Abstract
A novel species of Acidimicrobium appeared to be the predominant ferrous iron oxidizer in a mixed culture that effected the continuous, efficient extraction of nickel from a mineral concentrate at 49°C, but it was not isolated in pure culture. It outcompeted Acidimicrobium ferrooxidans, which was expected to have a major role in iron oxidation in reactors gassed with air, and was outnumbered at 49°C only by the sulfur-oxidizing Acidithiobacillus caldus. Sulfobacillus species were expected to compete with Acidimicrobium species when culture aeration was enriched with carbon dioxide, but they were a minor component of the populations with and without this enrichment. Sulfobacillus thermosulfidooxidans replaced the Acidimicrobium species and Acidithiobacillus caldus when the temperature was increased to 55°C.
Most commercial operations processing mineral sulfides in stirred-tank bioreactors have utilized mesophilic bacteria (22). However, thermoacidophiles can increase the rate and extent of mineral degradation (5, 17), and processes have been developed with bacteria at 50°C (15) and archaea at 78°C (3). The potential of moderate thermophiles to extract base metals from mineral sulfide concentrates during autotrophic growth was demonstrated (14) with organisms that were later incorporated into the Sulfobacillus genus (19). Sulfobacillus and Acidimicrobium species were noted as the principal iron-oxidizing acidophiles in a mixed enrichment culture used in the development of mineral sulfide concentrate processing at 50°C (10); characterization of the culture, which shared source material with the culture described in this paper, was not reported. Carbon dioxide availability was expected to be a major influence on the composition of the culture, because Acidimicrobium ferrooxidans appears to have a carbon dioxide-concentrating capacity whereas Sulfobacillus species require an enhanced concentration of carbon dioxide in culture aeration for extensive autotrophic growth (8, 16).
MATERIALS AND METHODS
Culture.
The enrichment culture used in bioreactors was originally established with acidic samples from various mineral sulfide mines, coal mines, spoil heaps, and natural geothermal areas. It was serially cultured for several years before these experiments. Pyrite (1%, wt/vol) was used as a substrate, with subculturing twice a year in a shaken flask at 48°C for a few days and interim maintenance as a static culture at room temperature.
Mineral sulfide concentrates.
A nickel concentrate was used in establishment of continuous cultures. This was replaced when a second concentrate that contained more nickel became available. The substitution was made after 119 days of continuous operation and 60 days before the first analyses of leached residues. The second concentrate was ground to pass a <75-μm-diameter sieve mesh and contained 10.5% (wt/wt) nickel (mostly in pentlandite and violarite) and 25.9% (wt/wt) iron (in pyrite, pyrrhotite, and the nickel sulfides).
Bioreactors and culture conditions.
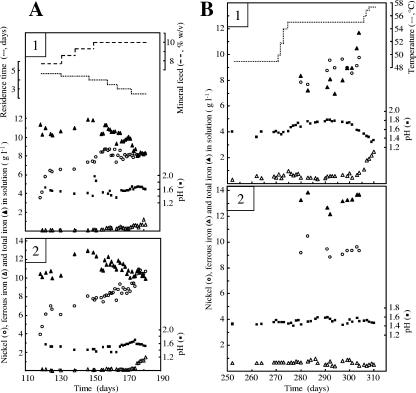
Two water-jacketed, baffled bioreactors were used with working culture volumes of 600 ml. Continuous feeds of sterilized medium contained the following (g liter−1): MgSO4·7H2O, 0.5; (NH4)2SO4, 0.4; and K2HPO4, 0.8. Overflows were through the reactors' side arms. Cultures were gassed (800 ml min−1) with air or with 5% (vol/vol) CO2 in air. Agitation was from overhead motors stirring flat, two-bladed paddles at 400 rpm. After inoculation, continuous flows were started with medium adjusted to pH 1.5 with H2SO4. The acidity of the feeds was reduced as stronger growth resulted in increased acid production, and no acid was added from day 150, with the influent medium at pH 8. The concentrates were added directly to reactors three times each day to avoid prior contact with the medium. Variations in dilution rates and mineral additions (expressed as the equivalent of a suspended-solids concentration) are indicated in Fig. 1A. After 150 days, a mineral feed equivalent to 10% (wt/vol) was maintained. Culture temperatures and aeration were adjusted (Table 1), with the indicated changes in temperature of reactor 1 made gradually (Fig. 1B).
FIG. 1.
Nickel sulfide concentrate biooxidation in two continuous cultures of moderately thermophilic acidophiles. (A) The reactors were operated at 49°C; one was gassed with CO2-enriched air (reactor 1) and one with air (reactor 2). The indicated mineral feed concentrations and residence times are for both reactors. The indicated time is that from inoculation of the reactors. (B) One of the reactors was maintained at 49°C (reactor 2), while the temperature in the other was increased gradually from 49 to 57.5°C (reactor 1). Both reactors were gassed with CO2-enriched air from day 270.
TABLE 1.
Temperatures and aerationa of two continuous cultures of mineral-processing moderate thermophiles
| Days | Reactor 1
|
Reactor 2
|
||
|---|---|---|---|---|
| Temp (°C)b | Culture gassing | Temp (°C) | Culture gassing | |
| 1−270 | 49 | CO2-air | 49 | Air |
| 271-275 | 49-55 | CO2-air | 49 | CO2-air |
| 306−308 | 55-57.5 | CO2-air | 49 | CO2-air |
Either air or 5% (vol/vol) CO2 in air.
The temperature shifts to the higher values in reactor 1 were made over the indicated days.
Microbial population analyses.
Centrifuged pellets from 20 ml of mineral, precipitates, and cells from reactors were washed by resuspension in dilute H2SO4 (pH 1.7) and distilled water before DNA extraction with the method of Barns et al. (2). Extraction buffer (10 ml) was added before a sequence of treatments with freeze-thaw cycles, lysozyme, proteinase K, phenol, and phenol-chloroform-isoamyl alcohol, followed by precipitation and washing of DNA. Amplifications of bacterial partial 16S rRNA genes from the extracted DNA were performed as described previously (7) using forward (F27) and reverse (R1492) primers. PCR products were cloned using Taq polymerase, the TOPO TA cloning kit (version J) vector (pCR2.1-TOPO), and a host Escherichia coli strain (Invitrogen, Carlsbad, CA). Plasmid DNA was extracted from E. coli by a standard alkaline lysis protocol. Digoxigenin nucleic acid labeling and chemiluminescence detection systems (Boehringer Mannheim) were used in analysis of DNA spotted onto nylon membranes (Hybond-N). The sequences of 16S rRNA oligonucleotide probes designed for dot blots and for fluorescent in situ hybridization (FISH) experiments are given in Table 2. Other eubacterial probes, R1492 and Eub338 (1), were also used. The sources of DNA for standards on dot blots and for testing probe specificity were Acidimicrobium ferrooxidans (DSMZ 10331T), Sulfobacillus thermosulfidooxidans (BC1) (19), Sulfobacillus acidophilus (DSMZ 10332T), and Acidithiobacillus caldus (DSMZ 8584T).
TABLE 2.
Sequences of 16S rRNA oligonucleotide probes designed for this study
| Probea | Sequence (5′→3′) | Target organism(s) |
|---|---|---|
| S:628 | CGTTTAGCCGCCCTCC | Sulfobacillus genus |
| St:989 | CTCACGAGCGTGTCCAGT | S. thermosulfidooxidans |
| Sa:988 | TCACGGCTCCCGTCTAGCC | S. acidophilus |
| Am:654 | CGATCYTCTACCGGACTC | Acidimicrobium group |
| Amf:995 | CTCTGCGGCTTTTCCCTCCATG | Acidimicrobium ferrooxidans |
| Am2:131 | GTTGTCCCGAACTATGGGGTAGG | “Acidimicrobium” species 2 |
| Atc:455 | AGACCGTATTCGGATCCGCC | Acidithiobacillus caldus |
Numbers indicate the first nucleotide of the corresponding E. coli sequence.
For FISH, 1-ml samples from bioreactors were centrifuged (1,000 × g for 1 min) to remove mineral particles and iron precipitates before free cells were pelleted, washed, and treated with paraformaldehyde. Hybridizations were performed at 37°C on gelatin-coated slides with fluorescently labeled probes that were synthesized with a 5′-Cy3 (indocarbocyanine) fluorochrome (Interactiva Biotechnologie GmbH, Ulm, Germany). Hybridization efficiencies and specificities of probes were estimated by monitoring the fluorescence intensities of whole-cell hybridizations with pure cultures of target and nontarget organisms. Concentrations of formamide were selected to give the strongest signal consistent with specificity. The Sulfobacillus- and Acidithiobacillus-targeting probes were specific for their target organisms with 20% (vol/vol) formamide. The Acidimicrobium-targeting probes Acm:654 and Acm2:131 were used with 30% formamide, and Acmf:995 was used with 45% formamide, to avoid nonspecific hybridization with Sulfobacillus and Acidithiobacillus, which occurred with 20% formamide. Cells were stained with DAPI (4′,6-diamidino-2-phenylindole) (7 μg ml−1) for 5 min at room temperature. Epifluorescence microscopy was performed with a Zeiss (Oberkochen, Germany) Axioskop light/fluorescence microscope.
Two solid media were used for isolation of bacteria from reactor samples. The first was made by mixing, at 60°C, equal volumes of sterilized solutions of Gelrite or Phytagel (Sigma) (0.8% [wt/vol] in distilled water) and mineral salts [in g liter−1: MgSO4·7H2O, 1.0; (NH4)2SO4, 0.8; KHSO4, and 0.2; K2HPO4, 0.01; acidified to pH 1.7 with sulfuric acid]. Ferrous sulfate (25 mM final concentration) was added to the mixed solutions before setting. Supplements of yeast extract (0.01% wt/vol) and/or tetrathionate (1 mM) were also tested. The second medium comprised ferrous-iron overlay agarose-gelled plates, kindly provided by D. B. Johnson. These plates have been described as being highly efficient in the successful assessment of mixed acidophile populations in tank bioleaching (21).
RESULTS
Mineral solubilization. (i) Influence of supplemental CO2.
Nickel solubilization (Fig. 1A) was essentially the same up to 160 days in cultures sparged with CO2-enriched air (reactor 1) or air (reactor 2). The nickel concentration in solution increased after 119 days of continuous operation because of the change to the concentrate with a higher nickel content. The reason for higher levels of nickel and iron in solution in reactor 2 than in reactor 1 between 160 and 260 days is not known. Some inadequate mixing and concentrate retention might have occurred in reactor 2, increasing the mineral concentration. Analysis of solid residues in effluents over three 24-h periods (days 178, 180, and 181) indicated similar degrees of mineral dissolution, with 23 and 20% of the nickel remaining in the residues from reactor 1 and reactor 2, respectively. The gradual decrease in total iron in solution in reactors 1 and 2 between 150 and 180 days reflected the rise in pH from just under 1.5 to over 1.6 and from about 1.4 to over 1.5, respectively, which resulted in incorporation of more hydrated ferric sulfates into the residues. An increase in ferrous iron in solution after a shorter residence time (dilution rate of 0.4 day−1 after 172 days) indicated that bacterial oxidation of ferrous iron did not keep pace with reduction of ferric iron by the mineral sulfides and solubilization of ferrous iron. The increase was greater in the absence of CO2 supplementation. The residence times were returned to 3 days in order to avoid culture washout.
(ii) Influence of temperature.
An increase in temperature to 55°C resulted in a transient reduction in acidity (Fig. 1B). This contributed to a loss of almost half of the iron from solution and a consequent increase in the amount of precipitated solids in the effluent (data not shown). Although microscopy showed a reduced planktonic cell concentration at 55°C (estimated as a 50% reduction), it is possible that nickel extraction was slightly enhanced with a lower nickel content (4.7%, wt/wt) in solid residue at 55°C than in residue (4.87% Ni, wt/wt) at 49°C. When iron oxidation was inhibited by the final temperature increase to 57.5°C (Fig. 1B), microscopy indicated a great reduction in cell numbers.
Culture analyses. (i) FISH and 16S rRNA gene clone banks.
Populations were analyzed (Fig. 2; Table 3) after 53 days of continuous culture at 49°C in reactors gassed with air and with CO2-enriched air and again after 301 days, when both reactors were gassed with CO2-enriched air but maintained at different temperatures (49°C and 55°C). Fluorescently labeled oligonucleotide probes targeted to 16S rRNA showed that populations in air-gassed and CO2-enriched cultures at 49°C were essentially the same (Table 3). The probe for the Acidimicrobium group (Acm:654) revealed cells that resembled Acidimicrobium ferrooxidans in morphology (Fig. 2), but they did not react with the species-specific Acidimicrobium ferrooxidans probe (Acm:995). However, colonies that were obtained from plating samples on ferrous iron-Phytagel and overlay plates were species of Sulfobacillus or Acidimicrobium ferrooxidans. Sulfobacillus species were a minor component of these populations, in which more than half of the cells were Acidithiobacillus caldus. After the temperature was increased to 55°C, Sulfobacillus species were predominant and relatively few Acidithiobacillus caldus or Acidimicrobium cells were seen with FISH (not shown). The population maintained throughout at 49°C was essentially the same at sampling after 53 days and 301 days of operation (Table 3), which was sampling before and after supplementation of the culture aeration with CO2. Clone banks of PCR-amplified 16S rRNA genes confirmed the dominance of Sulfobacillus species at 55°C (Table 3). The dominant cloned sequence had greater than 99% identity to that of S. thermosulfidooxidans. The clone bank of the 49°C sample could have been too small to represent the relatively low number of Sulfobacillus cells or could have been influenced by biases in DNA extraction and amplification. Acidimicrobium species and Acidithiobacillus caldus were again indicated as the prevalent organisms at 49°C.
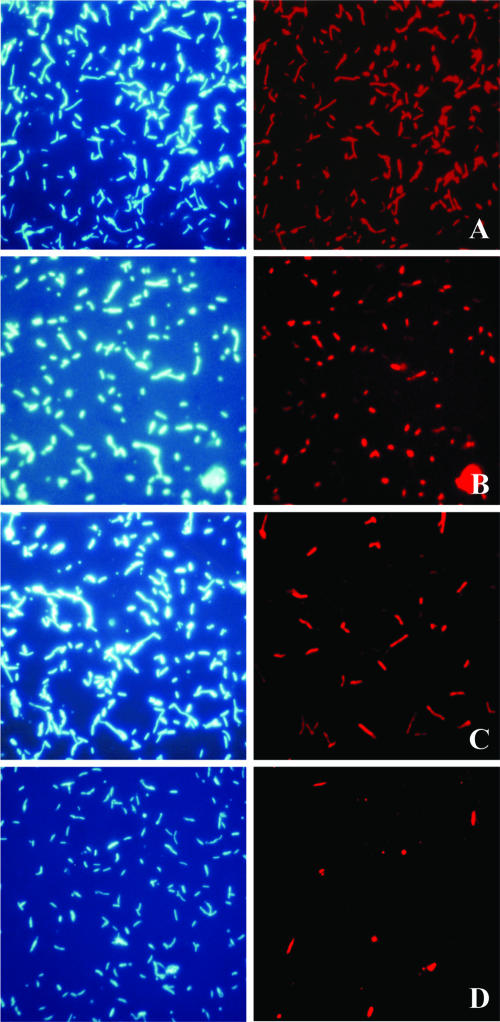
FIG. 2.
DAPI staining (left panels) and Cy3-labeled 16S rRNA-targeted FISH (right panels) of samples taken from a mineral-sulfide-oxidizing bioreactor after 53 days of continuous culture gassed with air (reactor 2). FISH probes and the targeted organisms were Eub338 (eubacteria) (A), Atc:455 (Acidithiobacillus caldus) (B), Am:654 (Acidimicrobium species) (C), and S:628 (Sulfobacillus species) (D).
TABLE 3.
Proportions of different cell types in continuous bioreactors as estimated by FISH with probes specific to the indicated target organisms (Table 2) and by rRNA gene clone bank analyses
| Probe or target | Fluorescent cells (% of DAPI-stained cells)a
|
Clones (%)b
|
||||
|---|---|---|---|---|---|---|
| Reactor 1, day 53, 49°C, air-CO2 | Reactor 2, day 53, 49°C, air | Reactor 1, day 301, 55°C, air-CO2 | Reactor 2, day 301, 49°C, air-CO2 | Reactor 1, day 301, 55°C | Reactor 2, day 301, 49°C | |
| Eubacterial 338 | 97.7, 98.1 | 91.6, 92.9 | 96.0 | 93.1 | 100 | 100 |
| Sulfobacillus genus | 3.7, 3.0 | 0.3, 0.9, 1.6 | 90.5 | 3.6 | 97.6 | 0 |
| S. thermosulfidooxidans | 2.1, 2.4 | 0.9, 0.7 | 87.1 | 1.3 | 91.7 | 0 |
| S. acidophilus | Not tested | Not tested | 3.4 | 1.3 | 5.9 | 0 |
| Acidimicrobium group | 19.0, 39.5, 23.7 | 24.9, 20.3 | 1.7 | 25.8 | 0 | 72.2 |
| Acidimicrobium ferrooxidans | 0 | 0 | 0 | 0 | 0 | 0 |
| Acidithiobacillus caldus | 59.0 | 60.2, 57.5 | 4.9 | 51.0 | 2.4 | 27.8 |
The mean number of DAPI-stained cells examined for specific fluorescence was 938 per count (range, 344 to 1,750) in up to three samples as indicated.
Eighty-four clones were examined from the 55°C sample and 296 from the 49°C sample. Five chimeras were excluded from the data.
The sequences of the cloned 16S rRNA genes that represented the Acidimicrobium group in the clone bank of the 49°C reactor culture sample were of one type (9) with 95% identity to that of Acidimicrobium ferrooxidans. The uncultured strain with this 16S rRNA gene sequence is referred to here as “Acidimicrobium” species 2. An enrichment culture that shared source material with that used in this work yielded an identical cloned sequence, which has been deposited in GenBank as belonging to uncultured bacterium clone JTC04 (GenBank accession number AY805539). There are many actinobacterial sequences in GenBank that are related to that of Acidimicrobium ferrooxidans. In phylogenetic analyses (data not shown), the “Acidimicrobium” species 2/clone JTC04 sequence was placed closest to that of Acidimicrobium ferrooxidans in a cluster which also contained sequences from two other bacteria shown to oxidize ferrous iron (“Ferrimicrobium acidiphilum” [GenBank accession number AF251436] and actinobacterium WJ25 [GenBank accession number AY495954]).
(ii) DNA dot blots.
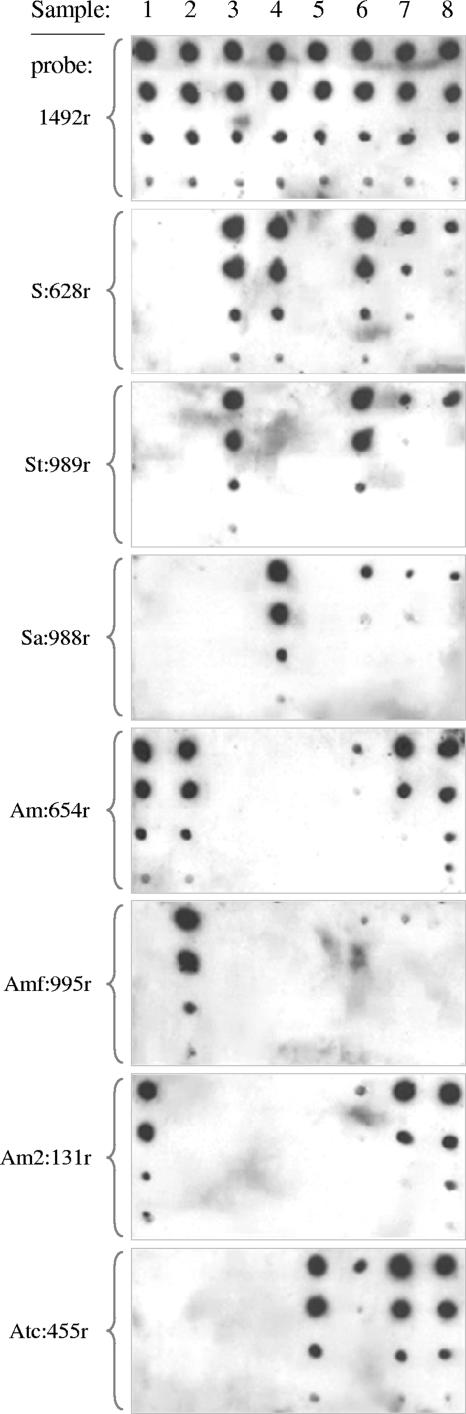
Dot blot hybridizations with the Sulfobacillus group probe (S:628) gave a stronger signal with the 301-day sample of DNA from reactor 2 than with the 53-day sample of DNA from reactor 1 (Fig. 3, columns 7 and 8, respectively), but stronger signals were obtained with Acidimicrobium and Acidithiobacillus caldus probes with these samples from 49°C cultures. In contrast, S. thermosulfidooxidans was clearly indicated as the dominant species in reactor 1 after the temperature was increased to 55°C (Fig. 3, column 6). There was a much smaller increase in the S. acidophilus signal at the higher temperature, while those representing other organisms that were present at 49°C greatly decreased.
FIG. 3.
Dot blot of DNA extracted from moderately thermophilic acidophiles and from mineral-sulfide-oxidizing cultures in continuous bioreactors. Eight 16S rRNA gene oligonucleotide probes were used (indicated to the left of the membranes; see Table 2 for target organisms). DNA was from “Acidimicrobium” species 2 (column 1), Acidimicrobium ferrooxidans (column 2), S. thermosulfidooxidans (column 3), S. acidophilus (column 4), Acidithiobacillus caldus (column 5), reactor 1 at 55°C and day 301 (column 6), reactor 2 at 49°C and day 301 (column 7), and reactor 1 at 49°C and day 53 (column 8). Columns of spotted DNA comprised 1 μg, 100 ng, 10 ng, and 1 ng of genomic DNA extracted from cultures, except for column 1, where spots comprised 10 ng, 1 ng, 100 pg, and 10 pg of plasmid DNA with the cloned 16S rRNA gene from “Acidimicrobium” species 2 (reading from top to bottom for each probe).
DNA from the Acidimicrobium group was more evident than that from Sulfobacillus species in samples from reactors at 49°C gassed with air or with CO2-supplemented air (Fig. 3, Acm:654 probe, columns 7 and 8). The poor signal with the probe designed to be specific for Acidimicrobium ferrooxidans (Acm:995) indicated that this species did not contribute most of the DNA responsible for the group probe signal. The 16S rRNA gene sequence of the unknown Acidimicrobium species that was obtained from a clone bank of DNA extracted from a reactor culture sample was used to design a probe that could differentiate between this “Acidimicrobium” species 2 and Acidimicrobium ferrooxidans. The new probe, Acm2:131, gave a strong signal with DNA from the 49°C reactors but not with Acidimicrobium ferrooxidans (Fig. 3).
Acidithiobacillus caldus was indicated to be a major component of the populations of the reactors at 49°C but was much less evident at 55°C (Fig. 3, probe Atc:455, columns 7 and 8 versus column 6).
DISCUSSION
Nickel extraction with moderately thermophilic bacteria in continuous culture was extensive, with 80% released from the mineral feed. The extraction was much closer to completion when second reactors were attached in series to the reactors described in this work (data not shown) to simulate a likely industrial process design. Development of a process at a pilot scale with mesophiles and a similar nickel concentrate indicated economic viability at a commercial scale, and the use of moderate thermophiles (including the strain referred to in the current work as “Acidimicrobium” species 2) was indicated to achieve higher metal recoveries at a given retention time (10). In the temperature range usually associated with moderately thermophilic, mineral-oxidizing acidophiles (45 to 50°C), the novel Acidimicrobium species appeared to be the most successful and potentially useful iron-oxidizing species over 10 months of continuous mineral processing in the current work. Some previous studies of populations in tank bioleaching of minerals using moderately thermophilic inocula have not included or have not recorded Acidimicrobium species (11, 21).
Limitations of the FISH analyses included assessment of only the planktonic population and the low sensitivity of some probes. The Acidimicrobium group probe (Acm:654) was the least sensitive of the probes in preliminary tests (data not shown), and it is possible that numbers of Acidimicrobium were underestimated in the FISH analysis, where cell counts with specific probes at 49°C were as low as 80% of the universal probe counts (Table 3). However, analysis of an rRNA gene clone bank did not indicate the presence of organisms that would have been unrecognized by the specific probes, and generally the use of RNA or rRNA gene oligonucleotide probes in whole-cell FISH, dot blots, and clone banks gave broadly similar pictures of the mixed culture composition and its change when the temperature was increased.
The supplanting of Acidimicrobium spp. and Acidithiobacillus caldus by Sulfobacillus species at 55°C was accompanied by an initial decrease in acidity, probably because of inhibition and partial washout of the sulfur-oxidizing acidithiobacilli. The sulfur-oxidizing capacity of S. thermosulfidooxidans is much weaker than that of some other sulfobacilli in autotrophic culture (19), but gradual restoration of the acidity at the higher temperature suggested some sulfur-oxidizing activity as its numbers increased. S. acidophilus, a stronger sulfur oxidizer than S. thermosulfidooxidans, was also detected more readily at 55°C than at 49°C (Table 3) but at a relatively low concentration.
Acidimicrobium species might be expected to have an advantage over Sulfobacillus species in a culture gassed with air because, unlike Sulfobacillus species, they enhance their CO2 uptake capacity when CO2 is limiting (8). It was unexpected that Sulfobacillus species did not compete more effectively when additional CO2 was provided. Acidimicrobium ferrooxidans appears to be more sensitive than S. thermosulfidooxidans to ferric iron end product inhibition of ferrous iron oxidation (18), but Acidimicrobium ferrooxidans was not the main Acidimicrobium species in the nickel extraction at 49°C. Although only one species of Acidimicrobium has been named, the existence of several others has been indicated from cloned rRNA gene sequences originating in acidic mine drainage and hot springs (4, 6, 12, 13). The novel “Acidimicrobium” species 2 is assumed to be an iron oxidizer because it greatly outnumbered other potential iron oxidizers while ferrous iron oxidation in the reactors was extensive and because the phylogenetic analysis placed it in a cluster of iron-oxidizing strains. Acidimicrobium ferrooxidans was the only Acidimicrobium species obtained from the reactor cultures using ferrous iron-supplemented solid medium, but it is not unusual for some strains of iron-oxidizing acidophiles to form colonies readily on solid media while others do not. Growth and activity of Acidimicrobium ferrooxidans during iron oxidation (8) and mineral dissolution (20) appeared stronger in mixed cultures with other bacteria, and, although capable of autotrophy, this organism has been considered to be heterotrophically inclined (20). The isolation of the novel Acidimicrobium species in pure culture has not yet proved possible but is required for definition of the characteristics which provide a competitive advantage over Acidimicrobium ferrooxidans and S. thermosulfidooxidans during mixed-culture mineral oxidation processes at the optimum temperature for these moderate thermophiles.
Acknowledgments
This work was supported by a BBSRC studentship and by Billiton International Development Ltd.
Footnotes
Published ahead of print on 27 April 2007.
REFERENCES
- 1.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barns, S. M., R. E. Fundyga, M. W. Jeffries, and N. R. Pace. 1994. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc. Natl. Acad. Sci. USA 91:1609-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batty, J. D., and G. V. Rorke. 2005. Development and commercial demonstration of the BioCOP™ thermophile process, p. 153-161. In S. T. L. Harrison, D. E. Rawlings, and J. Petersen (ed.), Proceedings of the 16th International Biohydrometallurgy Symposium. University of Capetown, Capetown, Republic of South Africa.
- 4.Bond, P. L., S. P. Smriga, and J. F. Banfield. 2000. Phylogeny of microorganisms populating a thick, subaerial, predominantly lithotrophic biofilm at an extreme acid mine drainage site. Appl. Environ. Microbiol. 66:3842-3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brierley, J. A., and C. L. Brierley. 1986. Microbial mining using thermophilic microorganisms, p. 279-305. In T. D. Brock (ed.), Thermophiles: general, molecular and applied microbiology. Wiley, New York, NY.
- 6.Brofft, J. E., J. V. McArthur, and L. J. Shimkets. 2002. Recovery of novel bacterial diversity from a forested wetland impacted by reject coal. Environ. Microbiol. 4:764-769. [DOI] [PubMed] [Google Scholar]
- 7.Burton, N. P., and P. R. Norris. 2000. Microbiology of acidic, geothermal springs of Montserrat: environmental rDNA analysis. Extremophiles 4:315-320. [DOI] [PubMed] [Google Scholar]
- 8.Clark, D. A., and P. R. Norris. 1996. Acidimicrobium ferrooxidans gen. nov. sp. nov.: mixed culture ferrous iron oxidation with Sulfobacillus species. Microbiology 141:785-790. [DOI] [PubMed] [Google Scholar]
- 9.Cleaver, A. A. 2000. Mineral sulphide oxidation, mixed cultures in bioreactors. Ph.D thesis. The University of Warwick, Warwick, United Kingdom.
- 10.Dew, D. W., C. van Buuren, K. McEwan, and C. Bowker. 1999. Bioleaching of base metal sulphide concentrates: a comparison of mesophile and thermophile bacterial cultures, p. 229-238. In R. Amils and A. Ballester (ed.), Biohydrometallurgy and the environment toward the mining of the 21st century, part A. Elsevier, Amsterdam, The Netherlands.
- 11.Dopson, M., and E. B. Lindström. 2004. Analysis of community composition during moderately thermophilic bioleaching of pyrite, arsenical pyrite, and chalcopyrite. Microb. Ecol. 48:19-28. [DOI] [PubMed] [Google Scholar]
- 12.Inskeep, W. P., R. E. Macur, G. Harrison, B. C. Bostick, and S. Fendorf. 2004. Biomineralization of As(V)-hydrous ferric hydroxide in microbial mats of an acid-sulfate-chloride geothermal spring, Yellowstone National Park. Geochim. Cosmochim. Acta 68:3141-3151. [Google Scholar]
- 13.Johnson, D. B., N. Okibe, and F. F. Roberto. 2003. Novel thermo-acidophilic bacteria isolated from geothermal sites in Yellowstone National Park: physiological and phylogenetic characteristics. Arch. Microbiol. 180:60-68. [DOI] [PubMed] [Google Scholar]
- 14.Marsh, R. M., and P. R. Norris. 1983. Mineral sulfide oxidation by moderately thermophilic acidophilic bacteria. Biotechnol. Lett. 5:585-590. [Google Scholar]
- 15.Miller, P. C. 1997. The design and operating practice of bacterial oxidation plant using moderate thermophiles (the BacTech Process), p. 81-102. In D. E. Rawlings (ed.), Biomining. Springer-Verlag, Berlin, Germany.
- 16.Norris, P. R. 1989. Factors affecting bacterial mineral oxidation: the example of carbon dioxide in the context of bacterial diversity, p. 3-14. In J. Salley, R. G. L. McCready, and P. L. Wichlacz (ed.), Biohydrometallurgy 1989. CANMET, Ontario, Canada.
- 17.Norris, P. R. 1997. Thermophiles and bioleaching, p. 247-258. In D. E. Rawlings (ed.), Biomining. Springer-Verlag, Berlin, Germany.
- 18.Norris, P. R. 2007. Acidophile diversity in mineral sulfide oxidation, p. 199-216. In D. E. Rawlings and D. B Johnson (ed.), Biomining. Springer-Verlag, Berlin, Germany.
- 19.Norris, P. R., D. A. Clark, J. P. Owen, and S. Waterhouse. 1996. Characteristics of Sulfobacillus acidophilus sp. nov. and other moderately thermophilic mineral-sulphide-oxidizing bacteria. Microbiology 142:775-783. [DOI] [PubMed] [Google Scholar]
- 20.Okibe, N., and D. B. Johnson. 2004. Biooxidation of pyrite by defined mixed cultures of moderately thermophilic acidophiles in pH-controlled bioreactors: significance of microbial interactions. Biotechnol. Bioeng. 87:574-583. [DOI] [PubMed] [Google Scholar]
- 21.Okibe, N., M. Gericke, K. B. Hallberg, and D. B. Johnson. 2003. Enumeration and characterization of acidophilic microorganisms isolated from a pilot plant stirred-tank bioleaching operation. Appl. Environ. Microbiol. 69:1936-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rawlings, D. E., D. Dew, and C. du Plessis. 2003. Biomineralization of metal-containing ores and concentrates. Trends Biotechnol. 21:38-44. [DOI] [PubMed] [Google Scholar]