Abstract
Enterobacter sakazakii causes a severe form of neonatal meningitis that occurs as sporadic cases as well as outbreaks. The disease has been epidemiologically associated with consumption of reconstituted, dried infant formulas. Very little information is available regarding pathogenicity of the organism and production of virulence factors. Clinical and environmental strains were screened for production of factors which have activity against Chinese hamster ovary (CHO) cells in tissue culture. Polymyxin B lysate and sonicate preparations but not culture supernatants from the strains caused “rounding” of CHO cells. Subsequent studies showed that the CHO cell-rounding factor is a proteolytic enzyme that has activity against azocasein. The cell-bound protease was isolated by using a combination of polymyxin B lysis, followed by sonication of cells harvested from tryptone broth. The protease was purified to homogeneity by sequential ammonium sulfate precipitation, gel filtration chromatography with Sephadex G-100, hydrophobic interaction chromatography with phenyl-Sepharose CL-4B, and a second gel filtration with Sephadex G-100. In addition to activity against azocasein, the purified protease also exhibits activity against azocoll and insoluble casein but not elastin. The protease has a molecular weight of 38,000 and an isoelectric point of 4.4. It is heat labile and for maximal activity against azocasein has an optimum temperature of 37°C and a pH range of 5 to 7. Proteolytic activity is inhibited by ortho-phenanthroline and Zincov but is not affected by phenylmethylsulfonyl fluoride, N-ethylmaleimide, and trypsin inhibitors, which demonstrates that the protease is a zinc-containing metalloprotease. The metalloprotease does not hemagglutinate chicken or sheep erythrocytes. Twenty-three to 27 of the first 42 N-terminal amino acid residues of the metalloprotease are identical to proteases produced by Serratia proteamaculans, Pectobacterium carotovorum, and Anabaena sp. PCR analysis using primers designed from a consensus nucleotide sequence showed that 135 E. sakazakii strains possessed the metalloprotease gene, zpx, and 25 non-E. sakazakii strains did not. The cloned zpx gene of strain 29544 consists of 1,026 nucleotides, and the deduced amino acid sequence of the metalloprotease has 341 amino acid residues, which corresponds to a theoretical protein size of 37,782 with a theoretical pI of 5.23. The sequence possesses three well-characterized zinc-binding and active-site motifs present in other bacterial zinc metalloproteases.
Enterobacter sakazakii is a member of the family Enterobacteriaceae and has recently been shown to be widespread in food processing plants and in households (17, 26). It can cause severe neonatal meningitis, septicemia, or necrotizing enterocolitis in premature babies and neonates (26). Though the incidences of these illnesses are low, the mortality rate has been reported to vary and to range from 10% to 80% (14). Meningitis occurs both as sporadic cases and as outbreaks, and contaminated dried infant formulas have been epidemiologically implicated as the source of the pathogen in many of these cases (26). In addition to causing disease in babies and neonates, E. sakazakii in adults also cause bacteremia, wound infections, and infections associated with indwelling catheters (7, 26). However, little is known about the mechanism(s) whereby the pathogen causes disease. Using suckling mice, Pagotto et al. (28) showed that some clinical and food strains were lethal when administered peritoneally, but only two strains caused death by the oral route. They also reported that some, but not all, E. sakazakii strains produced an enterotoxin which caused fluid accumulation in suckling mice, while other strains produced factors which lysed or rounded some tissue culture cells. In our laboratory, we have screened various clinical and environmental strains for the production of factors which have an effect on Chinese hamster ovary (CHO) cells in tissue culture. A qualitative and preliminary study showed that many of the strains produced factors which caused “rounding” of CHO cells. Rounding of tissue culture cells has been reported to be due to the action of various bacterial proteases (22). The rounding factor expressed by the E. sakazakii strains and described in this paper was purified and characterized for its physicochemical properties and was identified as a zinc-containing metalloprotease. The purpose of this paper is to describe the properties of this zinc-metalloprotease of E. sakazakii and to report the identity of the protease gene locus, zpx. Also described in this report are studies which have led to the development of a species-specific PCR detection assay for E. sakazakii, which is based on the presence of zpx.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
One hundred thirty-five strains of E. sakazakii obtained from clinical, food, environmental, and unknown sources, including representatives of 15 of 16 biotypes described by Farmer et al. (7, 15), were stored at −80°C in Trypticase soy broth (BBL, Cockeysville, MD) supplemented with 1% NaCl (TSB-S) and 25% glycerol. Routinely, frozen cultures were rapidly thawed and the cells were streaked onto plates containing Trypticase soy Agar (BBL) supplemented with 1% NaCl (TSA-S) and incubated at 37°C for 24 h.
Screening for protease production. (i) Broth method.
Twelve strains were initially screened for protease production in a liquid medium. Strains were grown in 5 ml Casamino Acids yeast extract broth (3% Casamino Acids, 0.4% yeast extract, 0.05% K2HPO4 [pH 7.4]) in a 50-ml flask overnight at 37°C on a rotary shaker at 100 rpm, and the supernatant was recovered after centrifugation (14,000 × g, 10 min, 4°C). Cells were washed once with saline, suspended in 200 μl of 0.02 M Tris-buffered saline containing polymyxin B (2 mg/ml), and incubated at 37°C for 1 h. After centrifugation (14,000 × g, 10 min, 4°C), both supernatants and lysates were assayed according to the procedure described by Kothary et al. (19) for “rounding” of CHO cells, which is frequently associated with the presence of proteases in culture fluids (22).
(ii) Agar method.
Strains were streaked onto tryptone-skim milk agar (2.5% tryptone-3.0% skim milk in 0.1 M Tris buffer, pH 7.6), and the cultures were incubated at 37°C for 3 to 4 days. Protease activity was indicated by a zone of clearing around or under the colony.
Purification of the protease from E. sakazakii.
Unless specified, all steps were carried out at 4°C. The work reported is from data obtained from both ATCC strains 51329 and 29544. Based on the report by Iversen et al. (15), strain 51329 is a member of the 16S rRNA gene cluster group 3, which is an atypical E. sakazakii strain. Strain 29544 is a member of the 16S rRNA gene cluster group 1, which contains most of the E. sakazakii strains causing disease.
(i) Stage 1: preparation of the lysate-sonicate.
Frozen E. sakazakii cells (ATCC 51329 or 29544) were thawed and streaked onto TSA-S and incubated at 37°C for 18 h. A seed culture suspension was prepared by suspending the cells in saline. Two flasks containing 3% tryptone broth (500 ml per flask) were each inoculated with 25 optical density at 650 nm units of the seed culture, and the flasks were incubated at 37°C on a rotary shaker at 100 rpm for 18 h. Cells were recovered by centrifugation, washed once with saline, and suspended in 200 ml of 0.02 M Tris-buffered saline, pH 7.5, containing polymyxin B (2 mg/ml) and incubated at 37°C on a rotary shaker at 100 rpm for 2 h. The cells were stored on ice and subjected to eight 15-s sonications with a 15-s rest between each consecutive sonication step by using a Tekmar sonicator equipped with a macrotip (Tekmar Co., Cincinnati, OH). The supernatant of the recovered cell lysate-sonicate was recovered by centrifugation (16,000 × g, 30 min) and represents the stage 1 preparation.
(ii) Stage 2: ammonium sulfate precipitation.
Ammonium sulfate was added to the stage 1 preparation to 70% saturation, and the precipitate was recovered by centrifugation (16,000 × g, 30 min) and dissolved in 5 ml of phosphate-buffered saline (PBS) (0.067 M Na2HPO4-0.077 M NaCl [pH 7.0]). This stage 2 preparation was centrifuged (25,000 × g, 15 min) to remove any insoluble residue.
(iii) Stage 3. Gel filtration chromatography.
The stage 2 preparation was applied to a Sephadex G-100 (GE Healthcare, Piscataway, NJ) column equilibrated with PBS. Fractions were collected and assayed for proteolytic activity toward azocasein. Active fractions were pooled and represent the stage 3 preparation.
(iv) Stage 4. Hydrophobic interaction chromatography.
Ammonium sulfate was added to the pooled stage 3 fractions to a final molarity of 1.0 and applied to a phenyl-Sepharose CL-4B column (GE Healthcare) equilibrated with PBS containing 1.0 M ammonium sulfate. The column was washed with the equilibrating buffer, and the bound proteins were eluted with a decreasing gradient of ammonium sulfate in PBS. Fractions having proteolytic activity were pooled and represent the stage 4 preparation. This preparation was concentrated to ca. 2.0 ml using a Centriprep 10 concentrator (Millipore Corp., Billerica, MA).
(v) Stage 5. Gel filtration chromatography.
The stage 4 concentrate was applied to a Sephadex G-100 column. Fractions were collected and assayed for proteolytic activity against azocasein, and active fractions were pooled to represent the stage 5 preparation.
SDS-PAGE and isoelectric focusing.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed using 8-to-25%-gradient gels in the PhastSystem (GE Healthcare). The molecular weight of the reduced and denatured protease was estimated by the relative mobility method of Weber et al. (34). Purified protease was analyzed by thin-layer isoelectric focusing using pH-3-to-9 gels in the PhastSystem. After electrophoresis, the gel was divided into two parts. One part of the gel was stained with Coomassie brilliant blue R-250, and the other part was soaked in 0.067 M Tris-buffered saline (pH 7.5) for 5 min and examined for protease activity of the focused band by a zymogram technique with an overlay of 1% soluble casein in agarose as described by Lyerly and Kreger (23).
Assays. (i) Protease activity.
Proteolytic activity of all stages during the purification of the enzyme was measured using azocasein (Sigma Chemical Co., St. Louis, Mo) as described by Kreger and Gray (20). The reaction mixture was incubated at 37°C for 30 min. One protease unit was defined as the amount that resulted in a mixture yielding an absorbance of 1 at 440 nm. Collagenolytic and elastolytic activities of the protease were determined using the azocoll (Sigma) method described by Chavira et al. (3) and a modification (18) of the elastin-Congo red (Sigma) method described by Sachar et al. (30), respectively.
(ii) Hemagglutination activity.
The purified metalloprotease preparation was assayed for hemagglutination activity using glutaraldehyde-stabilized chicken and fresh sheep erythrocytes according to the procedure described by Delston et al. (5).
(iii) Protein determination.
Protein was estimated by Bradford's method (2). The standard (bovine serum albumin) and the reagent were purchased from Bio-Rad Laboratories (Hercules, CA).
Inactivation studies.
The heat stability of the protease was examined by heating the preparation to 65 or 100°C for 15 min and measuring the residual proteolytic activity of the preparation. The pH stability of the protease was determined by diluting the enzyme in various buffers (pH 4 to 10) at 4°C and measuring the activity remaining after 16 h. The effects of various inhibitors on proteolytic activity were examined by incubating the protease with ortho-phenanthroline, Zincov, phenylmethylsulfonyl fluoride, N-ethylmaleimide, or soybean trypsin inhibitor for 30 min at 25°C before measuring activity against azocasein. Except for Zincov, all the inhibitors were purchased from Sigma. Zincov was purchased from CalBiochem, Inc. (San Diego, CA).
Temperature and pH optimization for proteolytic activity.
The activity of the protease was determined by incubating the enzyme with azocasein at 27, 32, 37, and 42°C for 5, 10, 15, 20, 25 and 30 min. The optimum pH for activity was determined by incubating the enzyme with the azocasein in 0.1 M Tris and 0.1 M Na2HPO4 buffers adjusted to values of 4 to 10. Activity at the lower pH values was confirmed using sodium acetate buffer adjusted to pH values of 4.6 to 5.6.
N-terminal amino acid sequence.
The N-terminal amino acid sequences of the purified protease preparation and excised Coomassie blue-stained bands from Western blots were determined by Edman degradation using a Procise Model 492 protein sequencer (Applied Biosystems, Foster City, CA). Western blots were prepared by separating the protein samples by SDS-PAGE and blotting onto Problott membrane (Applied Biosystems) in a transfer buffer (10 mM 3-[cyclohexylamino]-1-propane sulfonic acid containing 10% methanol, pH 11). Proteins on the membrane were visualized by staining with Coomassie brilliant blue R-250.
PCR analysis for presence of zinc-containing metalloprotease gene, zpx. (i) Primers designed from N-terminal amino acid sequence.
All primers used in the PCR experiments were prepared by Invitrogen Corp. (Carlsbad, CA) or Integrated DNA Technologies (Coralville, IA). The nucleotide sequence for degenerate primer BAM 55 (5′-GTNTCNGCNAARGGNGARCTNG-3′) was designed based on the zinc-metalloprotease (Zpx) N-terminal amino acid sequence, and the nucleotide sequence for degenerate primer BAM 57 (5′-CCAAASACATCRGAMARMGATTC-3′) was designed based on the nucleotide sequences of the proteases from Pectobacterium carotovorum (NCBI accession no. M36651.1) and Anabaena sp. strain PCC 7120 (NCBI accession no. NC_003272). Primers BAM 55 and 57 amplified a 422-bp fragment of the zpx gene using the Platinum PCR Supermix kit (Invitrogen), which supplied the 20-μl reaction mixtures with 1 U of Taq DNA polymerase, 1.5 mM MgCl2, and 200 μM of each deoxynucleoside triphosphate (dNTP). Primers were added at 1 μM each, and 1 μl of bacterial cell lysate served as the DNA template. The polymerase was activated by 2 min of incubation at 95°C, followed by 35 cycles of 30 s at 95°C, 30 s at 55°C, and 1 min at 72°C, followed by a final extension period of 7 min at 72°C. The assay was run in a Gene Amp 9700 (Applied Biosystems) thermal cycler.
(ii) PCR primers designed from zpx sequence to be used for detection of zpx.
The 422-bp product of primers BAM 55 and 57 from 14 different strains of E. sakazakii was cloned using the QIAGEN PCR Plus cloning kit according to the manufacturer's instructions (QIAGEN, Inc., Valencia, CA). Chromatographs of each of the 14 nucleotide sequences (sequenced by Amplicon Express, Pullman, WA) were edited, aligned, and compared with one another using ChromasPro software, version 1.2 (Technelysium Pty. Ltd., Tewantin, Australia). Based on 382 of the 422 nucleotides from this consensus protease gene sequence, the degenerate primer BAM 122 (5′-AWATCTATGACGCGCAGAACCG-3′) and primer BAM 123 (5′-AAAATAGATAAGCCCGGCTTCG-3′) were designed. Primers BAM 122 and 123 amplified a 350-bp fragment of the zpx gene by using the HotStarTaq Master Mix kit (QIAGEN, Inc.) which supplied the 20-μl reaction mixtures with 1 U of Taq DNA polymerase, 1.5 mM MgCl2, and 200 μM of each dNTP. Primers were added at 1 μM each, and 1 μl of bacterial cell lysate served as the DNA template. The hot start polymerase was activated by incubation for 15 min at 95°C, followed by 35 cycles of 1 min at 95°C, 1 min at 62°C, and 1 min at 72°C, followed by a final extension period of 7 min at 72°C. The assay was run in a DNA Engine thermal cycler (Bio-Rad).
Because primer BAM 122 used in the above PCR was degenerate, we redesigned primers, namely Es-ProF (5′-GAAAGCGTATAAGCGCGATTC-3′) and Es-ProR (5′-GTTCCAGAAGGCGTTCTGGT-3′), that would amplify a region within the 350-bp amplicon and tested their specificity. By using the HotStarTaq Master Mix kit (QIAGEN, Inc.) and PCR conditions described above for primers BAM 122 and BAM 123, except that the final extension period was increased to 10 min instead of the previously mentioned 7 min, the PCR with primers Es-ProF and Es-ProR resulted in the amplification of a 94-bp fragment of the zpx gene.
(iii) Cloning and sequencing of the E. sakazakii metalloprotease gene, zpx, of strain ATCC 29544.
Purified DNA for cloning and sequencing was isolated from ATCC strain 29544 using the Dneasy Plant Mini kit (QIAGEN, Inc.) according to the manufacturer's instructions and recommendations. PCR primers Esak 5P (5′-AACCAGTCACGTTATCCAACC-3′) and ES-R2 (5′-TCACAACACCCCTGTGGTT-3′) were designed from the partial genomic sequence of strain BAA 894 published by Washington University's Genome Sequencing Center (St. Louis, MO) (http://genome.wustl.edu/genome.cgi?GENOME=Enterobacter%20sakazakii). Primers Esak5P and ES-R2 amplified the zpx gene in 50-μl reaction mixtures using 1 μl of purified genomic DNA as a template. The reaction mixture consisted of 2× HotStarTaq Master Mix (QIAGEN, Inc.) containing 2.5 U of Taq DNA polymerase and 200 μM of each dNTP with an adjusted final concentration of 2 mM MgCl2 and 0.2 μM of each primer. Thermal cycling was conducted in a PTC-200 DNA Engine (Bio-Rad) and consisted of a 15-min incubation period at 95°C, followed by 30 cycles of 94°C for 30 s, 54°C for 45 s, 72°C for 45 s, and a final extension at 72°C for 10 min. The resulting 1,026-bp PCR product was confirmed by 1.5% agarose gel electrophoresis. The purified product was sequenced by using Amplicon Express.
For cloning and sequencing, a greater quantity of the 1,026-bp amplicon was needed and was produced by PCR as described above using primers Esak5P and Es-R2. The additional amplicons were purified using the QIAquick PCR purification kit (QIAGEN, Inc.) according to the manufacturer's instructions. The resulting gene was cloned into the pDrive vector using chemically competent cells (QIAGEN, Inc.). Cloning was performed as outlined by the manufacturer. Briefly, purified amplicon was ligated into pDrive (a component of the QIAGEN PCR Plus cloning kit) and an aliquot was added to the chemically competent cells. Cells were alternately incubated on ice and at 42°C, followed by plating on LB agar containing 100 μg/ml ampicillin. The colonies were allowed to grow overnight and were screened by lysing the cells and reamplifying the zpx gene as described above. The pDrive with zpx was purified and sequenced by Amplicon Express (Pullman, WA).
(iv) DNA template preparation.
For use in the PCR screening studies, DNA templates of each of the strains were prepared by inoculating a TSA-S plate from a −80°C frozen stock culture and incubating the plates at 30°C. A colony was transferred to 5 ml of TSB-S in a sterile 16-by-150-mm culture tube and grown on a shaker (100 rpm) for 18 to 24 h at 30°C. Cells from the cultures were harvested by centrifugation at 14,000 × g at 4°C for 10 min. Each cell pellet was suspended in 200 μl of sterile distilled water and boiled for 10 min. The lysed cell suspension was centrifuged again at 14,000 × g at 4°C for 10 min, and the supernatant was transferred to a second labeled tube for storage at −20°C.
(v) Nucleotide sequencing.
All products for nucleotide sequencing were sequenced utilizing Amersham Biosciences' ET Terminator chemistry using an ABI 377 DNA sequencer (Amplicon Express).
Nucleotide sequence accession number.
The zpx gene sequence of E. sakazakii ATCC 29544 has been submitted to GenBank (accession number EF061082).
RESULTS AND DISCUSSION
Bacterial metalloproteases, in which zinc is the basal metal ion for catalytic activity, are produced by a number of pathogenic bacteria and fungi which are infectious for a variety of animals (5, 8, 11, 12, 18, 24, 31) and plant species (35). In animals, these proteases are active against structural components of the tissue extracellular protein matrices, such as type IV collagen. They can also disassociate heme from host protein carriers and can digest various plasma proteins (24). Extracellular matrix protein degradation takes place during various pathological conditions, such as invasion of bacterial pathogens into otherwise aseptic tissues (24), i.e., meningitis (11) and periodontal disease (10). Animal studies have shown that these proteases can also possess vascular permeability-enhancing activity which leads to tissue necrosis, the most classic example of tissue damage caused by these exoproteins (24). It is also thought that specific degradation of extracellular matrix protein components, such as type IV collagen, may cause destruction of endothelial cell membranes of capillary vessels, leading to the leakage of blood components into surrounding tissues (24), thus enabling pathogens to cross the blood-brain barrier. Collectively, in this manner, bacterial metalloproteases are thought to play a role in disease by enhancing the dissemination of bacteria from local sites of infection into the systemic circulation (11, 24). Evidence presented in this report establishes that the protease expressed by E. sakazakii possesses properties that are similar to those found in other zinc-containing metalloproteases, while also possessing unique properties of its own.
Protease activity in broth and agar cultures.
Bacterial virulence factors of gram-negative organisms (e.g., enterotoxins, proteases, and hemolysins) may be actively secreted into the culture supernatant (8, 18, 20); others may be released only into the periplasmic space (13). In order to examine the effects of these proteins on cells in tissue culture, our laboratory utilizes CHO cells in a protocol (19) that routinely analyzes both culture supernatants and polymyxin B lysates of putative diarrheagenic organisms. Our initial observations with 12 strains showed that qualitatively, cell-rounding activity was present in only some of the lysates but not in any of the supernatants. Since proteases, such as those produced by marine vibrios, have been known to cause “rounding” of CHO cells (22), we analyzed samples from these strains for protease activity against azocasein. Results showed that the activity was below detectable limits (≤0.1 U/ml) in all of the supernatants and four of the lysates but was relatively higher in lysates from other strains. The lysate from strain 51329 had the maximum amount of protease activity observed among the strains tested (0.91 U/ml); lysates from seven other strains had values ranging from 0.2 to 0.34 U/ml. These results suggest that the protease is cell associated. Based on these results, we selected strain 51329 for additional studies. Results of these studies indicated that lysis of the cells followed by a sonication procedure increased the yield of the protease by about 50% (data not shown). Subsequently, the cells of this strain were routinely subjected to lysis and then sonication in order to isolate the cell-bound protease.
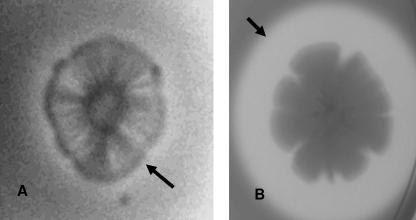
Alternatively, a screening assay using activity in skim milk agar plates showed that all 12 strains possessed proteolytic activity under and very close to the colony after an incubation period of 2 to 3 days at 37°C, and an example of the reaction is shown in Fig. 1A. In contrast, a wide zone of clearing was observed for a colony of a control strain, Vibrio vulnificus A9 (Fig. 1B). This result also suggests that the E. sakazakii protease is associated with the cell and is not secreted into the medium. These findings are different from that for other bacterial proteases, such as those expressed by Erwinia amylovora (35), V. vulnificus (18), Pseudomonas aeruginosa (20), and Vibrio cholerae (8), which are readily secreted into the culture medium. Testing of 74 additional E. sakazakii strains on skim milk agar plates showed that only 50% of the strains were positive in this assay (data not shown). It is quite likely that some strains do not qualitatively produce enough protease to degrade the milk casein in this medium even though PCR studies (see Table 4; also described later in Results) showed that they possess the protease gene. This screening method was not very useful in our studies.
FIG. 1.
Expression of protease on skim milk agar. (A) E. sakazakii strain 51329. (B) Vibrio vulnificus A9. Note slight zone of clearing, which is denoted by an arrow under the colony of E. sakazakii strain 51329, compared to a wide zone of clearing, also denoted by an arrow, irradiating away from the positive control colony of V. vulnificus.
TABLE 4.
Bacterial strains used in this study that were evaluated with degenerative zinc-containing metalloprotease PCR primers and zinc-containing metalloprotease-specific PCR primers
| Source group (na) | PCR resultb [no. of positive strains/no. tested (%)]
|
||
|---|---|---|---|
| BAM 55c and 57c | BAM 122c and 123 | EsProF and EsProR | |
| E. sakazakii strains | |||
| Clinical (31) | 30/31 | 31/31 | 31/31 |
| Food (51) | 34/34 | 21/21 | 51/51 |
| Environmental (42) | 29/29 | 31/31 | 42/42 |
| Unknown (11) | 11/11 | 11/11 | 11/11 |
| Total (135) | 104/105 (99) | 94/94 (100) | 135/135 (100) |
| Non-E. sakazakii strains | |||
| E. cloacae (9) | 0/5 | 0/5 | 0/9 |
| Enterobacter sp. (6) | NT | NT | 0/6 |
| Pantoea agglomerans (1) | NT | NT | 0/1 |
| Citrobacter sp. (2) | 0/1 | 0/2 | 0/2 |
| Other “enterics” (7) | 0/7 | 0/7 | 0/7 |
| Total (25) | 0/13 (0) | 0/13 (0) | 0/25 (0) |
n, no. of strains.
PCR product sizes are 422 bp for primers BAM 55 and BAM 57, 350 bp for primers BAM 122 and 123, and 94 bp for EsProF and EsProR, respectively.
Degenerate primer.
Molecular weight and pI.
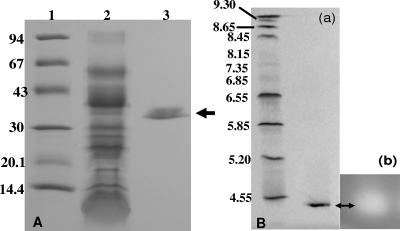
Sequential ammonium sulfate precipitation, gel filtration chromatography with Sephadex G-100, hydrophobic interaction chromatography with phenyl-Sepharose CL-4B, and a second gel filtration with Sephadex G-100 resulted in a purified preparation of the protease which was homogeneous by SDS-PAGE and isoelectric focusing (Fig. 2). The quantitative results of the purification of the E. sakazakii protease are summarized in Table 1. During the five-stage purification procedure, the specific activity of the protease increased from 5.7 to 450 U/mg of protein, and 12.6% of the protease present in the crude preparation was recovered in a purified form. The denatured and reduced protease has a molecular weight of ca. 38,000 (Fig. 2A), which is similar to the molecular sizes of proteases that are described for other bacterial pathogens (5, 21). However, it is 10 kDa smaller than the proteases produced by Erwinia amylovora (35) and V. vulnificus (24). Thin-layer isoelectric focusing in a pH-3-to-9 gel (Fig. 2B, panel a) revealed the presence of a single band with 4.4 as an isoelectric point of the native protein. This value is lower than pIs reported for various other proteases, such as those expressed by several marine vibrio species (5) and P. carotovorum (21). Zymogram analysis of the preparation separated by isoelectric focusing showed that proteolytic activity was associated with the Coomassie blue-stained band (Fig. 2B, panel b).
FIG. 2.
SDS-PAGE and isoelectric focusing gel of zinc-containing metalloprotease preparations from E. sakazakii strain 51329. (A) SDS-PAGE. Lanes: 1, molecular mass markers (values at left are in kilodaltons); 2, crude metalloprotease preparation (1.5 μg); 3, purified metalloprotease preparation (black arrow, 0.05 μg). Gel was stained with Coomassie brilliant blue R. (B) Analytical thin-layer isoelectric focusing. (a) Stained gel. Lanes: 1, pI markers; 2, purified metalloprotease preparation (0.4 μg). The gel was stained with Coomassie brilliant blue R. (b) Zymogram analysis of isoelectric focusing gel using an overlay of casein in 1% Noble agar.
TABLE 1.
Purification of E. sakazakii protease
| Stage | Volume (ml) | Protein concn (mg/ml) | Total amt of protein (mg) | Activity (U/ml) | Total activity (U) | Sp act (U/mg) | Recovery (%) |
|---|---|---|---|---|---|---|---|
| 1. Lysate/sonicate | 200 | 0.44 | 88 | 2.5 | 500 | 5.7 | 100 |
| 2. Ammonium sulfate precipitation | 11.5 | 1.35 | 15.5 | 38 | 437 | 28.1 | 87.4 |
| 3. Gel filtration chromatography (first) | 49 | 0.08 | 3.9 | 7.4 | 363 | 92.5 | 72.6 |
| 4. Hydrophobic interaction chromatography | 75 | 0.005 | 0.38 | 0.95 | 71 | 190 | 14.2 |
| 5. Gel filtration chromatography (second) | 35 | 0.004 | 0.14 | 1.8 | 63 | 450 | 12.6 |
Proteolytic activity.
The optimum temperature and pH range for activity against azocasein were determined to be 37°C and pHs 5 to 7, respectively. In addition to its activity against azocasein, the purified protease degraded azocoll and insoluble casein but not elastin and caused rounding of CHO cells; the minimum amount of protease required for 1 U of azocasein activity or CHO cell activity was 2.2 μg or 0.022 μg, respectively. Moreover, the E. sakazakii protease did not hemagglutinate chicken or sheep erythrocytes. The absence of elastase and hemagglutination activities suggest that the E. sakazakii protease is quite unique from other bacterial proteases, such as those expressed by marine vibrios, some of which possess hemagglutination activity (5, 8, 12, 18, 24).
Inactivation studies.
Results of the effects of various protease inhibitors, temperature, and pH on proteolytic activity are shown in Table 2. The protease lost 32 or 100% of its activity when heated for 15 min at 65 or 100°C, respectively, but was stable when incubated in buffers with pH values of 4 to 10 for 16 h at 4°C. The protease was inhibited by ortho-phenanthroline and Zincov but was not affected by N-ethylmaleimide, phenylmethylsulfonyl fluoride, or trypsin inhibitors. These results indicate that the protease is not a serine or cysteine protease but is a metalloprotease that contains zinc. Metalloproteases are the most diverse of the catalytic types of proteases and are characterized by the need for a divalent metal ion, such as zinc, for activity (29). These characteristics are similar to those of all known zinc-containing metalloproteases (29).
TABLE 2.
Inactivation of E. sakazakii protease
| Treatment | Residual activity (%) |
|---|---|
| None | 100 |
| ortho-Phenanthroline, 0.1 mM | 39 |
| Zincov, 0.1 mM | 39 |
| N-ethylmaleimide, 0.1 mM | 100 |
| Phenymethylsulfonylfluoride, 0.1 mM | 100 |
| Trypsin inhibitor, 10 μg | 100 |
| 65°C, 15 min | 68 |
| 100°C, 15 min | 0 |
| pH 4-10 at 4°C, 16 h | 100 |
N-terminal amino acid sequence.
Table 3 shows, in a comparative fashion, the N-terminal amino acid sequences of purified metalloproteases isolated from E. sakazakii strains 29544 and 51329, the deduced sequence from E. sakazakii strain BAA 894, and sequences obtained from BLAST analyses of the amino acid sequences of metalloproteases expressed by P. carotovorum and Serratia proteamaculans and that from Anabaena sp. Sequencing of both the purified preparation and the excised band from a Western blot gave identical N-terminal amino acid sequences for the metalloprotease expressed by E. sakazakii strain 51329. In comparison, 3 of the first 26 N-terminal amino acid residues of the E. sakazakii strain ATCC 25944 protease (obtained by sequencing a band excised from a Western blot of a crude metalloprotease preparation) were different from the N-terminal amino acid residues of the ATCC 51329 protease. Furthermore, the amino acid sequence from strain 29544 was identical to that of strain BAA 894. However, there were 6 amino acid residue differences found among the first 42 amino acid residues of the protease of strain 51329 and that of strain BAA 894. BLAST analysis of this amino acid sequence showed that the metalloprotease expressed by strain 51329 had strongest similarity to the proteases expressed by S. proteamaculans (65%), P. carotovorum (60%), and Pectobacterium atrosepticum (60%) and less similarity to other zinc-containing metalloproteases expressed by Arthrobacter sp. (58%), Xanthomonas sp. (55 to 57%), Myxococcus xanthus (55%), Frankia sp. (55%), Pseudomonas syringae pathovars syringae, tomato, and phaseolicola (50%-52%); and Bordetella subspecies parapertussis (50%) and bronchiseptica (50%). However, there was no similarity to the zinc-containing metalloprotease secreted by Erwinia amylovora (27, 35). Even though the zinc-containing metalloprotease expressed by E. sakazakii could degrade azocoll, it lacked elastinolytic activity and it shared no homology with elastin-degrading metalloproteases expressed by marine vibrios and Pseudomonas sp. (5, 8, 18, 20, 24).
TABLE 3.
Comparison between N-terminal amino acid sequences of E. sakazakii zinc-containing metalloproteases and those of Anabaena sp., P. carotovorum, and S. proteamaculans metalloproteases
| Source of metalloprotease | Sequence |
|---|---|
| E. sakazakii strains | |
| ATCC 29544a | -SAKGKLERDIYDAQNRETLPGELVRR--------------- |
| BAA 894b | VSAKGKLERDIYDAQNRETLPGELVRREGAPSNGDVAVDEAY |
| ATCC 51329c | VSAKGELERDIYDAQNREELPGKPVRREGEPSAGDVAVDEAY |
| Anabaena sp.d | SPSPGEKSRTIYDAQNSEQLPGKIVRVEGD------------ |
| P. caratovorume | KLPAGQANRSIHDAEQQQQLPGKLVRAEGQ------------ |
| S. proteamaculansf | TSTGGEVIRDIYDAENSTQLPGKQVRNEGQASNHDVAVDEAY |
Zinc-containing metalloprotease sequence obtained in this study.
These data were produced by the Genome Sequencing Center at Washington University School of Medicine in St. Louis, MO, and can be obtained from http://genome.wustl.edu/pub/organism/Microbes/Enteric_Bacteria/Enterobacter_sakazakii/assembly/Enterobacter_sakazakii-4.0/.
Metalloprotease sequence obtained in this study.
Metalloprotease sequence obtained from NCBI accession number NP_485026.1.
Metalloprotease sequence obtained from NCBI accession number AAD49575.
NCBI accession number 118067788.
PCR analysis and primer design for detection of the E. sakazakii species-specific metalloprotease gene, zpx.
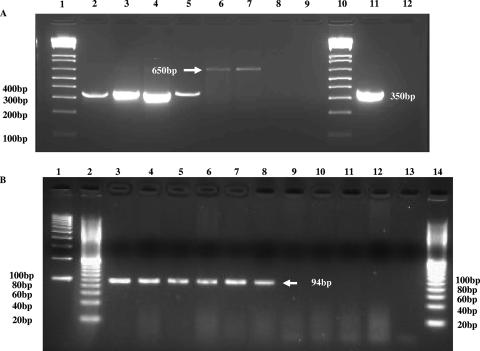
Using degenerate PCR primers, BAM 55 and BAM 57, positive PCR results (the presence of the 422-bp amplicon) were found for 104 of 105 (99%) E. sakazakii strains. The results of the PCR analysis are shown in Table 4. The single strain found to be negative with the degenerate PCR primers was strain 3523-75, a strain representative of biotype 15 according to Farmer et al.'s original biotyping scheme (7). At the time of these studies, the 16th biotype (15) was not available for analysis. Thirteen non-E. sakazakii control strains were negative in this PCR analysis. Results from experiments using PCR primers BAM 122 and BAM 123 (based on the consensus nucleotide sequence of 14 amplicons produced in the PCR using BAM 55 and BAM 57) detected the presence of zpx (the presence of the 350-bp amplicon) in all 94 E. sakazakii strains tested, including all 15 of 16 representatives of Farmer's biotypes. The same negative control strains tested previously were also negative with these primers. Results from experiments using PCR primers Es-ProF and Es-ProR (based on the sequence of the amplicon produced in the PCR using BAM 122 and BAM 123) detected the presence of zpx (the presence of the 94-bp amplicon) in all 135 E. sakazakii strains tested, including 15 of the 16 representatives of Farmer's biotypes. Twenty-five non-E. sakazakii/other Enterobacteriaceae strains, including members of Enterobacter cloacae, other Enterobacter spp., Escherichia hermannii, Citrobacter freundii, Escherichia coli, Klebsiella pneumoniae, Pantoea agglomerans, P. carotovorum, and Salmonella spp., were negative using these PCR primers. The 350-bp and 94-bp amplicons, the expected PCR products for the above PCRs, are shown in typical fashion in Fig. 3. Analysis of sequences of several of these PCR products showed that they matched the nucleotide sequence for zpx (data not shown). These results suggest that both sets of primers (BAM 122 and BAM 123 and Es-ProF and Es-ProR) detect a region of zpx that is specific for E. sakazakii and also suggest that zpx could be used as a species-specific gene target for the detection of E. sakazakii, similar to how gyrB and toxR gene targets for the detection of Vibrio hollisae (33) and that of vvhA for the detection of V. vulnificus (25) are used.
FIG. 3.
PCR results for E. sakazakii zpx using primers BAM 123 and BAM 122 (A) or Es-ProF and Es-ProR (B) for E. sakazakii and non-E. sakazakii strains. Panel A represents a typical 1% agarose gel after electrophoresis, showing the PCR amplification of the 350-bp amplicon. Lanes 1 and 10, Bio-Rad 100-bp molecular size DNA markers; lanes 2 to 5, E. sakazakii strains 11, 569, 573, and 665, respectively; lanes 6 to 9, non-E. sakazakii strains Enterobacter cloacae strain 23355, Citrobacter freundii strain 7, Klebsiella pneumoniae strain 8, and Escherichia coli strain 4, respectively; lane 11, E. sakazakii strain 29544; lane 12, distilled water was used instead of DNA template. Note that the 650-bp product was sequenced and was found not to be related to that of the metalloprotease gene. Panel B represents a typical 3% agarose gel showing the PCR amplification of the 94-bp band. Lanes 1 and 2, Bio-Rad 100-bp and 20-bp DNA molecular size markers, respectively; lanes 3 to 8, E. sakazakii strains 29544, 4963-71, 1121-73, 9363-75, 570, and 564, respectively; lane 9, distilled water; lanes 10 to 14, negative reactions of Enterobacter cloacae subspecies dissolvens ATCC strain 23373, Enterobacter gergoviae ATCC strain 33028, P. carotovorum strain EC15 (provided by Arvind Bhagwat, USDA, Beltsville, MD), Citrobacter freundii ATCC strain 43864, and 20-bp DNA molecular size marker, respectively.
Characterization and cloning of the metalloprotease gene, zpx.
Comparison of the nucleotide sequence of zpx obtained from E. sakazakii strain ATCC 29544 with that of the online-published BAA 894 sequence shown in Fig. 4 confirmed that the entire metalloprotease gene of strain 29544 consisted of 1,026 nucleotides. Further comparative analysis of the BAA 894 sequence with the sequence obtained from strain 29544 showed that a ribosome binding motif, AGGA, began 12 nucleotides upstream of the ATG start codon. This was confirmed by performing a PCR using primers Contig For (5′-CTTGCCAACCTGCGTGATG-3′) (designed from the BAA 894 DNA sequence) and Contig Rev (5′-TCTCTTTGGGCTGATGCG-3′) (designed from the 29544 DNA sequence), which produced a 1,019-bp PCR amplicon, which when sequenced gave a segment of nucleotide sequence spanning 875 bp upstream of the start codon. A single TGA stop codon was found at bp 1024 to 1026. Clustal X sequence alignment analysis, shown in Fig. 4, demonstrated that the nucleotide sequence identity between zpx genes possessed by both ATCC strain 29544 and strain BAA 894 was 97% (23 nucleotide differences found between these two sequences of zpx). In comparison, the nucleotide sequence of the metalloprotease gene possessed by S. proteamaculans was only 67% identical to the E. sakazakii zpx sequence. The homology between zpx and other protease genes, such as those of S. proteamaculans and P. carotovorum, was not equally distributed over the gene sequence and showed less homology at the N and C termini. However, the protein sequence homology among the proteases (Zpx of E. sakazakii versus others) was greater than the corresponding DNA sequence homologies.
FIG. 4.
CLUSTAL X alignment of nucleotide sequences of the E. sakazakii metalloprotease genes of strains 29544 and BAA 894 compared to sequence of S. proteamaculans gene. Gene sequences for E. sakazakii strain 29544 were obtained from this study; gene sequence for E. sakazakii strain BAA 894 was obtained from the genome center at Washington University, and gene sequence for S. proteamaculans came from NCBI accession number NZ43AAUN01000006. The Shine-Dalgarno motif and PCR primer sequences used in the study are indicated by underlining and subheadings (bold). Start and stop codons are also bold. Asterisks identify conserved nucleotides among the sequences.
The deduced amino acid sequences of Zpx for E. sakazakii strains 29544 and BAA 894 are shown in Fig. 5A. The metalloprotease of strain 29544 consists of 341 amino acid residues, which corresponds to a theoretical protein size of 37,782 with a theoretical pI of 5.23, which is similar to the metalloprotease (341 amino acids) possessed by strain BAA 894, with a theoretical molecular size of 37,818 and pI of 5.3, and is also similar to the findings reported here for the purified metalloprotease. Comparatively, the sequence of the metalloprotease of S. proteamaculans, also shown in Fig. 5A, has 243 (71%) of its 341 total amino acids identical to those in both that of E. sakazakii strain ATCC 29544 and that of BAA 894. The deduced amino acid sequence of the cloned zpx gene of strain 29544 and that for strain BAA 894 contained the sequence H-E-L-S-H, (amino acid residues 162 to 166), which is consistent with the signature consensus sequence of the zinc-binding motif HEXXH found in the metzincins of the metalloprotease family of peptidases (12). A comparison of 11 other microbial metalloprotease zinc-binding motifs is shown in Fig. 5B. Also, the putative zinc-binding signature as described by Jongeneel et al. (16), which consists of (uncharged amino acid)-(uncharged amino acid)-H-E-(uncharged amino acid)-(uncharged amino acid)-H-(uncharged amino acid)-(hydrophobic amino acid), was found, V-G-H-E-L-S-H-G-V (amino acid residues 160 to 168). In metalloproteases, the two histidine residues and the glutamic acid residue have been reported to be the putative zinc-binding and active-site residues, respectively (12). An additional feature found in the E. sakazakii metalloprotease deduced amino acid sequence which is also found in other metzincin proteases was a shortened and thus putative Met-turn signature (1), present as S-M-S (amino acid residues 228 to 230), which was located 60 amino acids downstream of the zinc-binding motif. A comparison of 11 other microbial metalloprotease Met-turn signature motifs is shown in Fig. 5B. A catalytic motif consisting of YFEQAGALNESLSDVFG (amino acid residues 177 to 193), similar to that described by Hase and Finkelstein (12), was also found downstream of the zinc-binding motif. Last, a second zinc catalytic motif, as described by both Sirakova et al. (31) and Hase and Finkelstein (12), was found, GGVHINSGIPNRAFY (amino acid residues 259 to 275), which was downstream of the Met-turn signature. A comparison of 11 other metalloprotease zinc-catalytic motifs is shown in Fig. 5B. Also, these results suggest that a larger preprotease form of the active protein, such as that found for V. cholerae and V. vulnificus, is not produced in E. sakazakii. In fact, the deduced amino acid sequence of the metalloprotease from strains 29544 and BAA 894 does not have an N-terminal amino acid signal peptide. This information was confirmed using the SignalP 3.0 server protein prediction software, SignalP-HMM, which uses neural networks and hidden Markov modeling of protein sequences according to the method described by Dyrløv-Bendtsen et al. (6) to find signal sequences (http://www.cbs.dtu.dk/services/SignalP/). Last, there was no evidence of a glycine-rich repeating motif, which has been implicated in Ca+2 binding and/or in presentation of a signal peptide to secretion machinery. Another unique finding for the E. sakazakii protease is that unlike other metalloproteases, such as those expressed by V. cholerae and P. aeruginosa, an indispensable secretion motif, constituting the four C-terminal amino acid residues (DXXX), in which the X represents a hydrophobic residue, was not found for Zpx. This motif is required for secretion of proteins via the type 1 secretion pathway (9). It is unclear at this point if the last four C-terminal amino acids of the protein, threonine, glycine, valine, and leucine, could serve in this manner. Nonetheless, these results support our finding that the protease was secreted poorly, if at all, into the culture supernatant and again signifies a unique property deviation observed for the E. sakazakii metalloprotease compared to other bacterial metalloproteases. There are many examples of toxins and proteins that never make it out of the periplasmic space via an active secretion process, for example, Shiga-like toxins I and II (SLT-I and SLT-II), produced by enterohemorrhagic E. coli strains (SLT-I more so than SLT-II [32]), the Salmonella enterotoxin (4), and the heat-labile enterotoxin produced by enterotoxigenic E. coli (13). Though we do not know whether the metalloprotease expressed by E. sakazakii gets out of the cell or not, we do show that molecularly it does not have specific known secretion motifs at its disposal. From these data, these results separate and distinguish this metalloprotease from various other bacterial proteases. Furthermore, Zpx shares a high degree of homology with the metalloproteases mentioned above in terms of structure and function and should be classified as a member of the metzincin family in the zincin superfamily of proteases (1).
FIG. 5.
(A) CLUSTAL X alignment of the deduced amino acid sequences for metalloproteases expressed by E. sakazakii strains 29544 and BAA 894 compared to that for S. proteamaculans. Numbers at the beginning and end of each line refer to the amino acid residue for each sequence. Arrowheads in bold font above the sequence for strain 29544 indicate the amino acid residues found by our analysis of the purified protease. (B) Comparison of the conservative motifs of E. sakazakii metalloprotease with other microbial metalloproteases. Numbers at the beginning and end of each sequence within each motif refer to the amino acid residue for each of the metalloprotease sequences. Amino acid residues common among the Zn-binding, catalytic, and Met turn motifs are indicated by shading. *, completely conserved residue; :, strongly conserved residue;., weakly conserved residue (as defined by Clustal X). Other microbial metalloprotease sequences came from the following GenBank references: S. proteamaculans, gi no. 118067788l; P. cartovorum, gi no. 1172650; P. atroseptica, gi no. 50122133; B. parapertussis 12822, gi no. 33597460; X. axonopodis pv. citri strain 306, gi no. 21240878; Anabaena sp. strain PCC 7120, gi no. 17228478; Anabaena variabilis ATCC 29413, gi no. 75909157; Arthrobacter sp. strain FB24, gi no. 116671175; S. coelicolor A3(2), gi no. 21220989; B. anthracis strain Ames, gi no. 30260755; S. aureus subsp. aureus MRSA252, gi no. 49242963.
The results of our investigation demonstrate that E. sakazakii produces a zinc-containing metalloprotease which shares some properties with those produced by other bacterial pathogens while possessing other properties which are unique. Evidence is presented which shows that it possesses collagenolytic activity but not elastinolytic activity. Taking advantage of unique nucleotide sequences, which were initially derived from the N-terminal amino acid sequence of the protein, a species-specific PCR assay was developed which has proven useful in confirming strains to be E. sakazakii. The role of this protease in disease has not been examined. Speculatively, it quite likely may be responsible singly or in combination with the recently described enterotoxin (28) for the necrosis and extensive cellular destruction in neonates with necrotizing enterocolitis (26). In addition, it may be involved in allowing the organism to cross the blood-brain barrier. The epidemiology and pathogenicity of E. sakazakii will be better understood only when the ecological nuances of the microbe-food-disease-host dynamics are elucidated.
Acknowledgments
We thank Lawrence Restaino, a true “gentleman scientist,” R&F Laboratories, Downers Grove, IL, for the many discussions about the research project from its infancy through to its completion and for contributing the many food-related strains. We also thank Eric Brown, CFSAN, FDA, and Christine Keys, CFSAN, FDA, for contributing bacterial strains which were used in the study. Special thanks goes to Caroline Mohr, Program Management Officer, Epidemiology and Laboratory Branch, Division of Healthcare Quality Promotion, Centers for Disease Control and Prevention (CDC), Atlanta, GA, who facilitated the transfer of the E. sakazakii strains which represented the original 15 biotypes described by James Farmer, CDC, from the Diagnostic Microbiology Section, Enteric Reference Lab, CDC, to the FDA. We thank Atin Datta, FDA, for his suggestions related to the manuscript.
Footnotes
Published ahead of print on 4 May 2007.
REFERENCES
- 1.Bode, W., F. X. Gomis-Rüth, and W. Stöker. 1993. Astacins, serralysins, snake venom and matrix metalloproteases exhibit identical zinc-binding environments (HEXXHXXGXXH and Met turn) and topologies and should be grouped into a common family, the metzincins. FEBS Lett. 331:134-140. [DOI] [PubMed] [Google Scholar]
- 2.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Chavira, R., Jr., T. J. Burnett, and J. H. Hageman. 1984. Assaying proteinases with azocoll. Anal. Biochem. 136:446-450. [DOI] [PubMed] [Google Scholar]
- 4.Chopra, A. K., C. W. Houston, J. W. Peterson, and J. J. Mekalanos. 1987. Chromosomal DNA contains the gene coding for Salmonella enterotoxin. FEMS Microbiol. Lett. 43:345-349. [Google Scholar]
- 5.Delston, R. B., M. H. Kothary, K. A. Shangraw, and B. D. Tall. 2003. Isolation and characterization of a zinc-containing metalloprotease expressed by Vibrio tubiashii. Can. J. Microbiol. 49:525-529. [DOI] [PubMed] [Google Scholar]
- 6.Dyrløv-Bendtsen, J., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 7.Farmer, J. J., III, M. A. Asbury, F. W. Hickman, D. J. Brenner, and the Enterobacteriaceae Study Group. 1980. Enterobacter sakazakii: a new species of “Enterobacteriaceae” isolated from clinical specimens. Int. J. Sys. Bacteriol. 30:569-584. [Google Scholar]
- 8.Finkelstein, R. A., M. Boesman-Finkelstein, and P. Holt. 1983. Vibrio cholerae hemagglutinin/lectin/protease hydrolyzes fibronectin and ovomucin: F. M. Burnet revisited. Proc. Natl. Acad. Sci. USA 80:1092-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghigo, J. M., and C. Wandersman. 1994. A carboxy-terminal four amino acid motif is required for secretion of the metalloprotease PrtG through the Erwinia chrysanthemi protease secretion pathway. J. Biol. Chem. 269:8979-8985. [PubMed] [Google Scholar]
- 10.Grenier, D., V. J. Uitto, and B. C. McBride. 1990. Cellular location of a Treponema denticola chymotrypsin-like protease and importance of the protease in migration through the basement membrane. Infect. Immun. 58:347-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harrington, D. J. 1996. Bacterial collagenases and collagen-degrading enzymes and their potential role in human disease. Infect. Immun. 64:1885-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hase, C. C., and R. A. Finkelstein. 1993. Bacterial extracellular zinc-containing metalloproteases. Microbiol. Rev. 57:823-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirst, T. R., L. L. Randall, and S. J. S. Hardy. 1984. Cellular location of heat-labile enterotoxin in Escherichia coli. J. Bacteriol. 157:637-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iversen, C., M. Lane, and S. J. Forsythe. 2004. The growth profile, thermotolerance and biofilm formation of Enterobacter sakazakii grown in infant formula milk. Lett. Applied. Microbiol. 38:378-382. [DOI] [PubMed] [Google Scholar]
- 15.Iversen, C., M. Waddington, J. J. Farmer III, and S. J. Forsythe. 2006. The biochemical differentiation of Enterobacter sakazakii genotypes. BMC Microbiol. 6:94-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jongeneel, C. V., J. Bouvier, and A. Bairoch. 1989. A unique signature identifies a family of zinc-dependent metallopeptidases. FEBS Lett. 242:211-214. [DOI] [PubMed] [Google Scholar]
- 17.Kandhai, M. C., M. W. Reij, L. G. Gorris, O. Guillaume-Gentil, and M. van Schothors. 2004. Occurrence of Enterobacter sakazakii in food production environments and households. Lancet 363:39-40. [DOI] [PubMed] [Google Scholar]
- 18.Kothary, M. H., and A. S. Kreger. 1985. Production and partial characterization of an elastolytic protease of Vibrio vulnificus. Infect. Immun. 50:534-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kothary, M. H., E. F. Claverie, M. D. Miliotis, J. M. Madden, and S. H. Richardson. 1995. Purification and characterization of a Chinese hamster ovary cell elongation factor of Vibrio hollisae. Infect. Immun. 63:2418-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kreger, A. S., and L. D. Gray. 1978. Purification of Pseudomonas aeruginosa proteases and microscopic characterization of pseudomonal protease-induced rabbit corneal damage. Infect. Immun. 19:630-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kyostio, S. R. M., C. L. Cramer, and G. H. Lacy. 1991. Erwinia carotovora subsp. carotovora extracellular protease: characterization and nucleotide sequence of the gene. J. Bacteriol. 173:6537-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lockwood, D. E., A. S. Kreger, and S. H. Richardson. 1982. Detection of toxins produced by Vibrio fluvialis. Infect. Immun. 35:702-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyerly, D., and A. Kreger. 1979. Purification and characterization of a Serratia marcescens metalloprotease. Infect. Immun. 24:411-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyoshi, S., and S. Shinoda. 2000. Microbial metalloproteases and pathogenesis. Microbes Infect. 2:91-98. [DOI] [PubMed] [Google Scholar]
- 25.Morris, J. G., Jr., A. C. Wright, D. M. Roberts, P. K. Woods, L. M. Simpson, and J. D. Oliver. 1987. Identification of environmental Vibrio vulnificus isolates with a DNA probe for the cytotoxin-hemolysin gene. Appl. Environ. Microbiol. 53:193-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nazarowec-White, M., and J. M. Farber. 1997. Enterobacter sakazakii: a review. Int. J. Food Microbiol. 34:103-113. [DOI] [PubMed] [Google Scholar]
- 27.Oh, C.-S., and S. V. Beer. 2005. Molecular genetics of Erwinia amylovora involved in the development of fire blight. FEMS. Microbiol. Lett. 253:185-192. [DOI] [PubMed] [Google Scholar]
- 28.Pagotto, F. J., M. Nazarowec-White, S. Bidawid, and J. M. Farber. 2003. Enterobacter sakazakii: infectivity and enterotoxin production in vitro and in vivo. J. Food Prot. 66:370-375. [DOI] [PubMed] [Google Scholar]
- 29.Rao, M. B., A. M. Tanksale, M. S. Ghatge, and V. V. Deshpande. 1998. Molecular and biotechnological aspects of microbial proteases. Microbiol. Mol. Biol. Rev. 62:597-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sachar, L. A., K. K. Winter, N. Sicher, and S. Frankel. 1955. Photometric method for estimation of elastase activity. Proc. Soc. Exp. Biol. Med. 90:323-326. [DOI] [PubMed] [Google Scholar]
- 31.Sirakova, T. D., A. Markaryan, and P. E. Kolattukudy. 1994. Molecular cloning and sequencing of the cDNA and gene for a novel elastinolytic metalloprotease from Aspergillus fumigatus and its expression in Escherichia coli. Infect. Immun. 62:4208-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strockbine, N. A., L. R. M. Marques, J. W. Newland, H. Williams-Smith, R. K. Holmes, and A. D. O'Brien. 1986. Two toxin-converting phages from Escherichia coli O157:H7 strain 933 encode antigenically distinct toxins with similar biologic activities. Infect. Immun. 53:135-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vuddhakul, V., T. Nakai, C. Matsumoto, T. Oh, T. Nishino, C.-H. Chen, M. Nishibuchi, and J. Okuda. 2000. Analysis of gyrB- and toxR-targeted PCR methods for isolation of Vibrio hollisae from the environment and its identification. Appl. Environ. Microbiol. 66:3506-3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weber, K., J. R. Pringle, and M. Osborn. 1972. Measurement of molecular weights by electrophoresis on SDS-acrylamide gel. Methods Enzymol. 26:3-27. [DOI] [PubMed] [Google Scholar]
- 35.Zhang, Y., D. D. Bak, H. Heid, and K. Geider. 1999. Molecular characterization of a protease secreted by Erwinia amylovora. J. Mol. Bacteriol. 289:1239-1251. [DOI] [PubMed] [Google Scholar]