Abstract
Isolates of Escherichia coli belonging to clonal group A (CGA), a recently described disseminated cause of drug-resistant urinary tract infections in humans, were present in four of seven sewage effluents collected from geographically dispersed areas of the United States. All 15 CGA isolates (1% of the 1,484 isolates analyzed) exhibited resistance to trimethoprim-sulfamethoxazole (TMP-SMZ), accounting for 19.5% of the 77 TMP-SMZ-resistant isolates. Antimicrobial resistance patterns, virulence traits, O:H serotypes, and phylogenetic groupings were compared for CGA and selected non-CGA isolates. The CGA isolates exhibited a wider diversity of resistance profiles and somatic antigens than that found in most previous characterizations of this clonal group. This is the first report of recovery from outside a human host of E. coli CGA isolates with virulence factor and antibiotic resistance profiles typical of CGA isolates from a human source. The occurrence of “human-type” CGA in wastewater effluents demonstrates a potential mode for the dissemination of this clonal group in the environment, with possible secondary transmission to new human or animal hosts.
Resistance to commonly prescribed antimicrobial agents is a matter of increasing concern. Along with respiratory infections, urinary tract infections (UTIs) are the most common bacterial infections in the United States requiring antimicrobial therapy. Escherichia coli is the most frequently isolated etiological agent of UTIs, and trimethoprim-sulfamethoxazole (TMP-SMZ) is one of the primary antibiotics empirically prescribed for the treatment of community-acquired UTIs (10). In the United States, there has been a notable increase in the isolation of uropathogenic E. coli strains resistant to TMP-SMZ (10). This finding is of particular interest since resistance to this drug is generally associated with resistance to additional antibiotics (15). Recent epidemiological studies have reported the widespread emergence of a single clonal group, provisionally designated clonal group A (CGA), among TMP-SMZ-resistant strains of uropathogenic E. coli (1, 4, 8, 9, 11). CGA has been reported to account for up to 50% of TMP-SMZ-resistant E. coli isolates from women with acute uncomplicated cystitis and pyelonephritis (8). Recent human CGA isolates have typically exhibited a number of traits that set them apart from other uropathogenic or drug-resistant E. coli isolates, including their characteristic virulence factor profile, several distinctive O antigens, the H18 flagellar antigen, and multidrug resistance, including resistance to ampicillin, chloramphenicol, streptomycin, sulfonamides, tetracycline, and TMP (8).
The widespread occurrence of E. coli CGA and the occurrence in one community of a seeming point-source outbreak of UTIs due to the same pulsotype of CGA (11) have raised questions regarding the mode of transmission of these organisms. Recent travel to areas with a high prevalence of TMP-SMZ-resistant E. coli strains has previously been cited as a risk factor for acquiring TMP-SMZ-resistant E. coli (12). This finding suggests that exposure to contaminated food or water may represent a means for the dissemination of these pathogenic E. coli strains (9, 11, 14, 17). The current study was designed to assess the prevalence and characteristics of TMP-SMZ-resistant E. coli and CGA strains in domestic sewage samples collected from various locations throughout the United States.
MATERIALS AND METHODS
Samples and primary cultures.
Two sewage effluent samples (primary and secondary) were collected prior to chlorination from each of seven wastewater treatment plants dispersed geographically in the following areas of the United States: San Jose, CA; Tallahassee, FL; Lewiston, ME; Cincinnati, OH; Virginia Beach, VA; Seattle, WA; and Milwaukee, WI. Samples were shipped on ice and analyzed within 24 h of collection. A membrane filtration procedure using mFC agar (BD Bioscience, Sparks, MD) and nutrient agar containing 4-methylumbelliferyl beta-d-glucuronide (BD Bioscience, Sparks, MD) (3) was used to detect E. coli. Approximately 200 E. coli colonies from each location were picked at random and streaked for purity on heart infusion agar (BD Bioscience, Sparks, MD).
Susceptibility testing.
Isolates were subcultured onto Mueller-Hinton agar (BD Bioscience, Sparks, MD) and screened for susceptibility to TMP-SMZ (BD Bioscience, Sparks, MD) by the disk diffusion method (13). E. coli strain 25922 (American Type Culture Collection) was used as a reference control strain. All TMP-SMZ-resistant isolates (zone diameter of ≤10 mm) were saved for further study, as was a group of TMP-SMZ-susceptible E. coli isolates (which were selected as described below as controls for geographically matched comparisons). All isolates were confirmed to be E. coli by using the API 20E test system (bioMerieux, Marcy'Etoile, France). Isolates were also tested for susceptibility to ampicillin (AM), chloramphenicol (CH), streptomycin (ST), and tetracycline (TE) by using Etest strips (AB Biodisk, Solna, Sweden), as directed by the manufacturer, to determine MICs. Isolates were classified as susceptible (s), intermediate (i), or resistant (r) based upon MIC criteria specified by the Clinical Laboratory and Standards Institute (CLSI) (13) or based on published precedent (8). The following MIC criteria were used (μg/ml): for AM, s = ≤8, i = 16, and r = ≥32; for CH, s = ≤8, i = 16, and r = ≥32; for ST, r = ≥8; for TE, s = ≤4, i = 8, and r = ≥16; and for TMP-SMZ, s = ≤2 and r = ≥4.
Molecular analysis.
All TMP-SMZ-resistant isolates were O:H serotyped by the E. coli Reference Center at the Pennsylvania State University (University Park, PA) and were phylotyped (for phylogenetic groups A, B1, B2, and D) using a multiplex PCR procedure (2). All TMP-SMZ-resistant isolates and a group of 77 geographically matched TMP-SMZ-susceptible isolates were tested for CGA status by using a gene-specific PCR procedure that detects a distinctive single-nucleotide polymorphism within fumC (7). All CGA isolates and a group of geographically matched non-CGA, TMP-SMZ-resistant control isolates (n = 22; the sample number chosen was 2:1 with respect to the CGA isolates, with the exception of the samples from California, where there were only 18 TMP-SMZ-resistant non-CGA isolates) were tested for 40 virulence factors of extraintestinal pathogenic E. coli by PCR as previously described (6). CGA isolates were further assessed by random amplified polymorphic DNA (RAPD) analysis using the arbitrary decamer primer 1254 (5′-CCGCAGCCAA-3′) (6) in comparison with CGA and non-CGA reference strains and by pulsed-field gel electrophoresis (PFGE) analysis of XbaI-digested total DNA (11).
Statistical analysis.
Comparisons of proportions were tested using Fisher's exact test (two-tailed). The criterion for statistical significance was a P value of <0.05. BioNumerics software (Applied Maths), which has a band tolerance of 1.15%, was used to assess the digital images of the PFGE profiles.
RESULTS
Distribution of TMP-SMZ resistance.
A total of 1,484 arbitrarily selected E. coli sewage isolates (for primary effluents, n = 732; for secondary effluents, n = 754) from seven locations were characterized, representing approximately 200 isolates from each location (Table 1). Seventy-seven (5%) of the isolates were resistant to TMP-SMZ. The prevalence of TMP-SMZ resistance varied among the various locales, with the highest values being observed for California (13%) and Wisconsin (7%), followed by Ohio, Washington (5%), Virginia (2%), and Maine (<1%). Six percent (n = 42) of the E. coli isolates from primary effluents and 5% (n = 35) of those from secondary effluents were resistant to TMP-SMZ.
TABLE 1.
Occurrence of TMP-SMZ resistance and Escherichia coli CGA among 1,484 E. coli sewage isolates from seven regions
| Region | Total no. of E. coli isolates | No. (%) of TMP-SMZ-resistant isolates | No. of CGA E. coli isolates (% of TMP-SMZ-resistant isolates)a |
|---|---|---|---|
| San Jose, CA | 226 | 30 (13) | 12 (40) |
| Tallahassee, FL | 210 | 5 (2) | 0 (<1) |
| Lewiston, ME | 204 | 1 (<1) | 0 (<1) |
| Cincinnati, OH | 219 | 10 (5) | 0 (<1) |
| Seattle, WA | 170 | 9 (5) | 1 (11) |
| Milwaukee, WI | 240 | 17 (7) | 1 (6) |
| Virginia Beach, VA | 215 | 5 (2) | 1 (20) |
| Total | 1,484 | 77 (5) | 15 (19.5) |
All TMP-SMZ-resistant isolates were screened for CGA status.
Prevalence of CGA isolates.
According to the CGA-specific single-nucleotide polymorphism assay, 15 (20%) of the 77 TMP-SMZ-resistant isolates were members of CGA. Of the 15 CGA isolates, 12 (80%) were from California, whereas 1 isolate each was from Virginia, Washington, and Wisconsin (P = 0.002 for percent CGA among isolates from California versus those from all other locales combined). Twenty percent (n = 8) of the E. coli isolates from primary effluents and 17% (n = 6) of the isolates from secondary effluents were members of CGA. In contrast, none of the 77 geographically matched TMP-SMZ-susceptible control isolates belonged to CGA.
O:H serotypes.
Serotyping of the 15 CGA isolates showed that 4 were O44:H18, 2 were O17,77:H18, and 1 each was O11:H18, O15:H18, O17:H18, O73:H18, O77:H18, and O86:H18, whereas 3 were O and H nontypeable. All 12 of the H-typeable CGA isolates were H18, and 6 (50%) of the 12 that were O typeable exhibited one or more of the O antigens characteristic of CGA (O11, O17, O73, and O77) (11). In contrast, among the 62 non-CGA, TMP-SMZ-resistant isolates, 43 different O:H serotypes were represented, and in only three instances did multiple isolates exhibit the same serotype, namely, O8:H12 (n = 2), O1:H+ (n = 2), and O2:H7 (n = 2). The only instance of a shared serotype among CGA and non-CGA isolates consisted of one isolate from each group exhibiting serotype O86:H18. Only one additional non-CGA isolate exhibited the H18 flagellar antigen (O1:H18), for an overall prevalence of 4.5% (in contrast to CGA isolates [80%]; P < 0.0001), and none exhibited any of the four CGA-associated O antigens (in contrast to CGA isolates [40%]; P < 0.0001). Nonetheless, while 33% of the environmental CGA isolates had serotypes typical of those reported for human clinical CGA isolates (i.e., O17,77:H18 and O11:H18), 67% had serotypes not previously described for human clinical or fecal CGA isolates.
Phylogenetic distribution.
As expected, all 15 CGA isolates belonged to phylogenetic group D (8). In contrast, the non-CGA, TMP-SMZ-resistant isolates (n = 62) were approximately evenly distributed over the four phylogenetic groups, with 18 (29%) group A, 15 (24%) group B1, 15 (24%) group B2, and 14 (23%) group D isolates.
Antimicrobial resistance profiles.
The CGA isolates and the TMP-SMZ-resistant non-CGA isolates differed according to their composite resistance phenotypes. Among the 15 CGA isolates, five resistance profiles were encountered. One isolate exhibited TMP-SMZ resistance only, 2 (13%) were resistant to two additional antimicrobials each (AM and ST or AM and TE), 11 (73%) were resistant to three additional antimicrobials each (AM, ST, and TE), and 1 exhibited resistance to all four additional antimicrobials tested (AM, CH, ST, and TE). In contrast, the 62 non-CGA, TMP-SMZ-resistant isolates exhibited 11 different multiple antimicrobial resistance patterns. In order of descending prevalence, 25 (40%) were resistant to two additional antimicrobials, 20 (32%) were resistant to three additional antimicrobials, 9 (14%) were resistant to four additional antimicrobials, 7 (11%) were resistant to one additional antimicrobial, and 1 (2%) was resistant only to TMP-SMZ. The four most commonly occurring associated resistance profiles among the non-CGA, TMP-SMZ-resistant isolates were resistance to AM, ST, and TE (n = 20); resistance to AM and TE (n = 13); resistance to AM and ST (n = 8); and resistance to AM, CH, ST, and TE (n = 8).
Virulence profiles.
The 15 CGA isolates were compared with 22 geographically matched, non-CGA, TMP-SMZ-resistant isolates for virulence traits, including alleles of papA (encoding the P fimbria structural subunit) (Table 2). The non-CGA isolates were selected to provide a distribution among the four phylogenetic groups, namely, A (n = 7), B1 (n = 4), B2 (n = 6), and D (n = 5). The CGA isolates were divided between the F10 (n = 9) and F16 (n = 6) papA alleles, whereas none of the non-CGA isolates exhibited either of these two papA alleles (P < 0.001 and P = 0.002, respectively). In contrast, three of the non-CGA isolates, but none of the CGA isolates, exhibited the F12 or F14 papA allele. Additionally, all CGA isolates, but only three (14%) of the non-CGA isolates, exhibited papEF (encoding P fimbria tip pilins) (P < 0.0001). The CGA and non-CGA isolates also differed significantly (P < 0.0001) according to the prevalence of five non-pap virulence traits, including iha (for adhesin/siderophore receptor), sat (for secreted autotransporter toxin), iutA (for aerobactin receptor), kpsM II (for the group II capsule), and ompT (for outer membrane protease T). afa/dra (encoding Dr antigen binding adhesins) was the only virulence trait which demonstrated a significant negative association with CGA.
TABLE 2.
Virulence factor profiles among CGA and non-CGA Escherichia coli sewage isolates
| Traita | Prevalence of associated trait [no. (%)]
|
P valuec | |
|---|---|---|---|
| CGA isolates (n = 15) | Non-CGA isolatesb (n = 22) | ||
| F10 papA allele | 9 (60) | 0 (0) | <0.0001 |
| F16 papA allele | 6 (40) | 0 (0) | 0.0022 |
| papEF | 15 (100) | 3 (14) | <0.0001 |
| papG allele II | 6 (40) | 1 (4) | 0.0113 |
| afa/dra | 0 (0) | 14 (64) | (<0.0001) |
| iha | 14 (93) | 3 (14) | <0.0001 |
| sat | 14 (93) | 4 (18) | <0.0001 |
| iutA | 14 (93) | 7 (32) | 0.0002 |
| kpsM II | 15 (100) | 10 (45) | 0.0004 |
| ompT | 15 (100) | 8 (36) | <0.0001 |
Only those traits for which prevalence differences between CGA and non-CGA isolates were statistically significant (Fisher's exact test; P < 0.05) are shown. Trait definitions: papA, P fimbria structural subunit gene; papEF, P fimbria tip pilus gene; papG allele II, P fimbriae adhesion variant II gene; afa/dra, Dr antigen binding adhesin gene; iha, adhesin/siderophore gene; sat, secreted autotransporter toxin gene; iutA, aerobactin receptor gene; kpsM II, group II capsule gene; and ompT, outer membrane protease T gene. Other traits present in CGA but not significantly different in overall prevalence compared with non-CGA isolates,, included papC (P fimbria assembly), fimH (type 1 fimbria adhesin), fyuA (yersiniabactin receptor), and traT (serum resistance-associated gene). Traits absent from CGA isolates but detected among non-CGA isolates included the F12 papA allele, papG allele III, iroN, K1 kpsM II variant, usp, iss, and malX; however, these differences (CGA versus non-CGA) were not statistically significantly by Fisher's exact test.
Non-CGA, TMP-SMZ-resistant control isolates were chosen in a 2:1 ratio to the CGA isolates within each locale, with the exception of San Jose, CA, where only 18 non-CGA, TMP-SMZ resistant isolates were recovered, all of which were used as controls.
The P value in parentheses is for a negative association with CGA.
Interestingly, although all CGA isolates exhibited papEF (for P fimbria tip pilins), only six (40%) also exhibited papA, papC (for P fimbria assembly), and papG allele II (for P fimbria adhesin variant II). Moreoever, all CGA isolates exhibiting the F10 papA allele (n = 9) had papEF as their only other pap element, whereas CGA isolates exhibiting the F16 papA allele (n = 6) contained a complete copy of the pap operon, including papA, papC, papEF, and papG (allele II).
RAPD and PFGE analysis.
The RAPD genomic profiles of the 15 CGA isolates were highly homogeneous and matched those of reference CGA isolates UMN026 and SEQ102, whereas they differed substantially from those of non-CGA controls (Fig. 1). Interestingly, CGA isolates containing the F10 papA allele typically exhibited a one-band RAPD profile difference in comparison with the CGA isolates containing the F16 papA allele, consistent with these two groups possibly representing different genomic subsets within CGA (Fig. 1). PFGE analysis (Fig. 2) demonstrated that although the profiles of the CGA isolates exhibited an obvious overall similarity, each isolate (lanes 2 to 9 and 11 to 17) had a distinct banding pattern, indicating that despite their considerable similarity (and, in several instances, common sample of origin), no two isolates were replicates of the same clone. The presence/absence of a single high-molecular-weight band (Fig. 2) corresponded closely with the split between the F10 and F16 papA alleles among CGA isolates.
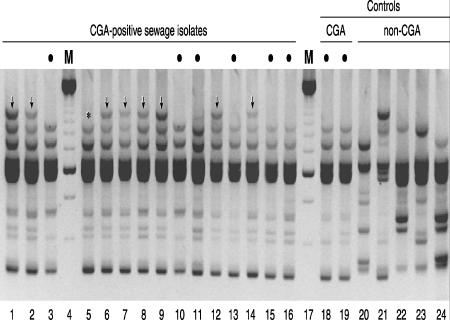
FIG. 1.
RAPD profiles of selected Escherichia coli isolates. Lane numbers are shown below the gel image. Lanes 4 and 17, 250-bp marker (M); lanes 1 to 3 and 5 to 16, CGA-positive sewage isolates; lanes 18 and 19, CGA positive controls (human cystitis isolates SEQ102 and UMN026, respectively); lanes 20 to 24, CGA negative controls (lanes 20 and 21, non-CGA sewage isolates [from phylogenetic groups B2 and D, respectively]; lane 22, strain CFT073 [from group B2]; and lanes 23 and 24, human-source non-CGA isolates [from groups D and B2, respectively]). Bullets above lanes indicate CGA isolates that exhibit the consensus CGA-associated virulence profile, including the F16 papA (P fimbria structural subunit) allele, and a characteristic CGA-associated RAPD profile. CGA isolates in lanes without bullets exhibit an atypical virulence profile that includes the F10 papA allele plus (except for strain 492 [lane 5]) an atypical RAPD profile that includes an extra, ∼2,000-bp band (vertical arrows; “*” for strain 492 [lane 5]). The profiles of all the CGA isolates are indistinguishable (within the reproducibility limits of RAPD analysis), except for the ∼2,000-bp band, and collectively, they are distinct from the profiles of the non-CGA isolates, each of which is unique.
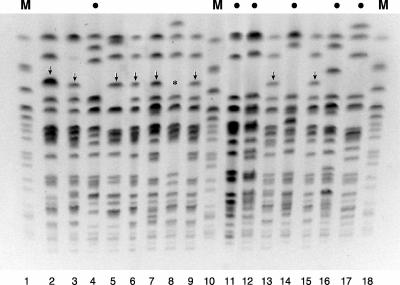
FIG. 2.
XbaI PFGE profiles of selected Escherichia coli isolates. Lane numbers are shown below the gel image. Lanes 1, 10, and 18, marker (M) lanes with E. coli O157:H7 strain G5244; lanes 2 to 9 and 11 to 17, CGA-positive sewage isolates. Bullets above lanes indicate CGA isolates that exhibit the consensus CGA-associated virulence profile, including the F16 papA (P fimbria structural subunit) allele. CGA isolates in lanes without bullets exhibit an atypical virulence profile that includes the F10 papA allele and (except for strain 518 [lane 8]) exhibit a large extra band in the PFGE profile (vertical arrows; “*” for strain 518). The profiles of the CGA isolates are all unique, yet they exhibit similarities that distinguish them as a group from the E. coli O157:H7 reference strain (lanes 1, 10, and 18).
DISCUSSION
Among E. coli isolates from wastewater effluents from seven U.S. geographic regions, the prevalence of CGA and of TMP-SMZ-resistant E. coli varied considerably by region; however, no significant difference was seen between the isolates from primary and secondary effluents. The highest percentage of CGA isolates was from California, which not only had the highest overall prevalence of TMP-SMZ resistance (39%) but also had the largest proportion of TMP-SMZ-resistant isolates accounted for by CGA (80%). It is of interest that California is a region that has previously reported a high proportion of CGA isolates (51%) among TMP-SMZ-resistant clinical isolates (11). The explanation for the observed variation in the prevalence of CGA and of TMP-SMZ-resistant E. coli in wastewater effluents is unknown. However, possible explanations include differences in processes between wastewater treatment plants, in population densities at each treatment plant, in the types of sewages found at each treatment plant (industrial, agricultural, or urban), and in the prevalence of CGA within the host population(s) served by each plant. Variation in the prevalence of CGA among clinical isolates from different regions has been documented (6, 8); it is possible that regional variation also exists among environmental samples.
The increasing occurrence of TMP-SMZ-resistant uropathogenic E. coli emphasizes the need to determine resistance prevalence levels within a community. Determining the prevalence of antimicrobial resistance in a given locale may require information beyond that obtained in clinical laboratory studies (5). Data on resistance rates collected only from patients with UTIs may not reflect the true prevalence of resistance in the local community (10, 19), since specimens are often submitted for culture and antibiotic susceptibility testing only after initial antimicrobial therapy has failed or because the patient has had recurrent episodes of UTIs or is otherwise considered at risk for having a resistant organism. The intestinal tract serves as the primary source of uropathogenic E. coli (10). E. coli found in domestic sewage comes predominantly from human fecal material (18). Thus, sewage isolates may serve as representatives of the strains of E. coli present within the human population in a given locale. Surveillance of antibiotic resistance in coliforms from sewage has previously been suggested as a means of monitoring changes in antibiotic resistance patterns in the general population (18). Seneviratne and Woods (16) specifically proposed that such a surveillance program would be helpful in providing therapeutic guidance for the treatment of urinary tract pathogens. The results of the present study suggest that such monitoring programs may also provide useful information on the occurrence and dissemination of specific clonal groups within a given community.
Fifteen (19.5%) TMP-SMZ-resistant isolates in this study belonged to CGA. This value is very similar to that reported in a recent national survey of clinical isolates, where CGA accounted for 15% of TMP-SMZ-resistant isolates from diverse locales across the United States (8). Among the sewage-source isolates, E. coli CGA isolates were distinct from geographically matched non-CGA, TMP-SMZ-resistant E. coli isolates according to a broad range of bacterial characteristics, as previously reported for human clinical isolates of CGA versus non-CGA E. coli (11).
Notwithstanding the overall similarity of the present sewage-source CGA isolates to previously described human CGA isolates, some differences were apparent. The environmental isolates exhibited a broader range of serogroups (O15, O44, and O86) beyond the O11, O17, O73, and O77 serogroups typically associated with clinical CGA isolates, a lower prevalence of CH resistance (7% versus >25% among clinical CGA isolates), and the unique occurrence of the F10 papA allele (60% versus 0%) (8). It is of interest that the F10 allele-positive isolates lacked most of the rest of the pap operon, so presumably could not express P fimbriae, which should make them less able to colonize or infect humans. Whether the F10 allele-positive subgroup represents an environmentally adapted variant of CGA, or perhaps an ancestral precursor, remains to be established. The detection of this interesting subgroup in multiple locales suggests that it is not an isolated entity but may be broadly prevalent in sewage effluents across the United States. These findings raise interesting questions regarding the ecology of these organisms outside the human host.
In summary, the sewage E. coli CGA isolates exhibited a wider diversity of resistance profiles and somatic antigens than that found in most previous characterizations of this clonal group (4). However, some of these isolates had virulence factor and antimicrobial resistance profiles typical of human-source CGA isolates. This is the first report of the recovery of E. coli CGA isolates with typical “human” pattern virulence and resistance profiles from outside the human host. The presence of CGA E. coli in sewage effluents may provide a means for dissemination of this clonal group into the environment. Sewage could serve as a vehicle for entering human and nonhuman hosts by direct contact or through contamination of drinking water supplies.
Acknowledgments
This material is based on work supported by the Office of Research and Development, Medical Research Service, Department of Veterans Affairs (J.R.J.).
Dave Prentiss (Minneapolis VA Medical Center) prepared the figures.
Any opinions expressed in this paper are those of the authors and do not necessarily reflect the official positions and policies of the USEPA. Any mention of products or trade names does not constitute recommendation for use by the USEPA.
Footnotes
Published ahead of print on 4 May 2007.
REFERENCES
- 1.Burman, W. J., P. E. Breese, B. E. Murray, K. V. Singh, H. A. Batal, T. D. MacKenzie, J. W. Ogle, M. L. Wilson, R. R. Reves, and P. S. Mehler. 2003. Conventional and molecular epidemiology of trimethoprim-sulfamethoxazole resistance among urinary Escherichia coli isolates. Am. J. Med. 115:358-364. [DOI] [PubMed] [Google Scholar]
- 2.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clesceri, L. S., A. E. Greenberg, and A. D. Eaton (ed.). 1998. Standard methods for the examination of water and wastewater, 20th ed. American Public Health Association, Washington, DC.
- 4.France, A. M., K. M. Kugeler, A. Freeman, C. A. Zalewski, M. Blahna, L. Zhang, C. F. Marrs, and B. Foxman. 2005. Clonal group and the spread of resistance to trimethoprim-sulfamethoxazole in uropathogenic Escherichia coli. Clin. Infect. Dis. 40:1101-1107. [DOI] [PubMed] [Google Scholar]
- 5.Gupta, K., T. M. Hooton, and W. E. Stamm. 2001. Increasing antimicrobial resistance and the management of uncomplicated community-acquired urinary tract infections. Ann. Intern. Med. 135:41-50. [DOI] [PubMed] [Google Scholar]
- 6.Johnson, J. R., A. R. Manges, T. T. O'Bryan, and L. W. Riley. 2002. A disseminated multidrug resistant clonal group of extraintestinal pathogenic Escherichia coli as a cause of pyelonephritis. Lancet 359:2249-2251. [DOI] [PubMed] [Google Scholar]
- 7.Johnson, J. R., K. Owens, A. R. Manges, and L. W. Riley. 2004. Rapid specific detection of Escherichia coli clonal group A by gene-specific PCR. J. Clin. Microbiol. 42:2618-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson, J. R., A. C. Murray, M. A. Kuskowski, S. Schubert, M. F. Prere, B. Picard, R. Colodner, R. Raz, and the Trans-Global Initiative for Antimicrobial Resistance Analysis (TIARA) Investigators. 2005. Distribution and characteristics of Escherichia coli clonal group A. Emerg. Infect. Dis. 11:141-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson, J. R., M. A. Kuskowski, K. Smith, T. T. O'Bryan, and S. B. Tatini. 2005. Antimicrobial-resistant and extraintestinal pathogenic Escherichia coli in retail foods. J. Infect. Dis. 191:1040-1049. [DOI] [PubMed] [Google Scholar]
- 10.Karlowsky, J. A., L. J. Kelly, C. Thornsberry, M. E. Jones, and D. F. Sahm. 2002. Trends in antimicrobial resistance among urinary tract infection isolates of Escherichia coli from female outpatients in the United States. Antimicrob. Agents Chemother. 46:2540-2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manges, A. R., J. R. Johnson, B. Foxman, T. T. O'Bryan, K. E. Fullerton, and L. W. Riley. 2001. Widespread distribution of urinary tract infections caused by a multidrug-resistant Escherichia coli clonal group. N. Engl. J. Med. 345:1007-1013. [DOI] [PubMed] [Google Scholar]
- 12.Murray, B. E., J. J. Mathewson, H. L. Dupont, C. D. Ericsson, and R. R. Reeves. 1990. Emergence of resistant fecal Escherichia coli in travelers not taking prophylactic antimicrobial agents. Antimicrob. Agents Chemother. 34:515-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.National Committee for Clinical Laboratory Standards. 2000. Supplemental tables: disk diffusion, p. 14-18. NCCLS document M100-S10. NCCLS, Wayne, PA.
- 14.Ramchandani, M., A. R. Manges, C. DebRoy, S. P. Smith, J. R. Johnson, and L. W. Riley. 2005. Possible animal origin of human-associated, multi-drug resistant, uropathogenic Escherichia coli. Clin. Infect. Dis. 40:251-257. [DOI] [PubMed] [Google Scholar]
- 15.Sahm, D. F., C. Thornsberry, D. C. Mayfield, M. E. Jones, and J. A. Karlowsky. 2001. Multidrug-resistant urinary tract isolates of Escherichia coli: prevalence and patient demographics in the United States in 2000. Antimicrob. Agents Chemother. 45:1402-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seneviratne, E. M. E., and G. Woods. 1976. Sensitivity patterns of urinary pathogens in the Southland community and a study of sewage flora. N. Z. Med. J. 84:15-18. [PubMed] [Google Scholar]
- 17.Stamm, W. E. 2001. An epidemic of urinary tract infections? N. Engl. J. Med. 345:1055-1057. [DOI] [PubMed] [Google Scholar]
- 18.Sturtevant, A. B., Jr., and T. W. Feary. 1969. Incidence of infectious drug resistance among lactose-fermenting bacteria isolated from raw and treated sewage. Appl. Microbiol. 18:918-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ti, T. Y., G. Kumarasinghe, M. B. Taylor, S. L. Tan, A. Ee, C. Chua, and A. Low. 2003. What is true community-acquired urinary tract infection? Comparison of pathogens identified in urine from routine outpatient specimens and from community clinics in a prospective study. Eur. J. Clin. Microbiol. Infect. Dis. 22:242-245. [DOI] [PubMed] [Google Scholar]