am. 35:275-290, viii. [DOI] [PubMed] [Google Scholar]
- 3.Atmar, R. L., F. H. Neill, J. L. Romalde, F. Le Guyader, C. M. Woodley, T. G. Metcalf, and M. K. Estes. 1995. Detection of Norwalk virus and hepatitis A virus in shellfish tissues with the PCR. Appl. Environ. Microbiol. 61:3014-3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beuret, C., D. Kohler, A. Baumgartner, and T. M. Luthi. 2002. Norwalk-like virus sequences in mineral waters: one-year monitoring of three brands. Appl. Environ. Microbiol. 68:1925-1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bon, F., P. Fascia, M. Dauvergne, D. Tenenbaum, H. Planson, A. M. Petion, P. Pothier, and E. Kohli. 1999. Prevalence of group A rotavirus, human calicivirus, astrovirus, and adenovirus type 40 and 41 infections among children with acute gastroenteritis in Dijon, France. J. Clin. Microbiol. 37:3055-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brugha, R., I. B. Vipond, M. R. Evans, Q. D. Sandifer, R. J. Roberts, R. L. Salmon, E. O. Caul, and A. K. Mukerjee. 1999. A community outbreak of food-borne small round-structured virus gastroenteritis caused by a contaminated water supply. Epidemiol. Infect. 122:145-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buesa, J., B. Collado, P. Lopez-Andujar, R. Abu-Mallouh, D. J. Rodriguez, D. A. Garcia, J. Prat, S. Guix, T. Llovet, G. Prats, and A. Bosch. 2002. Molecular epidemiology of caliciviruses causing outbreaks and sporadic cases of acute gastroenteritis in Spain. J. Clin. Microbiol. 40:2854-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duizer, E., P. Bijkerk, B. Rockx, A. De Groot, F. Twisk, and M. Koopmans. 2004. Inactivation of caliciviruses. Appl. Environ. Microbiol. 70:4538-4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fankhauser, R. L., S. S. Monroe, J. S. Noel, C. D. Humphrey, J. S. Bresee, U. D. Parashar, T. Ando, and R. I. Glass. 2002. Epidemiologic and molecular trends of “Norwalk-like viruses” associated with outbreaks of gastroenteritis in the United States. J. Infect. Dis. 186:1-7. [DOI] [PubMed] [Google Scholar]
- 10.Farkas, T., W. M. Zhong, Y. Jing, P. W. Huang, S. M. Espinosa, N. Martinez, A. L. Morrow, G. M. Ruiz-Palacios, L. K. Pickering, and X. Jiang. 2004. Genetic diversity among sapoviruses. Arch. Virol. 149:1309-1323. [DOI] [PubMed] [Google Scholar]
- 11.Foley, B., J. O'Mahony, S. M. Morgan, C. Hill, and J. G. Morgan. 2000. Detection of sporadic cases of Norwalk-like virus (NLV) and astrovirus infection in a single Irish hospital from 1996 to 1998. J. Clin. Virol. 17:109-117. [DOI] [PubMed] [Google Scholar]
- 12.Froggatt, P. C., I. Barry Vipond, C. R. Ashley, P. R. Lambden, I. N. Clarke, and E. O. Caul. 2004. Surveillance of norovirus infection in a study of sporadic childhood gastroenteritis in South West England and South Wales, during one winter season (1999-2000). J. Med. Virol. 72:307-311. [DOI] [PubMed] [Google Scholar]
- 13.Gallimore, C. I., M. A. Barreiros, D. W. Brown, J. P. Nascimento, and J. P. Leite. 2004. Noroviruses associated with acute gastroenteritis in a children's day care facility in Rio de Janeiro, Brazil. Braz. J. Med. Biol. Res. 37:321-326. [DOI] [PubMed] [Google Scholar]
- 14.Graham, D. Y., X. Jiang, T. Tanaka, A. R. Opekun, H. P. Madore, and M. K. Estes. 1994. Norwalk virus infection of volunteers: new insights based on improved assays. J. Infect. Dis. 170:34-43. [DOI] [PubMed] [Google Scholar]
- 15.Green, J., K. Henshilwood, C. I. Gallimore, D. W. Brown, and D. N. Lees. 1998. A nested reverse transcriptase PCR assay for detection of small round-structured viruses in environmentally contaminated molluscan shellfish. Appl. Environ. Microbiol. 64:858-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hafliger, D., P. Hubner, and J. Luthy. 2000. Outbreak of viral gastroenteritis due to sewage-contaminated drinking water. Int. J. Food Microbiol. 54:123-126. [DOI] [PubMed] [Google Scholar]
- 17.Hutson, A. M., R. L. Atmar, and M. K. Estes. 2004. Norovirus disease: changing epidemiology and host susceptibility factors. Trends Microbiol. 12:279-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jothikumar, N., J. A. Lowther, K. Henshilwood, D. N. Lees, V. R. Hill, and J. Vinje. 2005. Rapid and sensitive detection of noroviruses by using TaqMan-based one-step reverse transcription-PCR assays and application to naturally contaminated shellfish samples. Appl. Environ. Microbiol. 71:1870-1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katayama, K., H. Shirato-Horikoshi, S. Kojima, T. Kageyama, T. Oka, F. Hoshino, S. Fukushi, M. Shinohara, K. Uchida, Y. Suzuki, T. Gojobori, and N. Takeda. 2002. Phylogenetic analysis of the complete genome of 18 Norwalk-like viruses. Virology 299:225-239. [DOI] [PubMed] [Google Scholar]
- 20.Kawamoto, H., S. Hasegawa, S. Sawatari, C. Miwa, O. Morita, T. Hosokawa, and H. Tanaka. 1993. Small, round-structured viruses (SRSVs) associated with acute gastroenteritis outbreaks in Gifu, Japan. Microbiol. Immunol. 37:991-997. [DOI] [PubMed] [Google Scholar]
- 21.Kawamoto, H., K. Yamazaki, E. Utagawa, and T. Ohyama. 2001. Nucleotide sequence analysis and development of consensus primers of RT-PCR for detection of Norwalk-like viruses prevailing in Japan. J. Med. Virol. 64:569-576. [DOI] [PubMed] [Google Scholar]
- 22.Kirkwood, C. D., R. Clark, N. Bogdanovic-Sakran, and R. F. Bishop. 2005. A 5-year study of the prevalence and genetic diversity of human caliciviruses associated with sporadic cases of acute gastroenteritis in young children admitted to hospital in Melbourne, Australia (1998-2002). J. Med. Virol. 77:96-101. [DOI] [PubMed] [Google Scholar]
- 23.Kjeldsberg, E., G. Anestad, H. Greenberg, I. Orstavik, R. Pedersen, and E. Slettebo. 1989. Norwalk virus in Norway: an outbreak of gastroenteritis studied by electron microscopy and radioimmunoassay. Scand. J. Infect. Dis. 21:521-526. [DOI] [PubMed] [Google Scholar]
- 24.Koopmans, M., and E. Duizer. 2004. Foodborne viruses: an emerging problem. Int. J. Food Microbiol. 90:23-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koopmans, M., J. Vinje, M. de Wit, I. Leenen, W. van der Poel, and Y. van Duynhoven. 2000. Molecular epidemiology of human enteric caliciviruses in The Netherlands. J. Infect. Dis. 181(Suppl. 2):S262-S269. [DOI] [PubMed] [Google Scholar]
- 26.Kukkula, M., P. Arstila, M. L. Klossner, L. Maunula, C. H. Bonsdorff, and P. Jaatinen. 1997. Waterborne outbreak of viral gastroenteritis. Scand. J. Infect. Dis. 29:415-418. [DOI] [PubMed] [Google Scholar]
- 27.Kukkula, M., L. Maunula, E. Silvennoinen, and C. H. von Bonsdorff. 1999. Outbreak of viral gastroenteritis due to drinking water contaminated by Norwalk-like viruses. J. Infect. Dis. 180:1771-1776. [DOI] [PubMed] [Google Scholar]
- 28.Lawson, H. W., M. M. Braun, R. I. Glass, S. E. Stine, S. S. Monroe, H. K. Atrash, L. E. Lee, and S. J. Englender. 1991. Waterborne outbreak of Norwalk virus gastroenteritis at a southwest US resort: role of geological formations in contamination of well water. Lancet 337:1200-1204. [DOI] [PubMed] [Google Scholar]
- 29.Leclerc, H., L. Schwartzbrod, and E. Dei-Cas. 2002. Microbial agents associated with waterborne diseases. Crit. Rev. Microbiol. 28:371-409. [DOI] [PubMed] [Google Scholar]
- 30.Lodder, W. J., J. Vinjé, R. van de Heide, A. M. de Roda Husman, E. J. T. M. Leenen, and M. P. G. Koopmans. 1999. Molecular detection of Norwalk-like caliciviruses in sewage. Appl. Environ. Microbiol. 65:5624-5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marten, K., and S. Psarakos. 1994. Evidence of self-awareness in the bottlenose dolphin (Tursiops truncatus), p. 361-379. In S. Parker, M. Boccia, and R. Mitchell (ed.), Self-awareness in animals and humans: developmental perspectives. Cambridge University Press, New York, NY.
- 32.Martinez, N., C. Espul, H. Cuello, W. Zhong, X. Jiang, D. O. Matson, and T. Berke. 2002. Sequence diversity of human caliciviruses recovered from children with diarrhea in Mendoza, Argentina, 1995-1998. J. Med. Virol. 67:289-298. [DOI] [PubMed] [Google Scholar]
- 33.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Medici, M. C., M. Martinelli, F. M. Ruggeri, L. A. Abelli, S. Bosco, M. C. Arcangeletti, F. Pinardi, F. De Conto, A. Calderaro, C. Chezzi, and G. Dettori. 2005. Broadly reactive nested reverse transcription-PCR using an internal RNA standard control for detection of noroviruses in stool samples. J. Clin. Microbiol. 43:3772-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muscillo, M., G. La Rosa, A. Carducci, L. Cantiani, and C. Marianelli. 1999. Molecular and biological characterization of poliovirus 3 strains isolated in Adriatic seawater samples. Water Res. 33:3204-3212. [Google Scholar]
- 36.Nakata, S., S. Honma, K. K. Numata, K. Kogawa, S. Ukae, Y. Morita, N. Adachi, and S. Chiba. 2000. Members of the family Caliciviridae (Norwalk virus and Sapporo virus) are the most prevalent cause of gastroenteritis outbreaks among infants in Japan. J. Infect. Dis. 181:2029-2032. [DOI] [PubMed] [Google Scholar]
- 37.Ohyama, T., S. Yoshizumi, H. Sawada, Y. Uchiyama, Y. Katoh, N. Hamaoka, and E. Utagawa. 1999. Detection and nucleotide sequence analysis of human caliciviruses (HuCVs) from samples in non-bacterial gastroenteritis outbreaks in Hokkaido, Japan. Microbiol. Immunol. 43:543-550. [DOI] [PubMed] [Google Scholar]
- 38.O'Neill, H. J., C. McCaughey, D. E. Wyatt, F. Mitchell, and P. V. Coyle. 2001. Gastroenteritis outbreaks associated with Norwalk-like viruses and their investigation by nested RT-PCR. BMC Microbiol. 1:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pelosi, E., P. R. Lambden, E. O. Caul, B. Liu, K. Dingle, Y. Deng, and I. N. Clarke. 1999. The seroepidemiology of genogroup 1 and genogroup 2 Norwalk-like viruses in Italy. J. Med. Virol. 58:93-99. [PubMed] [Google Scholar]
- 40.Perriere, G., and M. Gouy. 1996. WWW-query: an on-line retrieval system for biological sequence banks. Biochimie 78:364-369. [DOI] [PubMed] [Google Scholar]
- 41.Pruss, A. 1998. Review of epidemiological studies on health effects from exposure to recreational water. Int. J. Epidemiol. 27:1-9. [DOI] [PubMed] [Google Scholar]
- 42.Queiroz, A. P., F. M. Santos, A. Sassaroli, C. M. Harsi, T. A. Monezi, and D. U. Mehnert. 2001. Electropositive filter membrane as an alternative for the elimination of PCR inhibitors from sewage and water samples. Appl. Environ. Microbiol. 67:4614-4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramirez, S., S. De Grazia, G. M. Giammanco, M. Milici, C. Colomba, F. M. Ruggeri, V. Martella, and S. Arista. 2006. Detection of the norovirus variants GGII.4 hunter and GGIIb/hilversum in Italian children with gastroenteritis. J. Med. Virol. 78:1656-1662. [DOI] [PubMed] [Google Scholar]
- 44.Rutjes, S. A., H. H. van den Berg, W. J. Lodder, and A. M. Roda Husman. 2006. Real-time detection of noroviruses in surface water by use of a broadly reactive nucleic acid sequence-based amplification assay. Appl. Environ. Microbiol. 72:5349-5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rzezutka, A., and N. Cook. 2004. Survival of human enteric viruses in the environment and food. FEMS Microbiol. Rev. 28:441-453. [DOI] [PubMed] [Google Scholar]
- 46.Schaub, S. A., and R. K. Oshiro. 2000. Public health concerns about caliciviruses as waterborne contaminants. J. Infect. Dis. 181(Suppl. 2):S374-S380. [DOI] [PubMed] [Google Scholar]
- 47.Schvoerer, E., F. Bonnet, V. Dubois, A. M. Rogues, J. P. Gachie, M. E. Lafon, and H. J. Fleury. 1999. A hospital outbreak of gastroenteritis possibly related to the contamination of tap water by a small round structured virus. J. Hosp. Infect. 43:149-154. [DOI] [PubMed] [Google Scholar]
- 48.Shieh, Y. C., R. S. Baric, J. W. Woods, and K. R. Calci. 2003. Molecular surveillance of enterovirus and Norwalk-like virus in oysters relocated to a municipal-sewage-impacted gulf estuary. Appl. Environ. Microbiol. 69:7130-7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Subekti, D. S., P. Tjaniadi, M. Lesmana, C. Simanjuntak, S. Komalarini, H. Digdowirogo, B. Setiawan, A. L. Corwin, J. R. Campbell, K. R. Porter, and B. A. Oyofo. 2002. Characterization of Norwalk-like virus associated with gastroenteritis in Indonesia. J. Med. Virol. 67:253-258. [DOI] [PubMed] [Google Scholar]
- 50.Sugieda, M., K. Nakajima, and S. Nakajima. 1996. Outbreaks of Norwalk-like virus-associated gastroenteritis traced to shellfish: coexistence of two genotypes in one specimen. Epidemiol. Infect. 116:339-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Traore, O., G. Belliot, C. Mollat, H. Piloquet, C. Chamoux, H. Laveran, S. S. Monroe, and S. Billaudel. 2000. RT-PCR identification and typing of astroviruses and Norwalk-like viruses in hospitalized patients with gastroenteritis: evidence of nosocomial infections. J. Clin. Virol. 17:151-158. [DOI] [PubMed] [Google Scholar]
- 53.Trujillo, A. A., K. A. McCaustland, D. P. Zheng, L. A. Hadley, G. Vaughn, S. M. Adams, T. Ando, R. I. Glass, and S. S. Monroe. 2006. Use of TaqMan real-time reverse transcription-PCR for rapid detection, quantification, and typing of norovirus. J. Clin. Microbiol. 44:1405-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ueki, Y., K. Akiyama, T. Watanabe, and T. Omura. 2004. Genetic analysis of noroviruses taken from gastroenteritis patients, river water and oysters. Water Sci. Technol. 50:51-56. [PubMed] [Google Scholar]
- 55.van den Berg, H., W. Lodder, W. van der Poel, H. Vennema, and A. M. de Roda Husman. 2005. Genetic diversity of noroviruses in raw and treated sewage water. Res. Microbiol. 156:532-540. [DOI] [PubMed] [Google Scholar]
- 56.Vennema, H., E. de Bruin, and M. Koopmans. 2002. Rational optimization of generic primers used for Norwalk-like virus detection by reverse transcriptase polymerase chain reaction. J. Clin. Virol. 25:233-235. [DOI] [PubMed] [Google Scholar]
- 57.Wang, J., X. Jiang, H. P. Madore, J. Gray, U. Desselberger, T. Ando, Y. Seto, I. Oishi, J. F. Lew, and K. Y. Green. 1994. Sequence diversity of small, round-structured viruses in the Norwalk virus group. J. Virol. 68:5982-5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Widdowson, M. A., A. Sulka, S. N. Bulens, R. S. Beard, S. S. Chaves, R. Hammond, E. D. Salehi, E. Swanson, J. Totaro, R. Woron, P. S. Mead, J. S. Bresee, S. S. Monroe, and R. I. Glass. 2005. Norovirus and foodborne disease, United States, 1991-2000. Emerg. Infect. Dis. 11:95-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zheng, D. P., T. Ando, R. L. Fankhauser, R. S. Beard, R. I. Glass, and S. S. Monroe. 2006. Norovirus classification and proposed strain nomenclature. Virology 346:312-323. [DOI] [PubMed] [Google Scholar]