Abstract
Genes homologous to avrBs3 of Xanthomonas were detected in 309 strains of Ralstonia solanacearum biovars 3, 4, and 5 but not biovar 1 or 2. A statistically significant association between the originating plant species and internal repeats of the gene was found. Sequences of repeats and variation between nearly clonal strains revealed evidence of frequent recombination.
Ralstonia solanacearum causes bacterial wilt in more than 200 plant species (8). Strains of this heterogenous species complex vary largely in host range and aggressiveness. Despite the complete genome sequencing of the strains GMI1000 (race 1, biovar 3) (17) and UW551 (race 3, biovar 2) (5), the genetic basis of this variation is still largely unknown (5). Host preferences seem to be determined by genes that change too rapidly to reflect the phylogenetic divergence of strains. Consequently, the attempt to subdivide R. solanacearum into races based on host range turned out to be of little taxonomic value (7), probably because the complex evolving plant defense system (10, 18) constantly selects for acquisition, mutation, elimination, or (reversible) inactivation of virulence/avirulence genes (6, 11).
Essential for pathogenicity of R. solanacearum and many other gram-negative phytopathogenic bacteria is a type III secretion system encoded by the hrp gene cluster that translocates effector proteins into the host cell (1, 2). Effectors are involved in causing disease in susceptible hosts or in eliciting a hypersensitive response in resistant or nonhost plants (2). Members of the avrBs3 effector family of Xanthomonas were shown to suppress nonspecific hypersensitive responses and expression of defense response genes in the plant (4). Recently, hrpB-regulated effector genes homologous to the avrBs3 family of Xanthomonas were discovered in the R. solanacearum biovar 1 strain GMI1000 (3) and in the biovar 4 strain RS1000 (14). These genes, designated brg11 and hpx17, are 98.9% identical in nucleotide sequence but differ by a deletion of 1,575 bp in the repeat domain of hpx17. Both gene products have less than 40% sequence identity with AvrBs3 family proteins of Xanthomonas, so functional analogy is uncertain. Repetitive DNA sequences are prone to increased frequencies of recombination. Such localized hypermutation can cause microdiverse populations that have enhanced chances of an adaptive response to evolving host factors (12, 13). Intragenic recombination within the repeat domain of the avrBs3 family genes pthA of Xanthomonas citri (20) and avrXa7 of Xanthomonas oryzae pv. oryzae (19) was shown to create new host specificities.
The distribution of avrBs3-like genes in strains of the R. solanacearum species complex is largely unknown. In contrast to GMI1000 and RS1000, homologous genes were not present in the genome of the biovar 2 strain UW551 (5). brg11 is located in an alternative codon usage region (17) and may thus be an accidental remnant of horizontal gene transfer in a few strains. It is also unknown whether avrBs3-like genes are involved in pathogenicity or modulate the host specificity of R. solanacearum. Mutants of GMI1000 with brg11 disrupted were still able to cause disease on tomato and Arabidopsis (3), which is analogous to observations with other avrBs3-like genes. The aim of this study was to investigate how common brg11 and hpx17 homologues are within the R. solanacearum species complex and whether the size of the repeat region is correlated with host preferences of the strains. Conserved brg11 and hpx17 sites flanking the central repeat domain were identified by comparison with Xanthomonas avrBs3 homologues and used to design primers hpx17f (5′-CG CTG CAT CTC ACA CCG CAG CAG GT-3′) and hpx17r (5′-C CTT CAC CGG CAA CCC CTG CCT GAC-3′) to amplify the repeat region. PCR conditions were 7 min at 94°C, 10 cycles consisting of 30 s at 94°C, 30 s at 65°C − 1°C/cycle, and 120 s at 72°C and 20 cycles consisting of 30 s at 94°C, 30 s at 56°C, and 120 s at 72°C. PCR mixtures contained 0.2 mM of each deoxynucleoside triphosphate, 2.5 mM MgCl2, 0.2 μM of both primers, and 1.25 U Stoffel fragment in 25 μl Stoffel buffer (Applied Biosystems, Foster City, CA).
A diverse collection of 319 R. solanacearum strains from 15 provinces of China and 30 strains from 19 other countries was recently characterized (Y. N. Yin, Q. Y. Xue, L. L. Xu, J. H. Guo, and K. Smalla, unpublished data). The strains originated from 18 plant species and comprised five races and five biovars (Table 1). They were grouped into 29 clusters of closely related or clonal strains by BOX-PCR genomic fingerprinting (Yin et al., unpublished), a highly discriminatory technique to determine the taxonomic diversity and phylogenetic structure of bacterial populations (16). We screened all strains for the presence of brg11 and hpx17 homologues and analyzed the correlation between the repeat size and the originating host plant. All strains gave either a single PCR product or no product. The size of the PCR products was either 0.3 kb (as expected from hpx17), 1.9 kb (like that from brg11), 1.7 kb, 1.8 kb, or 2.0 kb (Fig. 1A). The latter sizes could be explained by the presence of 15.8, 16.8, or 18.8 repeats, respectively. Interestingly, very similar strains sharing the same type of BOX fingerprint could have different sizes of brg11 or hpx17 homologues. This is evidence of a high rate of mutation by recombination relative to the genome. For instance, BOX fingerprint type 23 was shared by 99 isolates from Zingiber officinale with a 300-bp PCR product of the repeat region and by 27 strains from other plants with a 1,875-bp product, which coincided with a change in the race of the strains (Table 1). The gene could not be detected in any of the five biovar 1 strains or the 29 biovar 2 strains analyzed. These results suggested that avrBs3-like genes are preferentially distributed among R. solanacearum strains of the Asiaticum and not the Americanum division (15).
TABLE 1.
Sizes and AluI digestion types of the repeat regions of brg11 and hpx17 gene homologues and characteristics of R. solanacearum strains used in this study
| Repeat size (kb) | AluI typea | Biovarb | Host plant | Raceb | BOX typeb | Geographical origin | Strain(s) (sourcec) |
|---|---|---|---|---|---|---|---|
| 0.3 | a | 3 | Solanum melongena | 1 | 21 | Hunan | HN517(A) |
| 0.3 | a | 3 | Solanum melongena | 1 | 25 | Hunan | HN521 (A) |
| 0.3 | a | 4 | Capsicum annuum | 1 | 22 | Jiangsu | JS52, JS54 (A) |
| 0.3 | a | 4 | Capsicum annuum | 1 | 29 | Hubei | HB51, 52, 55, 57, 510 (A) |
| 0.3 | a | 4 | Ipomoea batatas | 1 | 25 | Fujian | FJ1989B2 (C), FJ2001B5, FJ2002B3, FJ2003B2, FJ2003B4, FJ2004B1 (E), FJ516-520 (A) |
| 0.3 | a | 4 | Zingiber officinale | 4 | 18 | Henan | HeN1994Z8 (C) |
| 0.3 | a | 4 | Zingiber officinale | 4 | 23 | Shandong | 99 of type SD58 (A) |
| 0.3 | a | 3-1 | Solanum melongena | 1 | 25 | Hunan | HN531, HN532 (A) |
| 0.3 | a | 3-1 | Capsicum annuum | 1 | 25 | Hunan | HN515 (A) |
| 1.7 | b1 | 4 | Zingiber officinale | 4 | 11 | Shandong | ICPM11119-20 (B) |
| 1.7 | b1 | 4 | Zingiber officinale | 4 | NDd | Philippines | UW197 (D), Zo4-1986 (C) |
| 1.7 | b2 | 4 | Arachis hypogaea | 1 | 26 | Guangxi | GX526-527 (A) |
| 1.8 | c1 | 3 | Capsicum annuum | 1 | 9 | Guangdong | GD52 (A) |
| 1.8 | c1 | 3 | Lycopersicon esculentum | 1 | 24 | Hubei | HB512(A) |
| 1.8 | c1 | 3 | Solanum melongena | 1 | 24 | Guangxi | GX517-518 (A) |
| 1.8 | c1 | 3-1 | Lycopersicon esculentum | 1 | 9 | Fujian | FJ47 (A) |
| 1.8 | c2 | 3 | Lycopersicon esculentum | 1 | 6 | Guangdong | GD45 (A) |
| 1.8 | c2 | 3 | Solanum melongena | 1 | 6 | Guangdong | GD43 (A) |
| 1.8 | c2 | 3 | Solanum tuberosum | 1 | 12 | Shandong | SD1991Po1 (C) |
| 1.9 | d1 | 3 | Arachis hypogaea | 1 | 6 | Guangxi | ICPM11106-07 (B), GX523 (A) |
| 1.9 | d1 | 3 | Capsicum annuum | 1 | 6 | Guangxi | GX54 (A) |
| 1.9 | d1 | 3 | Capsicum annuum | 1 | 6 | Hunan | HN51-53 (A) |
| 1.9 | d1 | 3 | Capsicum annuum | 1 | 8 | Guangdong | GD51 (A) |
| 1.9 | d1 | 3 | Capsicum annuum | 1 | 9 | Guangxi | GX510-511, 55, 56, 58 (A) |
| 1.9 | d1 | 3-1 | Capsicum annuum | 1 | 9 | Hunan | HN56 (A) |
| 1.9 | d1 | 3 | Capsicum annuum | 1 | 17 | Hunan | HN55 (A) |
| 1.9 | d1 | 3 | Capsicum annuum | 1 | 23 | Guizhou | GZ52-53 (A) |
| 1.9 | d1 | 3 | Capsicum annuum | 1 | 9 | Hunan | HN57, HN511, HN512 (A) |
| 1.9 | d1 | 3 | Lycopersicon esculentum | 1 | 6 | Hunan | HN538 (A) |
| 1.9 | d1 | 3 | Lycopersicon esculentum | 1 | 6 | Jiangsu | JS528 (A) |
| 1.9 | d1 | 3 | Lycopersicon esculentum | 1 | 6 | Sichuan | SC1983Tm10 (C) |
| 1.9 | d1 | 3 | Lycopersicon esculentum | 1 | 8 | Guizhou | GZ58 (A) |
| 1.9 | d1 | 3 | Lycopersicon esculentum | 1 | 23 | Guizhou | GZ511-516 (A) |
| 1.9 | d1 | 4 | Nicotiana tabacum | 1 | 6 | Guangxi | GX1994Tb28 (C) |
| 1.9 | d1 | 3 | Nicotiana tabacum | 1 | 8 | Guangdong | GD1993Tb3 (C), GD48 (A) |
| 1.9 | d1 | 3 | Semen ricini | 1 | 6 | Fujian | FJ1986Bd1 (C) |
| 1.9 | d1 | 3 | Solanum melongena | 1 | 6 | Guizhou | GZ54, GZ55 (A) |
| 1.9 | d1 | 3 | Solanum melongena | 1 | 6 | Hunan | HN516, HN518 (A) |
| 1.9 | d1 | 3 | Solanum melongena | 1 | 9 | Fujian | FJ45 (A) |
| 1.9 | d1 | 3 | Solanum melongena | 1 | 9 | Hunan | HN522, HN524, HN527-528, HN533, HN535, HN537 (A) |
| 1.9 | d1 | 3 | Solanum melongena | 1 | 17 | Hunan | HN519-520, HN523, HN525-526, HN529, HN534 (A) |
| 1.9 | d1 | 3 | Solanum melongena | 1 | 23 | Guizhou | GZ56 (A) |
| 1.9 | d1 | 3 | Solanum melongena | 1 | 25 | Hunan | HN530 (A) |
| 1.9 | d1 | 3 | Solanum melongena | 1 | 27 | Hunan | HN536 (A) |
| 1.9 | d1 | 4 | Arachis hypogaea | 1 | 6 | Guangxi | GX528(A) |
| 1.9 | d1 | 4 | Arachis hypogaea | 1 | 6 | Hubei | HB1986P3 (C) |
| 1.9 | d1 | 4 | Arachis hypogaea | 1 | 9 | Fujian | FJ49 (A) |
| 1.9 | d1 | 4 | Capsicum annuum | 1 | 6 | Jiangsu | JS414, JS510 (A) |
| 1.9 | d1 | 4 | Casuarina equisetifolia | 1 | 6 | Guangdong | GD1986C2 (C) |
| 1.9 | d1 | 4 | Lycopersicon esculentum | 1 | 6 | Guangdong | GD1995Tm1 (C), UW364 (D) |
| 1.9 | d1 | 4 | Lycopersicon esculentum | 1 | 23 | Guizhou | GZ517-518 (A) |
| 1.9 | d1 | 4 | Solanum melongena | 1 | 20 | Guangxi | GX519 (A) |
| 1.9 | d1 | 4 | Solanum melongena | 1 | 23 | Guizhou | GZ57 (A) |
| 1.9 | d1 | 4 | Solanum melongena | 1 | ND | Brazil | UW89 (D) |
| 1.9 | d1 | 3-1 | Solanum melongena | 1 | 6 | Sichuan | SC1986E4 (C) |
| 1.9 | d1 | ND | Lycopersicon esculentum | 1 | ND | Africa | UW393 (D) |
| 1.9 | d2 | 3 | Boehmeria nivea | 1 | 6 | Zhejiang | ZJ1993Bn1 (A) |
| 1.9 | d3 | 3 | Capsicum annuum | 1 | 6 | Fujian | FJ43 (A) |
| 1.9 | d3 | 3 | Capsicum annuum | 1 | 6 | Guangxi | GX1993Pe1 (C) |
| 1.9 | d3 | 3 | Capsicum annuum | 1 | 9 | Fujian | FJ42 (A) |
| 1.9 | d3 | 3 | Capsicum annuum | 1 | 9 | Guangxi | GX51-52 (A) |
| 1.9 | d3 | 3 | Capsicum annuum | 1 | 23 | Guangxi | GX57, GX59 (A) |
| 1.9 | d3 | 3 | Capsicum annuum | 1 | 23 | Hubei | HB53-54, HB56 (A) |
| 1.9 | d3 | 3 | Casuarina equisetifolia | 1 | 6 | Guangdong | ICPM11270 (B) |
| 1.9 | d3 | 3 | Lycopersicon esculentum | 1 | 6 | Guangxi | GX1994Tm2 (C), UW363 (D) |
| 1.9 | d3 | 3 | Lycopersicon esculentum | 1 | 14 | Guizhou | GZ59-510 (A) |
| 1.9 | d3 | 3 | Nicotiana tabacum | 1 | 14 | Guizhou | GZ520-521 (A) |
| 1.9 | d3 | 3 | Solanum melongena | 1 | 9 | Guangxi | GX514 (A) |
| 1.9 | d3 | 3 | Solanum melongena | 1 | 14 | Jiangsu | JS520-521 (A) |
| 1.9 | d3 | 4 | Arachis hypogaea | 1 | 9 | Guangxi | GX524-525 (A) |
| 1.9 | d3 | 4 | Capsicum annuum | 1 | 6 | Hunan | HN54 (A) |
| 1.9 | d3 | 4 | Capsicum annuum | 1 | 14 | Jiangsu | JS55 (A) |
| 1.9 | d3 | 4 | Capsicum annuum | 1 | 15 | Jiangsu | JS412 (A) |
| 1.9 | d3 | 4 | Capsicum annuum | 1 | 28 | Jiangsu | JS511 (A) |
| 1.9 | d3 | 4 | Lycopersicon esculentum | 1 | 14 | Jiangsu | JS527 (A) |
| 1.9 | d3 | 4 | Lycopersicon esculentum | 1 | 23 | Hunan | HN539 (A) |
| 1.9 | d3 | 4 | Solanum melongena | 1 | 9 | Fujian | UW356 (D) |
| 1.9 | d3 | 4 | Solanum melongena | 1 | 9 | Guangxi | GX513, 515, 516 (A) |
| 1.9 | d3 | 4 | Solanum melongena | 1 | 28 | Jiangsu | JS422 (A) |
| 1.9 | d3 | 4 | Zingiber officinale | 4 | 15 | Shandong | SD53 (A) |
| 1.9 | d3 | 4 | Zingiber officinale | 4 | 26 | Shandong | ICPM11121 (B) |
| 1.9 | d3 | 3-1 | Capsicum annuum | 1 | 6 | Guangxi | GX53 (A) |
| 1.9 | d3 | 3-1 | Capsicum annuum | 1 | 23 | Hunan | HN508-510, HN513-514 (A) |
| 1.9 | d3 | ND | Lycopersicon esculentum | ND | ND | Indonesia | UW391 (D) |
| 1.9 | d4 | 3 | Arachis hypogaea | 1 | 17 | Fujian | FJP16 (C), FJ48 (A) |
| 1.9 | d4 | 3 | Capsicum annuum | 1 | 17 | Fujian | FJ41 (A) |
| 1.9 | d4 | 3 | Lycopersicon esculentum | 1 | ND | Australia | UW143 (D) |
| 1.9 | d4 | 3 | Lycopersicon esculentum | 1 | ND | Guyana | GMI1000 (D) |
| 1.9 | d4 | 3 | Nicotiana tabacum | 1 | 17 | Yunnan | YN49-414 (A) |
| 1.9 | d4 | 3 | Nicotiana tabacum | 1 | ND | Australia | UW147 (D) |
| 1.9 | d4 | 3 | Rapistrum rugosum | 1 | ND | Australia | UW148 (D) |
| 1.9 | d4 | 3 | Solanum mauritianum | 1 | ND | Australia | UW289 (D) |
| 1.9 | d4 | 3 | Solanum melongena | 1 | ND | Australia | UW293 (D) |
| 1.9 | d4 | 3 | Solanum nigrum L. | 1 | 17 | Fujian | FJ1994Sn1 (C) |
| 1.9 | d4 | 3 | Solanum tuberosum | 1 | ND | Australia | UW152 (D) |
| 1.9 | d4 | 4 | Arachis hypogaea | 1 | 17 | Fujian | FJ48 (A) |
| 1.9 | d4 | 4 | Zingiber officinale | 1 | ND | Australia | UW141 (D) |
| 1.9 | d5 | 3 | Capsicum annuum | 1 | 6 | Jiangsu | JS59 (A) |
| 1.9 | d5 | 3 | Capsicum annuum | 1 | 7 | Zhejiang | ZJ51 (A) |
| 1.9 | d5 | 3 | Capsicum annuum | 1 | 13 | Jiangsu | JS55-58, JS513 (A) |
| 1.9 | d5 | 3 | Casuarina equisetifolia | 1 | 6 | Guangdong | ICPM11110 (B) |
| 1.9 | d5 | 3 | Lycopersicon esculentum | 1 | 6 | Jiangsu | JS526 (A) |
| 1.9 | d5 | 3 | Lycopersicon esculentum | 1 | 7 | Zhejiang | ZJ53, ZJ55 (A) |
| 1.9 | d5 | 3 | Lycopersicon esculentum | 1 | 13 | Jiangsu | JS430 (A) |
| 1.9 | d5 | 3 | Lycopersicon esculentum | 1 | 16 | Zhejiang | ZJ54 (A) |
| 1.9 | d5 | 3 | Lycopersicon esculentum | 1 | 18 | Jiangsu | ICPM11113 (B) |
| 1.9 | d5 | 3 | Lycopersicon esculentum | 1 | 23 | Hubei | HB511 (A) |
| 1.9 | d5 | 3 | Solanum melongena | 1 | 6 | Jiangsu | ICPM11271 (B) |
| 1.9 | d5 | 3 | Solanum melongena | 1 | 10 | Jiangsu | ICPM11117 (B) |
| 1.9 | d5 | 3 | Solanum melongena | 1 | 13 | Jiangsu | JS517-519, JS523 (A) |
| 1.9 | d5 | 3 | Solanum melongena | 1 | 16 | Zhejiang | ZJ52 (A) |
| 1.9 | d5 | 4 | Arachis hypogaea | 1 | 6 | Guangxi | GX522 (A) |
| 1.9 | d5 | 4 | Capsicum annuum | 1 | 6 | Jiangsu | JS51, JS53 (A) |
| 1.9 | d5 | 4 | Capsicum annuum | 1 | 18 | Jiangsu | ICPM11112 (B) |
| 1.9 | d5 | 4 | Capsicum annuum | 1 | 19 | Jiangsu | ICPM11111 (B) |
| 1.9 | d5 | 4 | Lycopersicon esculentum | 1 | 6 | Jiangsu | JS529 (A) |
| 1.9 | d6 | 3 | Sesamum indicum | 1 | 6 | Guangxi | GX1993Ssp1 (C) |
| 1.9 | d6 | 4 | Lycopersicon esculentum | 1 | 4 | Taiwan | UW264 (D) |
| 1.9 | d7 | 3 | Capsicum annuum | 1 | 6 | Guizhou | GZ51 (A) |
| 1.9 | d7 | 3 | Nicotiana tabacum | 1 | 6 | Guizhou | GZ519 (A) |
| 1.9 | d7 | 3 | Nicotiana tabacum | 1 | 23 | Guizhou | GZ522-524 (A) |
| 1.9 | d7 | 4 | Zingiber officinale | 4 | 3 | Shandong | SD54 (A) |
| 2.0 | e | 5 | Morus alba | 5 | 5 | Guangdong | UW360-361, UW373 (D) |
| 1 | Lycopersicon esculentum | 1 | ND | USA | K60 (D) | ||
| 1 | Musa sp. | 2 | ND | Honduras | UW40 (D) | ||
| 1 | Musa sp. | 2 | ND | Venezuela | UW41 (D) | ||
| 1 | Nicotiana tabacum | 1 | ND | Mexico | UW278 (D) | ||
| 1 | Solanum tuberosum | 1 | ND | Kenya | UW134 (D) | ||
| 2 | Casuarina equisetifolia | 3 | 2 | Guangdong | GD1993C1 (C) | ||
| 2 | Lycopersicon esculentum | 3 | ND | France | UW523 (D) | ||
| 2 | Nicotiana tabacum | 3 | 2 | Taiwan | UW265 (D) | ||
| 2 | Pelargonium capitatum | 3 | ND | USA | I35 (D) | ||
| 2 | Solanum melongena | 3 | 2 | Fujian | FJE1 (C) | ||
| 2 | Solanum tuberosum | 3 | 2 | Beijing | BJ1987Po17 (C) | ||
| 2 | Solanum tuberosum | 3 | 2 | Guangdong | GD1994Po75 (C) | ||
| 2 | Solanum tuberosum | 3 | 2 | Guizhou | GZ1988Po58 (C) | ||
| 2 | Solanum tuberosum | 3 | 2 | Hebei | HeB1992Po46, B2005Po2k5 (C) | ||
| 2 | Solanum tuberosum | 3 | 2 | Hunan | HN1983Po33, HN1990Po77 (C) | ||
| 2 | Solanum tuberosum | 3 | 2 | Shandong | SD1992Po84 (C) | ||
| 2 | Solanum tuberosum | 3 | 2 | Sichuan | SC1986Po88 (C) | ||
| 2 | Solanum tuberosum | 3 | 2 | Yunnan | YN1994Po3 (C), YN41-47 (A) | ||
| 2 | Solanum tuberosum | 3 | ND | Australia | UW420 (D) | ||
| 2 | Solanum tuberosum | 3 | ND | Ceylon | UW51 (D) | ||
| 2 | Solanum tuberosum | 3 | ND | Colombia | UW37 (D) | ||
| 2 | Solanum tuberosum | 3 | ND | Colombia | UW80 (D) | ||
| 2 | Solanum tuberosum | 3 | ND | Europe | IPO1609 (F) | ||
| 2 | Solanum tuberosum | 3 | ND | Israel | UW24 (D) | ||
| 2 | Solanum tuberosum | 3 | ND | Peru | UW260 (D) | ||
| 3 | Arachis hypogaea | 1 | 1 | Guangxi | UW368 (D) | ||
| ND | Lycopersicon esculentum | 1 | ND | Brazil | UW88 (D) | ||
| ND | Capsicum annuum | 1 | ND | Panama | UW76 (D) | ||
| ND | Lycopersicon esculentum | 1 | ND | Nigeria | UW386 (D) | ||
| 4 | Nicotiana tabacum | 1 | 1 | Guangxi | GX1994Tb23 (C), GX532 (A) |
See Fig. 1.
Data from Y. N. Yin, Q. Y. Xue, L. L. Xu, J. H. Guo, and K. Smalla (unpublished data).
Strains sources are indicated as follows: A, Department of Plant Pathology, Nanjing Agricultural University, Nanjing, China; B, International Collection of Micro-organisms from Plants (ICMP), Auckland, New Zealand; C, L. Y. He, Chinese Academy of Agricultural Sciences, Beijing, China; D, C. Allen, University of Wisconsin, Madison; E, T. Lu, Fujian Academy of Agricultural Sciences, Fuzhou, China; F, K. Smalla, BBA, Braunschweig, Germany.
ND, not determined.
FIG. 1.
Agarose gel electrophoresis of PCR products (A) and AluI restriction patterns (B) from brg11- and hpx17-like genes of R. solanacearum strains. Lanes: M, size marker; 0, no product; a to e, representative strains of the five size classes.
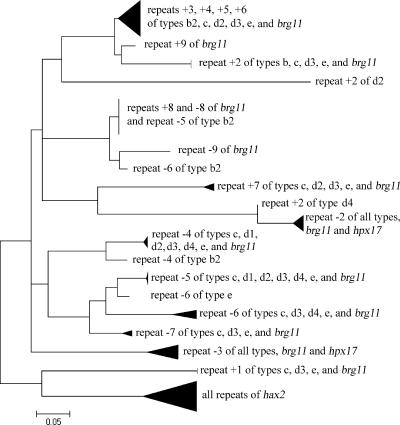
To investigate whether the brg11 or hpx17 type affected the preference of R. solanacearum strains for host plants in the field, the association between the originating host plant and the size of the PCR product was analyzed by Fisher's exact test. The association was found to be highly significant (P < 0.0001). However, the composition of the strain collection may be biased by the sampling of identical strains from the same host several times. Thus, a nonredundant set of strains was tested in which all 100 isolates differed in either their BOX genomic fingerprint, their geographical origin, or the host from which they were isolated (Table 2). Also, with this data set, the sizes of the brg11 or hpx17 repeat regions of the strains and their originating hosts were significantly correlated (P = 0.046). This indicated that the host preference of R. solanacearum in the field is modulated by recombination of the brg11 or hpx17 repeats. The PCR products were further analyzed by AluI digestion and sequencing. The five size classes could be further differentiated into 13 AluI patterns, reflecting the sequence diversity of the repeat region (Fig. 1B; Table 1). Pattern type b1 was exclusively linked to Zingiber officinale and b2 to Arachis hypogaea. It remains to be investigated whether the number of repeats or rather the sequence of some repeats determines specific interactions with host cells. Sequencing of 81 repeats from nine strains (representing gene types a, b2, c1, d1, d2, d3, d4, and e) revealed high similarities to the repeats of brg11 and hpx17 (Fig. 2). The 300-bp fragment of strain SD58 was identical to hpx17 in sequence and size. Also, the other sequences showed 98 to 100% sequence conservation in the nucleotides homologous to hpx17, while the repeats not present in hpx17 were more variable. Strain GD52 (1.8-kb repeat) and five strains with a 1.9-kb repeat fragment, which represented different AluI restriction patterns, showed 97.7 to 98.5% sequence identity to brg11 in their common repeats, whereas strain GX526 (1.7-kb repeat) showed only 90% identity (Fig. 2). Neighboring repeats tended to be highly similar. Duplications and deletions also gave evidence for frequent recombination events (Fig. 2). Typically, the repeats comprised 35 codons, but a length of 34 codons was also observed. Among the Xanthomonas avrBs3 gene family, the repeats of hax2 from Xanthomonas campestris pv. armoraciae (9) were most similar to those of R. solanacearum, especially to the first repeat of the region. In contrast to hax2, the R. solanacearum repeats cluster according to their position in the first or second half of the repeat region, which implies a modular structure of the internal repeats.
TABLE 2.
Association between originating plant species and amplicon size of the avrBs3 homologue of nonredundant R. solanacearum isolatesa
| Host plant | % of strains with amplicon of indicated size (bp)b
|
||||
|---|---|---|---|---|---|
| 300 (7) | 1,665 (3) | 1,770 (7) | 1,875 (82) | 1,980 (1) | |
| Arachis hypogaea | 33 | 6 | |||
| Boehmeria nivea | 1 | ||||
| Capsicum annuum | 29 | 14 | 24 | ||
| Casuarina equisetifolia | 1 | ||||
| Ipomoea batatas | 14 | ||||
| Lycopersicon esculentum | 43 | 24 | |||
| Morus alba | 100 | ||||
| Nicotiana tabacum | 9 | ||||
| Rapistrum rugosum | 1 | ||||
| Ricinus communis | 1 | ||||
| Sesamum indicum | 1 | ||||
| Solanum sp. | 29 | 43 | 26 | ||
| Zingiber officinale | 29 | 67 | 5 | ||
Fisher's exact test; P < 0.05.
Numbers in parentheses are numbers of strains.
FIG. 2.
Amino acid neighbor-joining tree of single internal repeats of brg11, hpx17, hax2, and homologues of R. solanacearum strains SD58 (type a), GX526 (type b2), GD52 (type c), FJ49 (type d1), ZJ1993Bn1 (type d2), GX57 and ICPM11121 (type d3), FJP16 (type d4), and UW360 (type e). In total, 123 repeats were analyzed. Similar repeats were grouped. Repeats were numbered according to the position in brg11 +1 to + 9 (from the 5′ end) and −1 to −9 (from the 3′ end). Bootstrap consensus tree, n = 100, PAM.
In conclusion, conservation of the internal repeats of brg11-like genes among strains of R. solanacearum and association of repeat types with originating host plants implied a specific function in virulence. Frequent recombinatorial changes of the internal repeats and the presence of a single type per strain are in accordance with the assumption of an interaction with the plant defense system.
Nucleotide sequence accession numbers.
Sequence data have been submitted to the DDBJ/EMBL/GenBank databases under accession numbers EF435034 to EF435042.
Acknowledgments
This research was supported by a grant-in-aid for science research from the Chinese 863 High-Tech Program (2006AA10Z431) and Sino-Germany Cooperation Project on Agricultural Science and Technology (2004-Z17).
Footnotes
Published ahead of print on 27 April 2007.
REFERENCES
- 1.Casper-Lindley, C., D. Dahlbeck, E. T. Clark, and B. J. Staskawicz. 2002. Direct biochemical evidence for type III secretion-dependent translocation of the AvrBs2 effector protein into plant cells. Proc. Natl. Acad. Sci. USA 99:8336-8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cornelis, G. R., and F. Van Gijsegem. 2000. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54:735-774. [DOI] [PubMed] [Google Scholar]
- 3.Cunnac, S., A. Occhialini, P. Barberis, C. Boucher, and S. Genin. 2004. Inventory and functional analysis of the large Hrp regulon in Ralstonia solanacearum: identification of novel effector proteins translocated to plant host cells through the type III secretion system. Mol. Microbiol. 53:115-128. [DOI] [PubMed] [Google Scholar]
- 4.Fujikawa, T., H. Ishihara, J. E. Leach, and S. Tsuyumu. 2006. Suppression of defense response in plants by the avrBs3/pthA gene family of Xanthomonas spp. Mol. Plant-Microbe Interact. 19:342-349. [DOI] [PubMed] [Google Scholar]
- 5.Gabriel, D. W., C. Allen, M. Schell, T. P. Denny, J. T. Greenberg, Y. P. Duan, Z. Flores-Cruz, Q. Huang, J. M. Clifford, G. Presting, E. T. Gonzalez, J. Reddy, J. Elphinstone, J. Swanson, J. Yao, V. Mulholland, L. Liu, W. Farmerie, M. Patnaikuni, B. Balogh, D. Norman, A. Alvarez, J. A. Castillo, J. Jones, G. Saddler, T. Walunas, A. Zhukov, and N. Mikhailova. 2006. Identification of open reading frames unique to a select agent: Ralstonia solanacearum race 3 biovar 2. Mol. Plant-Microbe Interact. 19:69-79. [DOI] [PubMed] [Google Scholar]
- 6.Genin, S., and C. Boucher. 2004. Lessons learned from the genome analysis of Ralstonia solanacearum. Annu. Rev. Phytopathol. 42:107-134. [DOI] [PubMed] [Google Scholar]
- 7.Hayward, A. C. 1964. Characteristics of Pseudomonas solanacearum. J. Appl. Bacteriol. 27:265-277. [Google Scholar]
- 8.Hayward, A. C. 1991. Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu. Rev. Phytopathol. 29:65-87. [DOI] [PubMed] [Google Scholar]
- 9.Kay, S., J. Boch, and U. Bonas. 2005. Characterization of AvrBs3-like effectors from a Brassicaceae pathogen reveals virulence and avirulence activities and a protein with a novel repeat architecture. Mol. Plant-Microbe Interact. 18:838-848. [DOI] [PubMed] [Google Scholar]
- 10.Li, Z. K., M. Arif, D. B. Zhong, B. Y. Fu, J. L. Xu, J. Domingo-Rey, J. Ali, C. H. M. Vijayakumar, S. B. Yu, and G. S. Khush. 2006. Complex genetic networks underlying the defensive system of rice (Oryza sativa L.) to Xanthomonas oryzae pv. oryzae. Proc. Natl. Acad. Sci. USA 103:7994-7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Metzgar, D., and C. Wills. 2000. Evolutionary changes in mutation rates and spectra and their influence on the adaptation of pathogens. Microbes Infect. 2:1513-1522. [DOI] [PubMed] [Google Scholar]
- 12.Moxon, E. R., and D. S. Thaler. 1997. The tinkerer's evolving tool-box. Nature 387:659-662. [DOI] [PubMed] [Google Scholar]
- 13.Moxon, R., C. Bayliss, and D. Hood. 2006. Bacterial contingency loci: the role of simple sequence DNA repeats in bacterial adaptation. Annu. Rev. Genet. 40:307-333. [DOI] [PubMed] [Google Scholar]
- 14.Mukaihara, T., N. Tamura, Y. Murata, and M. Iwabuchi. 2004. Genetic screening of Hrp type III-related pathogenicity genes controlled by the HrpB transcriptional activator in Ralstonia solanacearum. Mol. Microbiol. 54:863-875. [DOI] [PubMed] [Google Scholar]
- 15.Poussier, S., P. Vandewalle, and J. Luisetti. 1999. Genetic diversity of African and worldwide strains of Ralstonia solanacearum as determined by PCR-restriction fragment length polymorphism analysis of the hrp gene region. Appl. Environ. Microbiol. 65:2184-2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rademaker, J. L. W., B. Hoste, F. J. Louws, K. Kersters, J. Swings, L. Vauterin, P. Vauterin, and F. J. de Bruijn. 2000. Comparison of AFLP and rep-PCR genomic fingerprinting with DNA-DNA homology studies: Xanthomonas as a model system. Int. J. Syst. Evol. Microbiol. 50:665-677. [DOI] [PubMed] [Google Scholar]
- 17.Salanoubat, M., S. Genin, F. Artiguenave, J. Gouzy, S. Mangenot, M. Arlat, A. Billault, P. Brottier, J. C. Camus, L. Cattolico, M. Chandler, N. Choisne, C. Claudel-Renard, S. Cunnac, N. Demange, C. Gaspin, M. Lavie, A. Moisan, C. Robert, W. Saurin, T. Schiex, P. Siguier, P. Thebault, M. Whalen, P. Wincker, M. Levy, J. Weissenbach, and C. A. Boucher. 2002. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415:497-502. [DOI] [PubMed] [Google Scholar]
- 18.Schornack, S., A. Meyer, P. Romer, T. Jordan, and T. Lahaye. 2006. Gene-for-gene-mediated recognition of nuclear-targeted AvrBs3-like bacterial effector proteins. J. Plant Physiol. 163:256-272. [DOI] [PubMed] [Google Scholar]
- 19.Yang, B., A. Sugio, and F. F. White. 2005. Avoidance of host recognition by alterations in the repetitive and C-terminal regions of AvrXa7, a type III effector of Xanthomonas oryzae pv. oryzae. Mol. Plant-Microbe Interact. 18:142-149. [DOI] [PubMed] [Google Scholar]
- 20.Yang, Y., and D. W. Gabriel. 1995. Intragenic recombination of a single plant pathogen gene provides a mechanism for the evolution of new host specificities. J. Bacteriol. 177:4963-4968. [DOI] [PMC free article] [PubMed] [Google Scholar]