Abstract
Enzymes expressed in response to vinyl chloride, ethene, and epoxyethane by Nocardioides sp. strain JS614 were identified by using a peptide mass fingerprinting (PMF) approach. PMF provided insight concerning vinyl chloride biodegradation in strain JS614 and extends the use of matrix-assisted laser desorption-ionization time of flight mass spectrometry as a tool to enhance characterization of biodegradation pathways.
Vinyl chloride (VC), a known human carcinogen (2) and groundwater contaminant (21), is often generated in groundwater by the incomplete reduction of chlorinated solvents. Diverse bacterial genera, including Mycobacterium (3, 12), Nocardioides (3, 18), Ochrobactrum (5), Pseudomonas (5, 22), and Ralstonia (9), use both VC and ethene as carbon and energy sources. Several strains appear to use the same enzymes to metabolize both VC and ethene (4, 12, 18). Alkene monooxygenase (AkMO) oxidizes VC to chlorooxirane (12, 22) and ethene to epoxyethane (3, 7, 8, 18). Epoxyalkane:coenzyme M transferase (EaCoMT) participates in further metabolism of both epoxyethane (4, 6, 18) and chlorooxirane (4). An unknown number of enzymatic steps catalyze the conversion of these epoxides to acetyl coenzyme A (acetyl-CoA) (7). Elucidating the remaining enzymes and intermediates of aerobic VC and ethene biodegradation will facilitate development of molecular tools for detecting and differentiating VC- and ethene-assimilating bacteria in the environment. The completion of the Nocardioides sp. strain JS614 genome sequence (http://genome.ornl.gov/microbial/noca/) provides opportunities to use new approaches to identify enzymes involved in VC and ethene biodegradation. In this study, proteomic techniques were used to rapidly and accurately identify enzymes expressed in response to VC, ethene, and epoxyethane in strain JS614.
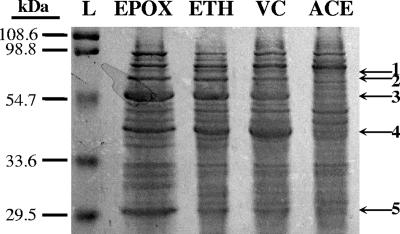
Chemicals, media, growth conditions, and protein extraction methods are described elsewhere (18). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis was performed by the method of Laemmli (15) with extracts (10 to 50 μg protein) from VC-, ethene-, epoxyethane-, and acetate-grown cells. Polyacrylamide gels were stained with Bio-Safe Coomassie blue (Bio-Rad Laboratories, Inc.). Visual inspection (Fig. 1) revealed several polypeptides expressed in response to ethene, epoxyethane, and VC that were not expressed in response to acetate, suggesting that they were directly involved in VC, ethene, and epoxyethane metabolism. Polypeptide bands from all lanes in each numbered section of the gel were excised and digested with bovine trypsin (Promega Corp.). The resulting monoisotopic peptide fragment masses were analyzed with a Bruker BiflexIII matrix-assisted laser desorption-ionization time of flight (MALDI-TOF) mass spectrometer in positive-ion/reflector mode, using an α-cyano-4-hydroxycinnamic acid matrix (14, 19). Peptide mass fingerprints (PMFs) of digested polypeptide bands (Fig. 2) were compared to the PMF of a control gel fragment that had no contact with cell extracts, and matching masses within a 0.3-kDa mass tolerance were excluded from analysis. The remaining peptide masses were compared to those of all bacteria in the NCBInr database, using the following parameters in Mascot (20): zero missed cleavages, carbamidomethyl (fixed modification), methionine oxidation (variable modification), and a 0.3-kDa mass tolerance. Results were evaluated using the Mowse probability-based scoring algorithm (19).
FIG. 1.
Representative results of SDS-PAGE with cell extracts from epoxyethane (EPOX)-, vinyl chloride (VC)-, ethene (ETH)-, and acetate (ACE)-grown JS614 cultures and a protein ladder (L). Numbers and arrows indicate sections of the gel that were excised for MALDI-TOF MS analysis and correspond to results presented in Table 1.
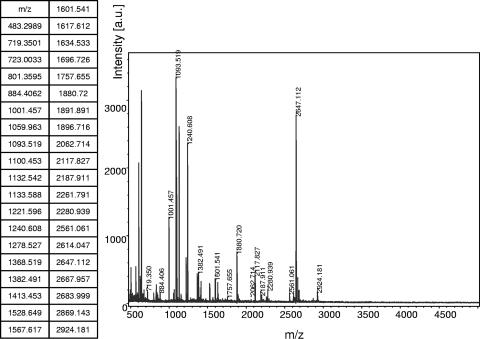
FIG. 2.
A representative PMF of the probable AkMO alpha subunit (Noc4807) measured from a protein extract of a VC-grown JS614 culture (a.u., atomic units). The mass spectrum and a complete list of peptide masses that make up the PMF are shown.
PMF analysis revealed seven polypeptides expressed in response to VC, ethene, and epoxyethane with statistically significant Mowse scores (P < 0.05) from at least two independent PMF analyses of a single growth condition (Table 1). None of these polypeptides were observed in extracts from acetate-grown JS614 cultures (data not shown). However, our PMF data do not rule out the possibility that proteins reported in Table 1 are present in acetate-grown cells at very low abundances. Eleven additional polypeptides expressed in response to VC, ethene, and/or epoxyethane were identified (P < 0.05) either in just one PMF analysis or only once under a particular growth condition (see Table S1 in the supplemental material).
TABLE 1.
JS614 proteins expressed in response to VC, ethene, and epoxyethane with statistical relevance (P < 0.05), identified by using MALDI-TOF MS PMF analysisl
| Excised gel sectiona | Growth substrateb | Significant Mascot hitsc | Predicted mass (kDa)d | Observed mass (kDa)e | Mowse scoref | E valueg | % Coverageh | NBMi | MM/TPMSj | Gene no.k |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Epoxyethane | FAD-dependent pyridine nucleotide-disulfide oxidoreductase | 60.47 | 74 | 142 | 1.6E−08 | 37 | 61 | 14/49 | 4827 |
| 1 | VC | FAD-dependent pyridine nucleotide-disulfide oxidoreductase | 60.47 | 79 | 101 | 2.5E−04 | 28 | 58 | 12/55 | 4827 |
| 1 | Ethene | FAD-dependent pyridine nucleotide-disulfide oxidoreductase | 60.47 | 73 | 141 | 7.5E−07 | 30 | 60 | 14/41 | 4827 |
| 2 | Epoxyethane | Probable alkene monooxygenase alpha subunit | 57.78 | 62 | 178 | 1.5E−06 | 44 | 59 | 19/70 | 4807 |
| 2 | VC | Probable alkene monooxygenase alpha subunit | 57.78 | 65 | 142 | 1.5E−08 | 36 | 75 | 19/66 | 4807 |
| 2 | Ethene | Probable alkene monooxygenase alpha subunit | 57.78 | 65 | 126 | 9.0E−03 | 33 | 56 | 13/56 | 4807 |
| 3 | Epoxyethane | Alcohol/acetaldehyde dehydrogenase | 51.78 | 54 | 138 | 9.5E−08 | 46 | 60 | 15/72 | 4833 |
| 3 | VC | Alcohol/acetaldehyde dehydrogenase | 51.78 | 55 | 119 | 3.3E−06 | 41 | 61 | 14/77 | 4833 |
| 3 | Ethene | Alcohol/acetaldehyde dehydrogenase | 51.78 | 53 | 115 | 1.2E−04 | 41 | 62 | 14/68 | 4833 |
| 3 | Epoxyethane | Bifunctional protein: alcohol/acetaldehyde dehydrogenase | 50.02 | 54 | 149 | 3.0E−06 | 42 | 59 | 14/52 | 4822 |
| 3 | VC | Bifunctional protein: alcohol/acetaldehyde dehydrogenase | 50.02 | 55 | 129 | 5.7E−03 | 40 | 59 | 14/66 | 4822 |
| 3 | Ethene | Bifunctional protein: alcohol/acetaldehyde dehydrogenase | 50.02 | 53 | 112 | 6.5E−04 | 35 | 64 | 14/61 | 4822 |
| 4 | Epoxyethane | Epoxyalkane:coenzyme M transferase | 40.68 | 45 | 166 | 1.9E−08 | 50 | 60 | 13/40 | 4810 |
| 4 | VC | Epoxyalkane:coenzyme M transferase | 40.68 | 46 | 163 | 2.4E−08 | 49 | 60 | 13/46 | 4810 |
| 4 | Ethene | Epoxyalkane:coenzyme M transferase | 40.68 | 45 | 150 | 2.1E−04 | 45 | 62 | 13/47 | 4810 |
| 5 | Epoxyethane | Short-chain dehydrogenase/reductase | 25.85 | 31 | 140 | 3.4E−07 | 51 | 63 | 11/46 | 4814 |
| 5 | VC | Short-chain dehydrogenase/reductase | 25.85 | 33 | 158 | 1.5E−09 | 59 | 64 | 12/40 | 4814 |
| 5 | Ethene | Short-chain dehydrogenase/reductase | 25.85 | 31 | 145 | 2.0E−06 | 53 | 61 | 10/35 | 4814 |
| 5 | Epoxyethane | Short-chain dehydrogenase/reductase | 26.20 | 31 | 149 | 1.2E−07 | 59 | 61 | 12/45 | 4841 |
| 5 | VC | Short-chain dehydrogenase/reductase | 26.20 | 33 | 170 | 1.2E−09 | 61 | 64 | 12/40 | 4841 |
| 5 | Ethene | Short-chain dehydrogenase/reductase | 26.20 | 31 | 163 | 1.5E−09 | 60 | 63 | 12/38 | 4841 |
Gel sections were excised as shown in Fig. 1.
Growth conditions and methods are described in reference 18.
Most significant Mascot hits to current gene annotations in the NCBInr database. FAD, flavin adenine dinucleotide.
Theoretical mass reported by Mascot.
Estimated from results of SDS-PAGE (Fig. 1).
Mowse score reported by Mascot.
E value reported by Mascot.
Percent coverage of amino acids to matched peptides reported by Mascot.
iMowse score of the next best Mascot match (NBM) not associated with JS614.
MM, mass matches; TPMS, total number of peptide masses searched.
Gene number in finished JS614 genome sequence corresponding to Mascot hit. All NBMs, with the exception of one unknown lipoprotein match to Noc4827 (Mowse score, 80), were not significant. A putative translation elongation factor (encoded by Noc3922; Mowse score, 162) was the only peptide consistently identified in acetate-grown cells (Fig. 1).
All proteins shown were found in at least two independent analyses, and the results reported are averaged values.
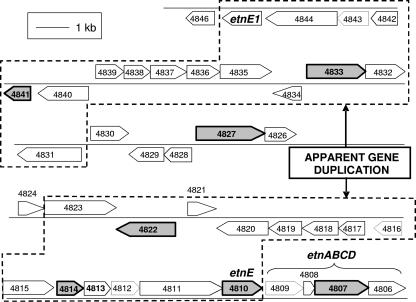
The polypeptides reported in Table 1 mapped to a plasmid-encoded region of the JS614 genome sequence (Fig. 3) that contained known VC/ethene biodegradation genes (18), suggesting that they are likely to participate in VC/ethene metabolism. Identification of AkMO subunit (EtnC), EaCoMT (EtnE), putative CoM biosynthesis protein ComA, and a probable CoA transferase in cell extracts from VC- and ethene-grown cells is consistent with previous reports (17, 18), confirming the robustness of this proteomics approach. Five proteins not previously reported to be involved in VC/ethene metabolism were also identified. Reverse transcription (RT)-PCR assays were conducted to provide independent confirmation of proteomics results. Total RNA was extracted and purified, and RT-PCR was performed as previously described (17, 18), using 0.1 ng RNA template. JS614 genomic DNA, extracted as described previously (3), was used as the positive RT-PCR control. Sterile water was the negative RT-PCR control, the 16S rRNA gene was the positive RNA control, and the RpoB gene was the positive mRNA control. Using the primers described in Table 2, RT-PCR products of the expected size were observed with RNA extracted from ethene-, epoxyethane-, and VC-grown cells but not acetate-grown cells (data not shown). RT-PCR products were observed in all RNA extracts when 16S rRNA gene and rpoB primers were used, and no products from control PCRs with the RNA template and no reverse transcriptase were observed (data not shown).
FIG. 3.
Diagram of plasmid-encoded region containing ethene/VC biodegradation genes in Nocardioides sp. strain JS614. Genes expressed and translated into protein in response to epoxyethane, VC, or ethene as identified by MALDI-TOF MS and PMF analysis are shaded gray. Genes 4831 to 4844 and the second EaCoMT allele, etnE1, appear to be the result of a duplication of genes 4810 to 4823, evidenced by high amino acid similarities and the conservation of gene orientation. For example, genes 4814 and 4822 share 90 and 86% amino acid similarities, respectively, with genes 4841 and 4833.
TABLE 2.
Oligonucleotides used for RT-PCR
| Oligonucleotide (5′ to 3′) | Sequence (5′ to 3′) | Gene no.a | Expected product size (bp) | Reference |
|---|---|---|---|---|
| CoM-F3 | GCTCTCAAGATGTGCTTCTGCCAACCA | Noc4810 | 831 | 18 |
| CoM-R3 | CGGTGCGTCCGACCTCGTAGTTCAG | Noc4810 | 831 | 18 |
| etnC_F | CTTGAAACCGTCCACGAGAAGAG | Noc4807 | 1,481 | 18 |
| etnC_R | AGCGGGTCCTTGATCTCGTACTT | Noc4807 | 1,481 | 18 |
| scdehd_F | GCCATAGGCGTACTTGACCTG | Noc4814 | 560 | Present study |
| scdehd_R | ATGGGGTACTTCACGAGTGTCT | Noc4814 | 560 | Present study |
| carb_F | GGAGTACGGCTCCTTCTACCA | Noc4827 | 832 | Present study |
| carb_R | GTTGAGGAACAACTCATTCATCTC | Noc4827 | 832 | Present study |
| dehy_F | ATCACGTCAATACTGGGGATGT | Noc4833 | 703 | Present study |
| dehy_R | AGGACAAGGTCGTCAAGATCAA | Noc4833 | 703 | Present study |
| 27f | AGAGTTTGATC(C/A)TGGCTCAG | N/A | 1,482 | 16 |
| 1492r | TACGG(C/T)TACCTTGTTACGACTT | N/A | 1,482 | 16 |
| rpoB_F | GCTTCGGGTTGAAGTAGTAGTTGT | Noc0711 | 802 | 17 |
| rpoB_R | GCAAAGATCACCGAACCTCTC | Noc0711 | 802 | 17 |
Gene from the JS614 genome sequence that was used to design the oligonucleotide. N/A, not applicable.
Our JS614 genome-assisted proteomics approach has led us to reformulate hypotheses concerning the VC/ethene biodegradation pathway in strain JS614. Previously, we proposed that a putative reductase (encoded by Noc4815; GenBank accession no. ZP_00655559) participated in regeneration of free CoM (18). In this study, we identified a protein (encoded by Noc4827; GenBank accession no. ZP_00655570) expressed in response to ethene, VC, and epoxyethane with 53% amino acid identity (E = 2e−156) to the 2-oxopropyl-CoM reductase/carboxylase from the propene-oxidizing Xanthobacter sp. strain Py2 (GenBank accession no. Q56839) (1) (Table 1 and Fig. 1). By analogy to the strain Py2 propene biodegradation pathway, the Noc4827 product could participate in carboxylating a VC/ethene pathway metabolic intermediate with concomitant regeneration of free CoM. Based on BLAST homologies, we propose that the Noc4815 product is a CoA reductase rather than a previously predicted CoM reductase. We also previously hypothesized that a putative short-chain dehydrogenase (encoded by Noc4814; GenBank accession no. ZP_00655558) carried out a two-step dehydrogenation of 2-hydroxyethyl-CoM to 2-carboxymethyl-CoM (18). Here, we provide circumstantial evidence (Fig. 1 and Table 1) that a putative dual-function alcohol/acetaldehyde dehydrogenase encoded by Noc4822 (GenBank accession no. ZP_00655566) and/or Noc4833 (GenBank accession no. ZP_00655575) participates in the JS614 VC/ethene pathway. Detailed biochemical studies are required to determine the role of these proteins in VC/ethene metabolism.
Previously, we reported that JS614 harbors two alleles of the EaCoMT gene etnE (18). Bioinformatic analysis of the JS614 genome sequence has revealed a cluster of 14 additional genes that appear to have been recently duplicated, including Noc4841 (GenBank accession no. ZP_00655584; 90% amino acid identity with Noc4814) and Noc4833 (86% amino acid identity with Noc4822) (Fig. 3). Theoretical trypsin protein digests obtained from ExPASy PeptideMass (10, 23) showed several peptide masses unique to each protein (Table 3; see Table S2 in the supplemental material). Significant Mowse scores were obtained when these unique masses were input to Mascot (data not shown), indicating that PMF can distinguish between these highly similar proteins.
TABLE 3.
Comparison of peptide masses resulting from theoretical trypsin digestion of closely related deduced protein sequences (Noc4814/Noc4841 and Noc4822/Noc4833) in Nocardioides sp. strain JS614e
| Gene no. | Total no. of peptide massesa | No. of unique peptide massesb | Shared peptide massesc | % Coverage (aa)d |
|---|---|---|---|---|
| Noc4814 | 21 | 9 | 12 | 46 |
| Noc4841 | 23 | 11 | ||
| Noc4822 | 39 | 21 | 18 | 15 |
| Noc4833 | 39 | 21 |
Total peptide masses generated from an in silico trypsin digestion.
Number of unique peptide masses resulting from an in silico trypsin digestion.
Peptide masses shared between theoretical digests of either Noc4814 and Noc4841 or Noc4822 and Noc4833.
Percent coverage of either protein by shared peptide masses. The analysis did not include peptide masses that resulted from variable modifications (e.g., methionine oxidation). aa, amino acids.
See Table S2 in the supplemental material for more details.
Although MALDI-TOF mass spectrometry (MS) is increasingly being used in microbiology, there are limited publications demonstrating its application in biodegradation research (11, 13). Here, we used MALDI-TOF MS to identify seven proteins expressed in response to VC and ethene by strain JS614. Unique peptide masses were used to distinguish between mixtures of highly similar expressed proteins. To our knowledge, this is the first extensive application of PMF in conjunction with a completed genome sequence to identify enzymes translated in response to a xenobiotic pollutant. The ability to monitor and distinguish microbial involvement in pollutant degradation is a significant obstacle for bioremediation. An understanding of aerobic VC and ethene biodegradation pathways and the ability to detect the involved expressed proteins are important initial steps toward monitoring VC and ethene biodegradation in the environment.
Supplementary Material
Acknowledgments
This work was supported by the Engineering Research Centers Program of the National Science Foundation under NSF award EEC-0310689 and by University of Iowa start-up funds for T.E.M. Support for A.S.C. was provided by the NSF GRFP.
We thank Yalan Li and Lynn Teesch of the UI Mass Spectrometry Center for their technical assistance.
Footnotes
Published ahead of print on 4 May 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Allen, J. R., and S. A. Ensign. 1997. Characterization of three protein components required for functional reconstitution of the epoxide carboxylase multienzyme complex from Xanthobacter strain Py2. J. Bacteriol. 179:3110-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bucher, J. R., G. Cooper, J. K. Haseman, C. W. Jameson, M. Longnecker, F. Kamel, R. Maronpot, H. B. Matthews, R. Melnick, R. Newbold, R. W. Tennant, C. Thompson, and M. Waalkes. 2005. Report on carcinogens, 11th ed. National Toxicology Program, Public Health Service, U.S. Department of Health and Human Services.
- 3.Coleman, N. V., T. E. Mattes, J. M. Gossett, and J. C. Spain. 2002. Phylogenetic and kinetic diversity of aerobic vinyl chloride-assimilating bacteria from contaminated sites. Appl. Environ. Microbiol. 68:6162-6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coleman, N. V., and J. C. Spain. 2003. Epoxyalkane:coenzyme M transferase in the ethene and vinyl chloride biodegradation pathways of Mycobacterium strain JS60. J. Bacteriol. 185:5536-5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Danko, A. S., M. Luo, C. E. Bagwell, R. L. Brigmon, and D. L. Freedman. 2004. Involvement of linear plasmids in aerobic biodegradation of vinyl chloride. Appl. Environ. Microbiol. 70:6092-6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Danko, A. S., C. A. Saski, J. P. Tomkins, and D. L. Freedman. 2006. Involvement of coenzyme M during aerobic biodegradation of vinyl chloride and ethene by Pseudomonas putida strain AJ and Ochrobactrum sp. strain TD. Appl. Environ. Microbiol. 72:3756-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Bont, J. A. M., and W. Harder. 1978. Metabolism of ethylene by Mycobacterium E 20. FEMS Microbiol. Lett. 3:89-93. [Google Scholar]
- 8.de Bont, J. A. M., M. M. Atwood, S. B. Primrose, and W. Harder. 1979. Epoxidation of short-chain alkenes in Mycobacterium E20: the involvement of a specific monooxygenase. FEMS Microbiol. Lett. 6:183-188. [Google Scholar]
- 9.Elango, V. K., A. S. Liggenstoffer, and B. Z. Fathepure. 2006. Biodegradation of vinyl chloride and cis-dichloroethene by a Ralstonia sp. strain TRW-1. Appl. Microbiol. Biotechnol. 72:1270-1275. [DOI] [PubMed] [Google Scholar]
- 10.Gasteiger, E., C. Hoogland, A. Gattiker, S. Duvaud, M. R. Wilkins, R. D. Appel, and A. Bairoch. 2005. Protein identification and analysis tools on the ExPASy server, p. 571-607. In J. M. Walker (ed.), The proteomics protocols handbook. Humana Press, Totowa, NJ.
- 11.Halden, R. U., D. R. Colquhoun, and E. S. Wisniewski. 2005. Identification and phenotypic characterization of Sphingomonas wittichii strain RW1 by peptide mass fingerprinting using matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 71:2442-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartmans, S., and J. A. M. de Bont. 1992. Aerobic vinyl chloride metabolism in Mycobacterium aurum L1. Appl. Environ. Microbiol. 58:1220-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hickey, W. J., and G. Sabat. 2001. Integration of matrix-assisted laser desorption ionization-time of flight mass spectrometry and molecular cloning for the identification and functional characterization of mobile ortho-halobenzoate oxygenase genes in Pseudomonas aeruginosa strain JB2. Appl. Environ. Microbiol. 67:5648-5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensen, O. N., A. V. Podtelejnikov, and M. Mann. 1997. Identification of the components of simple protein mixtures by high-accuracy peptide mass mapping and database searching. Anal. Chem. 69:4741-4750. [DOI] [PubMed] [Google Scholar]
- 15.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 16.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 177-203. In E. Stackbrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, Chichester, United Kingdom.
- 17.Mattes, T. E., N. V. Coleman, A. S. Chuang, A. J. Rogers, J. C. Spain, and J. M. Gossett. 2007. Mechanism controlling the extended lag period associated with vinyl chloride starvation in Nocardioides sp. strain JS614. Arch. Microbiol. 187:217-226. [DOI] [PubMed] [Google Scholar]
- 18.Mattes, T. E., N. V. Coleman, J. C. Spain, and J. M. Gossett. 2005. Physiological and molecular genetic analyses of vinyl chloride and ethene biodegradation in Nocardioides sp. strain JS614. Arch. Microbiol. 183:95-106. [DOI] [PubMed] [Google Scholar]
- 19.Pappin, D. J., P. Hojrup, and A. J. Bleasby. 1993. Rapid identification of proteins by peptide-mass fingerprinting. Curr. Biol. 3:327-332. [DOI] [PubMed] [Google Scholar]
- 20.Perkins, D. N., D. J. Pappin, D. M. Creasy, and J. S. Cottrell. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551-3567. [DOI] [PubMed] [Google Scholar]
- 21.Squillace, P. J., M. J. Moran, W. W. Lapham, C. V. Price, R. M. Clawges, and J. S. Zogorski. 1999. Volatile organic compounds in untreated ambient groundwater of the United States, 1985-1995. Environ. Sci. Technol. 33:4176-4187. [Google Scholar]
- 22.Verce, M. F., R. L. Ulrich, and D. L. Freedman. 2000. Characterization of an isolate that uses vinyl chloride as a growth substrate under aerobic conditions. Appl. Environ. Microbiol. 66:3535-3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilkins, M. R., I. Lindskog, E. Gasteiger, A. Bairoch, J. C. Sanchez, D. F. Hochstrasser, and R. D. Appel. 1997. Detailed peptide characterization using PEPTIDEMASS—a World-Wide-Web-accessible tool. Electrophoresis 18:403-408. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.