Abstract
The molecular diversity of rumen methanogens in feedlot cattle and the composition of the methanogen populations in these animals from two geographic locations were investigated using 16S rRNA gene libraries prepared from pooled PCR products from 10 animals in Ontario (127 clones) and 10 animals from Prince Edward Island (114 clones). A total of 241 clones were examined, with Methanobrevibacter ruminantium accounting for more than one-third (85 clones) of the clones identified. From these 241 clones, 23 different 16S rRNA phylotypes were identified. Feedlot cattle from Ontario, which were fed a corn-based diet, revealed 11 phylotypes (38 clones) not found in feedlot cattle from Prince Edward Island, whereas the Prince Edward Island cattle, which were fed potato by-products as a finishing diet, had 7 phylotypes (42 clones) not found in cattle from Ontario. Five sequences, representing the remaining 161 clones (67% of the clones), were common in both herds. Of the 23 different sequences, 10 sequences (136 clones) were 89.8 to 100% similar to those from cultivated methanogens belonging to the orders Methanobacteriales, Methanomicrobiales, and Methanosarcinales, and the remaining 13 sequences (105 clones) were 74.1 to 75.8% similar to those from Thermoplasma volcanium and Thermoplasma acidophilum. Overall, nine possible new species were identified from the two clone libraries, including two new species belonging to the order Methanobacteriales and a new genus/species within the order Methanosarcinales. From the present survey, it is difficult to conclude whether the geographical isolation between these two herds or differences between the two finishing diets directly influenced community structure in the rumen. Further studies are warranted to properly assess the differences between these two finishing diets.
Enteric methane emission is a major source of greenhouse gas in agriculture (9), and the methane is formed in the rumen through a process called enteric fermentation. During this normal digestive process, hydrogen is released by other microbes during fermentation of forage and is used by methanogenic archaea (i.e., methanogens) to reduce carbon dioxide to methane.
In 2002, enteric fermentation from livestock accounted for 32% of all agricultural greenhouse gas emissions in Canada (3). Despite a 20% decline in the number of dairy cows from 199,000 in 1990 to 160,600 in 2000 (26), enteric emissions in Canada have increased 11% since 1990, mainly due to increased beef production (3). Reducing enteric methane emissions has been identified as one way of lowering global methane emissions. However, the effectiveness of any strategy that will reduce greenhouse gas emissions and also increase production or nutritional efficiency will likely depend upon having an understanding of the numbers and/or distribution of methanogen species among ruminant livestock.
In the present study, one 16S rRNA gene library was constructed from pooled rumen digesta samples from 10 feedlot cattle in Ontario, Canada, and another 16S rRNA gene library was constructed from pooled rumen digesta samples from 10 feedlot cattle from Prince Edward Island, Canada. The two libraries were examined to determine the molecular diversity of methanogens from feedlot cattle and to compare the compositions of the methanogen populations in these animals from two different provinces.
MATERIALS AND METHODS
Sources of samples and processing.
Approximately 50 ml of rumen digesta was collected from 10 Hereford-Cross cattle in Ontario and 10 Hereford cattle from Prince Edward Island immediately after slaughter from a large commercial abattoir (Better Beef Limited, Guelph, Canada) in June 2004. The 20 samples were maintained on ice for approximately 60 min while being transported to the laboratory for processing. Rumen digesta was aliquoted into two 1.5-ml Eppendorf tubes. The remaining rumen sample was fixed with an equal volume of 100% ethanol and stored. The feedlot cattle from Ontario were fed a corn-based diet, whereas the feedlot cattle from Prince Edward Island were fed potato by-products as a finishing diet.
DNA extraction, PCR amplification, and clone library construction.
DNA was extracted using a commercially available kit according to the manufacturer's instructions (QIAGEN DNeasy plant kit; QIAGEN, CA). The PCR amplification was performed in a PTC-100 thermal cycler (MJ Research, MA) using the methanogen-specific forward and reverse primers Met86F and Met1340R (30), following stringent parameters (33). A 16S rRNA gene library was constructed from the pooled PCR products from each group of 10 animals. Cloning of the 1.3-kb PCR products, selection of positive clones, and reamplification of the PCR products were by the protocol of Wright et al. (31, 33).
The final PCR product was digested with HaeIII endonuclease (30) according the to manufacturer's specifications. The digested DNA was separated on a 4% molecular screening agarose gel running at 100 V for at least 2 h. Restriction fragment length polymorphisms were grouped according to their riboprint patterns and compared to a riboprint database for identification (30). Clones resembling the HaeIII riboprint patterns for Methanobrevibacter smithii and Methanobrevibacter strains SM9 and M6 were further differentiated using the restriction endonuclease Sau3AI (32). A clone representing each new riboprint pattern and a clone from each known riboprint were sequenced in both directions for confirmation. Sequencing was performed with an ABI Prism 3730 48 capillary sequencer (Applied Biosystems Inc., CA) using BigDye Terminator (version 3) and Taq FS with two forward and two reverse methanogen 16S rRNA gene primers (30). Contigs were assembled using SEQMAN (DNASTAR Inc., WI).
Clones from cattle in Ontario, Canada, were designated ON-CAN to indicate the location of the herd, followed by a clone number. Similarly, clones from cattle from Prince Edward Island, Canada, were designated PE-CAN to indicate the location of the herd, followed by a clone number.
The sampling effort in each library was evaluated by calculating the coverage (C) according to the equation C = 1 − (n/N), where n is the number of sequences represented by a single clone (Tables 1 and 2) and N is the total number of clones analyzed in the library (7). The Shannon index (H) (19) was also used to characterize species diversity between the two clone libraries.
TABLE 1.
16S rRNA clones from pooled samples from 10 corn-fed feedlot cattle from Ontario, Canada
| 16S rRNA phylotype | No. of clonesa | Size (bp) | GenBank accession no. | Nearest valid taxon | % Sequence similarity |
|---|---|---|---|---|---|
| ON-CAN.01 | 47 | 1,260 | DQ123873 | Methanobrevibacter ruminantium | 98.8 |
| ON-CAN.02 | 25 | 1,256 | DQ123874 | Thermoplasma volcanium | 74.7 |
| ON-CAN.03 | 16 | 1,256 | DQ123875 | Thermoplasma acidophilum | 75.3 |
| ON-CAN.04 | 14 | 1,260 | DQ123876 | Methanobrevibacter ruminantium | 100.0 |
| ON-CAN.05 | 12 | 1,258 | DQ123877 | Methanimicrococcus blatticola | 89.8 |
| ON-CAN.08 | 2 | 1,262 | DQ123878 | Methanobrevibacter thaueri | 98.4 |
| ON-CAN.09 | 2 | 1,257 | DQ123879 | Thermoplasma volcanium | 74.5 |
| ON-CAN.10 | 1 | 1,262 | DQ123880 | Methanobrevibacter smithii | 100.0 |
| ON-CAN.11 | 1 | 1,260 | DQ123881 | Methanobrevibacter thaueri | 98.5 |
| ON-CAN.12 | 1 | 1,261 | DQ123882 | Methanobrevibacter thaueri | 98.1 |
| ON-CAN.13 | 1 | 1,265 | DQ123883 | Methanosphaera stadtmanae | 95.8 |
| ON-CAN.14 | 1 | 1,256 | DQ123884 | Thermoplasma acidophilum | 75.6 |
| ON-CAN.15 | 1 | 1,256 | DQ123885 | Thermoplasma acidophilum | 75.4 |
| ON-CAN.16 | 1 | 1,256 | DQ123886 | Thermoplasma acidophilum | 75.5 |
| ON-CAN.17 | 1 | 1,256 | DQ123887 | Thermoplasma volcanium | 74.5 |
| ON-CAN.18 | 1 | 1,256 | DQ123888 | Thermoplasma acidophilum | 75.2 |
A total of 127 clones were examined.
TABLE 2.
16S rRNA clones from pooled samples from 10 potato-fed feedlot cattle from Prince Edward Island, Canada
| 16S rRNA phylotype | No. of clonesa | Size (bp) | GenBank accession no. | Nearest valid taxon | % Sequence similarity |
|---|---|---|---|---|---|
| PE-CAN.01 | 38 | 1,256 | DQ123861 | Thermoplasma volcanium | 74.7 |
| PE-CAN.02 | 15 | 1,256 | DQ123862 | Thermoplasma acidophilum | 75.8 |
| PE-CAN.03 | 22 | 1,261 | DQ123863 | Methanobrevibacter smithii | 96.2 |
| PE-CAN.04 | 19 | 1,260 | DQ123864 | Methanobrevibacter ruminantium | 98.8 |
| PE-CAN.05 | 7 | 1,262 | DQ123865 | Methanobrevibacter thaueri | 98.4 |
| PE-CAN.06 | 5 | 1,260 | DQ123866 | Methanobrevibacter ruminantium | 100.0 |
| PE-CAN.07 | 3 | 1,260 | DQ123867 | Methanobrevibacter thaueri | 98.5 |
| PE-CAN.08 | 1 | 1,256 | DQ123868 | Thermoplasma volcanium | 74.3 |
| PE-CAN.09 | 1 | 1,256 | DQ123869 | Thermoplasma volcanium | 74.1 |
| PE-CAN.10 | 1 | 1,256 | DQ123870 | Thermoplasma volcanium | 74.2 |
| PE-CAN.11 | 1 | 1,260 | DQ123871 | Methanobrevibacter thaueri | 98.1 |
| PE-CAN.12 | 1 | 1,256 | DQ123872 | Thermoplasma acidophilum | 74.9 |
A total of 114 clones were examined.
Phylogenetic analysis.
One hundred nine additional 16S rRNA gene sequences representing the orders Methanobacteriales, Methanomicrobiales, Methanosarcinales, Methanococcales, Halobacteriales, and Thermoplasmatales were included in the phylogenetic analysis. All sequences were globally aligned using the Dedicated Comparative Sequence Editor program (2) and further refined by eye. Three members of the Crenarchaeota (Pyrolobus fumarius, Sulfolobus acidocaldarius, and Thermosphaera aggregans) were used as the outgroup. The online chimeric detection program BELLEROPHON (8) was used to identify chimeric sequences, which were then excluded from analysis. The phylogenetic software package PHYLIP (version 3.62C) (5) was used to calculate the evolutionary distances between pairs of nucleotide sequences using the Kimura two-parameter correction model (12). A distance-matrix tree was then constructed using the neighbor-joining method (18) and bootstrap resampled (4) 1,000 times.
Nucleotide sequence accession numbers.
A total of 28 nucleotide sequences from both 16S clone libraries have been deposited in the GenBank database under accession numbers DQ123861 to DQ123888.
RESULTS AND DISCUSSION
The 16S rRNA gene clones generated from this study were similar to 16S rRNA gene clones previously identified from the bovine rumen in Alberta, Canada (29); Japan (27); and New Zealand (22) and from the ovine rumen in Australia (32, 33) and New Zealand (23) (Fig. 1). In the first 16S rRNA gene clone library, 130 clones were examined from the pooled PCR products from 10 Hereford-cross cattle in Ontario fed a corn-based finishing diet (Table 1). These 130 clones revealed 19 different sequences, or phylotypes, but 3 of these (1 clone each) were identified as chimeras and excluded from the data. Of the remaining 127 clones, the 16 phylotypes were very similar to clones (i.e., those beginning with ARC [Fig. 1]) generated from five Holstein dairy cattle in Alberta, Canada, that were also fed a predominantly corn-based diet (30% corn silage and 17.8% corn concentrate) (29). In that study, Whitford et al. (29) found that 59% of sequences (24 of 41) grouped with Methanobrevibacter ruminantium. In the current study, Methanobrevibacter ruminantium-like clones accounted for almost 50% (61 of 127 clones) of the clones from corn-fed cattle in Ontario (Table 1) but for only 21% (24 of 114 clones) of the clones from potato-fed cattle in Prince Edward Island (Table 2). Whitford et al. (29) also found several sequences that were related to Methanosphaera stadtmanae. Similarly, we found a sequence (ON-CAN.13) in cattle from Ontario that was 95.8% similar to that of Methanosphaera stadtmanae and 99.8% similar to that of their clone ARC29. Methanogens similar to Methanosphaera stadtmanae have also been reported to occur in pasture-fed dairy cattle (22).
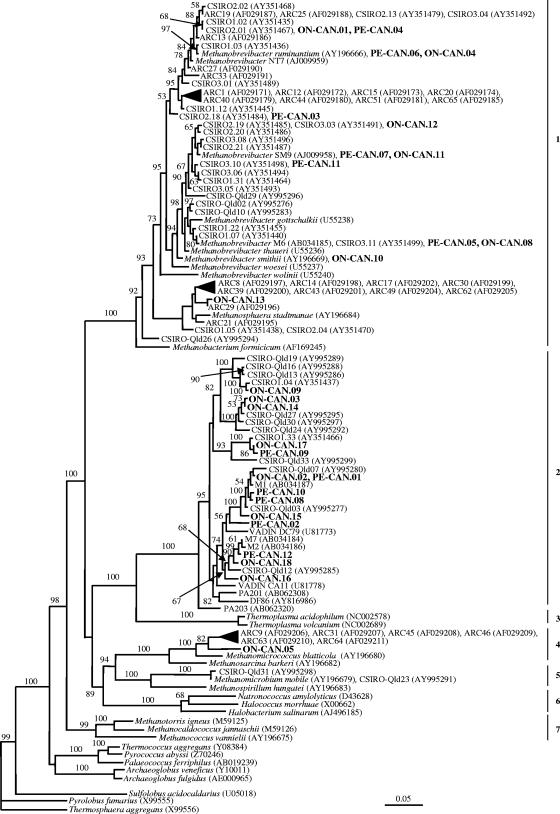
FIG. 1.
Phylogenetic relationships within the methanogenic archaea, derived from 16S evolutionary distances produced by the Kimura two-parameter correction model (12) and constructed using the neighbor-joining method (18). The tree was bootstrap resampled (4) 1,000 times. Only bootstrap values greater than 50% are shown on the internal nodes. The scale bar equals an average of five nucleotide substitutions per 100 positions. The GenBank accession numbers for these nucleotide sequences are given in parentheses. Higher taxonomic groupings are indicated as follows: 1, Methanobacteriales; 2, novel group of uncultivated archaea; 3, Thermoplasmatales; 4, Methanosarcinales; 5, Methanomicrobiales; 6, Halobacteriales; 7, Methanococcales.
In the second clone library, 114 clones were examined from the pooled PCR products from 10 feedlot cattle from Prince Edward Island, which were fed potato by-products. Sequence examination of these 114 clones revealed 12 phylotypes (Table 2). Using a 97% similarity cutoff value (25), clone library coverages were estimated at 92.9% and 95.6% for cattle from Ontario and Prince Edward Island, respectively, indicating that the libraries were very well sampled for the diversity that they contained (7). However, in contrast to the case for the first library, 50% of the clones from the second library belonged within a clade consisting solely of clone sequences of uncultured archaea (6, 24, 27, 32, 33). Recently, Wright et al. (32) reported that as many as 80% of the total clones analyzed from sheep in Queensland also belonged to this clade, whose sister group is a distant branch consisting of Thermoplasma acidophilum and Thermoplasma volcanium.
In total, 241 clones were examined across the two 16S rRNA gene clone libraries, revealing 28 phylotypes. However, five phylotypes (PE-CAN.01/ON-CAN.02, PE-CAN.04/ON-CAN.01, PE-CAN.05/ON-CAN.08, PE-CAN.06/ON-CAN.04, and PE-CAN.07/ON-CAN11) were found in cattle from both provinces. Eleven phylotypes were unique to cattle from Ontario (38 clones), and seven phylotypes (42 clones) were unique to cattle from Prince Edward Island. Pairwise distance data (not shown) of the 23 distinct phylotypes revealed that the average genetic divergence over all possible pairs of sequences was 19.7%, with the greatest genetic distance being 33.2% between ON-CAN.08/PE-CAN.05 and PE-CAN.09.
Overall, 51.5% of the clones (124 of 241 clones) belonged within the order Methanobacteriales (Fig. 1). These findings are in agreement with previous studies indicating species belonging to the genus Methanobrevibacter as the major methanogens in the rumen or feces of most domesticated ruminants (10, 14, 20, 22, 23, 28, 29, 33). However, a few studies have reported other methanogens, not affiliated with Methanobrevibacter, as the most prevalent in the rumen (21, 27, 32). Approximately 42% (101 of 241 clones) of the clones revealed a high degree of sequence similarity to three methanogen species whose names have been validly published: Methanobrevibacter smithii (100%), Methanobrevibacter ruminantium (98.8 to 100%), and Methanobrevibacter thaueri (98.1 to 98.5%). Like other investigations (22, 29), we did not find any sequences that were closely related to Methanobacterium, Methanomicrobium, or Methanosarcina. However, various species from these three genera have been identified previously (11, 17, 21, 27). In total, nine sequences were identified as new taxa based on a similarity criterion of <97% (25), and these accounted for more than 55% of the 241 clones. Of these nine new taxa, six (ON-CAN.03, ON-CAN.09, PE-CAN.01 [= ON-CAN.02], PE-CAN.02, PE-CAN.09, and PE-CAN.12) belong to the distant group of uncultured archaea, two (ON-CAN.13 and PE-CAN.03) belong to the order Methanobacteriales, and one (ON-CAN.05) represents a new genus within the order Methanosarcinales.
From our survey, it is difficult to draw any conclusions about whether the geographical isolation between these two herds of cattle or differences between the two diets directly influenced community structure in the rumen. However, if there were a geographical effect, then there should be unique phylogenetic groupings of methanogens that have been identified from sheep in Australia, Canada, Japan, New Zealand, Scotland, and Venezuela; rather, they are scattered throughout the tree. Also, there does not appear to be distinct phylogenetic lineages of “ovine” or “bovine” methanogens. Furthermore, although potato by-products and corn-based diets are commonly used in cattle feedlot diets in the United States and Canada (1, 15, 16), very little is known about the effect, if any, of these two different finishing diets on methanogen diversity. Interestingly, Kocherginskaya et al. (13) reported that corn-fed steers displayed more diverse bacterial populations than hay-fed animals. Though it would appear that the corn-fed cattle in this study exhibited more methanogen diversity than potato-fed cattle, the Shannon diversity index values for these clone libraries were very close, calculated as 1.89 and 1.86, respectively.
Reducing enteric methane emissions has been identified as one way of lowering global methane emissions, and the rumen environment holds the key to developing new techniques capable of raising the level of production of food in an ecologically sustainable way. Our study has identified a number of new taxa of methanogenic archaea, and this information adds to our understanding of the community structure of the rumen methanogens in feedlot cattle. It also indicates that further studies are warranted to properly assess whether differences between the two finishing diets influenced community structure in the rumen.
Acknowledgments
We thank Serhiy Hlamazda (Better Beef Ltd., Guelph, Canada) for permission to collect fresh samples from the abattoir. We also thank Chris McSweeney, Mark Morrison, Belinda Norris, and Stuart Denman (all at CSIRO Livestock Industries, Brisbane, Australia) for their critical comments on the manuscript.
Footnotes
Published ahead of print on 4 May 2007.
REFERENCES
- 1.Beauchemin, K. A., and S. M. McGinn. 2005. Methane emissions from feedlot cattle fed barley or corn diets. J. Anim. Sci. 83:653-661. [DOI] [PubMed] [Google Scholar]
- 2.de Rijk, P., and R. de Wachter. 1993. DCSE, an interactive tool for sequence alignment and secondary structure research. Comput. Applic. Biol. Sci. 9:735-740. [DOI] [PubMed] [Google Scholar]
- 3.Environment Canada. 2004. Canada's greenhouse gas inventory 1990-2000. Government of Canada EPS 5 /AP/ 8. Environment Canada, Ottawa, Canada.
- 4.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 5.Felsenstein, J. 2004. PHYLIP (Phylogeny Inference Package) documentation files, version 3.62c. Department of Genetics, University of Washington, Seattle.
- 6.Godon, J.-J., E. Zumstein, P. Dabert, F. Habouzit, and R. Moletta. 1997. Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl. Environ. Microbiol. 63:2802-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Good, I. J. 1953. The population frequency of species and the estimation of population parameters. Biometrika 40:237-262. [Google Scholar]
- 8.Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319. [DOI] [PubMed] [Google Scholar]
- 9.Intergovernmental Panel on Climate Change. 2001. Climate change 2001: a scientific basis. University Press, Cambridge, United Kingdom.
- 10.Irbis, C., and K. Ushida. 2004. Detection of methanogens and proteobacteria from a single cell of rumen ciliate protozoa. J. Gen. Appl. Microbiol. 50:203-212. [DOI] [PubMed] [Google Scholar]
- 11.Jarvis, G. N., C. Strompl, D. M. Burgess, L. C. Skillman, E. R. Moore, and K. N. Joblin. 2000. Isolation and identification of ruminal methanogens from grazing cattle. Curr. Microbiol. 40:327-332. [DOI] [PubMed] [Google Scholar]
- 12.Kimura, M. 1980. A simple method of estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 13.Kocherginskaya, S. A., R. I. Aminov, and B. A. White. 2001. Analysis of the rumen bacterial diversity under two different diet conditions using denaturing gradient gel electrophoresis, random sequencing, and statistical ecology approaches. Anaerobe 7:119-134. [Google Scholar]
- 14.Miller, T. L., and M. J. Wolin. 1986. Methanogens in human and animal digestive tracts. Syst. Appl. Microbiol. 7:223-229. [Google Scholar]
- 15.Nelson, M. L., J. R. Busboom, J. D. Cronrath, L. Falen, and A. Blankenbaker. 2000. Effects of graded levels of potato by-products in barley- and corn-based beef feedlot diets. I. Feedlot performance, carcass traits, meat composition, and appearance. J. Anim. Sci. 78:1829-1836. [DOI] [PubMed] [Google Scholar]
- 16.Pen, B., T. Iwama, M. Ooi, T. Saitoh, K. Kida, T. Iketaki, J. Takahashi, and H. Hidari. 2006. Effect of potato by-products based silage on rumen fermentation, methane production and nitrogen utilization in Holstein steers. Asian-Aust. J. Anim. Sci. 19:1283-1290. [Google Scholar]
- 17.Regensbogenova, M., N. R. McEwan, P. Javorsky, S. Kisidayova, T. Michalowski, C. J. Newbold, J. H. P. Hackstein, and P. Pristas. 2004. A re-appraisal of the diversity of the methanogens associated with the rumen ciliates. FEMS Microbiol. Lett. 238:307-313. [DOI] [PubMed] [Google Scholar]
- 18.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for constructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 19.Shannon, C. E., and W. Weaver. 1949. The mathematical theory of communication. University of Illinois Press, Urbana.
- 20.Sharp, R., C. J. Ziemer, D. S. Marshall, and D. A. Stahl. 1998. Taxon-specific associations between protozoal and methanogen populations in the rumen and a model system. FEMS Microbiol. Ecol. 26:71-78. [Google Scholar]
- 21.Shin, E. C., B. R. Choi, W. J. Lim, S. Y. Hong, C. I. An, K. M. Cho, Y. K. Kim, J. M. An, J. M. Kang, S. S. Lee, H. Kim, and H. D. Yun. 2004. Phylogenetic analysis of archaea in three fractions of cow rumen based on the 16S rDNA sequence. Anaerobe 10:313-319. [DOI] [PubMed] [Google Scholar]
- 22.Skillman, L. C., P. N. Evans, C. Strompl, and K. N. Joblin. 2006. 16S rDNA directed PCR primers and detection of methanogens in the bovine rumen. Lett. Appl. Microbiol. 42:222-228. [DOI] [PubMed] [Google Scholar]
- 23.Skillman, L. C., P. N. Evans, G. E. Naylor, B. Morvan, G. N. Jarvis, and K. N. Joblin. 2004. 16S ribosomal DNA-directed PCR primers for ruminal methanogens and identification of methanogens colonising young lambs. Anaerobe 10:277-285. [DOI] [PubMed] [Google Scholar]
- 24.Snell-Castro, R., J. J. Godon, J. P. Delgenes, and P. Dabert. 2005. Characterisation of the microbial diversity in a pig manure storage pit using small subunit rDNA sequence analysis. FEMS Microbiol. Ecol. 52:229-242. [DOI] [PubMed] [Google Scholar]
- 25.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA:DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 26.Statistics Canada. 2003. Livestock statistics: fourth quarter 2002 (catalog no. 23-603-XIE). Agriculture Division, Livestock and Animal Products Section, Ottawa, Canada.
- 27.Tajima, K., T. Nagamine, H. Matsui, M. Nakamura, I. Rustam, and R. I. Aminov. 2001. Phylogenetic analysis of archaeal 16S rRNA libraries from the rumen suggests the existence of a novel group of archaea not associated with known methanogens. FEMS Microbiol. Lett. 200:67-72. [DOI] [PubMed] [Google Scholar]
- 28.Tokura, M., I. Chagan, K. Ushida, and Y. Kojima. 1999. Phylogenetic study of methanogens associated with rumen ciliates. Curr. Microbiol. 39:123-128. [DOI] [PubMed] [Google Scholar]
- 29.Whitford, M. F., R. M. Teather, and R. J. Forster. 2001. Phylogenetic analysis of methanogens from the bovine rumen. BMC Microbiol. 1:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wright, A.-D. G., and C. Pimm. 2003. Improved strategy for presumptive identification of methanogens using 16S riboprinting. J. Microbiol. Methods 55:337-349. [DOI] [PubMed] [Google Scholar]
- 31.Wright, A.-D. G., K. Tajima, and R. I. Aminov. 2005. 16S/18S rDNA clone library construction and analysis, p. 163-174. In H. P. S. Makkar, and C. S. McSweeney (ed.), Methods in gut microbial ecology for ruminants. Springer Publishers, Dordrecht, The Netherlands.
- 32.Wright, A.-D. G., A. F. Toovey, and C. L. Pimm. 2006. Molecular identification of methanogenic archaea from sheep in Queensland, Australia reveal more uncultured novel archaea. Anaerobe 12:134-139. [DOI] [PubMed] [Google Scholar]
- 33.Wright, A.-D. G., A. J. Williams, B. Winder, C. Christophersen, S. Rodgers, and K. Smith. 2004. Molecular diversity of rumen methanogens from sheep in Western Australia. Appl. Environ. Microbiol. 70:1263-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]