Abstract
Identification and classification of Vibrio species have relied upon band pattern methods (e.g., amplified fragment length polymorphism) and DNA-DNA hybridization. However, data generated by these methods cannot be used to build an online electronic taxonomy. In order to overcome these limitations, we developed the first standard multilocus sequence scheme focused on the ubiquitous and pathogenic Vibrio harveyi species group (i.e., V. harveyi, V. campbellii, V. rotiferianus, and a new as yet unnamed species). We examined a collection of 104 isolates from different geographical regions and hosts using segments of seven housekeeping genes. These two species formed separated clusters on the basis of topA, pyrH, ftsZ, and mreB gene sequences. The phylogenetic picture obtained by the other three loci, i.e., gyrB, recA, and gapA, was more complex though. V. campbellii appeared nested within V. harveyi in the recA trees, whereas V. harveyi formed a tight nested cluster within V. campbellii by gapA. The gyrB gene had no taxonomic resolution and grouped the two species together. The fuzziness observed in these three genes seems not be related to recombination but to low divergence due to the accumulation of only a few substitutions. In spite of this, the concatenated sequences provided evidence that the two species form two separated clusters. These clusters did not arise by recombination but by accumulation of point mutations. V. harveyi and V. campbellii isolates can be readily identified through the open database resource developed in this study (http://www.taxvibrio.lncc.br/). We argue that the species should be defined by evolutionary criteria. Strains of the same species will share at least 95% concatenated sequence similarity using the seven loci, and, most importantly, cospecific strains will form cohesive readily recognizable phylogenetic clades.
The species Vibrio harveyi and V. campbellii are widespread in the marine environment and among the main species responsible for disease in many wild and reared aquatic organisms, most notably peneid shrimp, several fish species, and mollusks (2). Luminous vibrios related to V. harveyi have been implicated principally in disease outbreaks in shrimp larviculture facilities and in grow-out ponds worldwide. More recently V. harveyi has been associated with infections in corals (20). Indeed recent studies have shown that vibrios, including the V. harveyi group, are abundant in the mucus of corals and may cause infections during periods of environmental imbalances (15).
In the past, the identification of V. harveyi and related species isolated from the marine environment has been imprecise and represents hard work as it involves performing many biochemical and/or physiological tests (14). Presumptive V. harveyi isolates grow on thiosulfate-citrate-bile salts-sucrose agar are motile, ferment glucose, and are oxidase positive and sensitive to the vibriostatic agent 0/129 at 150 μg. They are arginine dihydrolase negative and lysine and ornithine decarboxylase positive. Most presumptive V. harveyi isolates are luminescent, utilize d-gluconate, l-glutamate, d-glucuronate, heptanoate, d-galactose, and sucrose, and grow at 40°C. These isolates do not utilize l-histidine and l-arabinose (1). Notably a number of diagnostic laboratories still rely on the presumptive identification of the V. harveyi species group by means of phenotypic tests (1). Because V. campbellii and V. harveyi share nearly 100% 16S rRNA gene sequence similarity and around 70% DNA-DNA hybridization (DDH) similarity (14), discriminating these species remains a hard task for taxonomy. Also, infections by phages may contribute to changing phenotypes in V. harveyi (19).
Examples are abundant in the literature illustrating the difficulties of correctly identifying strains of V. campbellii, V. harveyi, and V. rotiferianus and the related species V. alginolyticus, V. natriegens, and V. parahaemolyticus. These studies point out several instances of ambiguous identifications, possibly due to the limitations of the identification methods. An alternative explanation for the difficulty of identifying these strains is the plasticity of the vibrio genomes with hybridization events in the marine environment, leading to soft species boundaries (5). In a most detailed study using molecular fingerprinting methods and DDH, we showed that presumptive V. harveyi isolates, identified by phenotypic features, belonged in fact to the species V. campbellii (8). Molecular fingerprinting methods (e.g., amplified fragment length polymorphism and repetitive extragenic palindrome-PCR [rep-PCR]) and DDH are very reliable for species identification, but they are restricted to a few reference laboratories and, most importantly, are not readily available through the Internet to many end users of taxonomy. Thus, researchers still lack a standard taxonomic scheme for the identification and classification of the Vibrio harveyi species group. An alternative to these shortcomings is the development of an electronic online taxonomy based on segments (ca. 500 nucleotides [nt]) of housekeeping genes (17).
Here we report on the first standard multilocus sequence analysis (MLSA) scheme for the V. harveyi species group based on sequences of genes encoding recombination repair protein (recA), urydilate kinase (pyrH), glyceraldehyde 3-phosphate dehydrogenase (gapA), an actin-like cytoskeleton protein (mreB), a cell division protein (ftsZ), DNA gyrase B subunit (gyrB), and topoisomerase I (topA). This standard multilocus scheme has indeed a high resolution for resolving closely related vibrio species. We show that the widespread marine bacterium V. harveyi and its sister species, V. campbellii, form separated gene clusters and that the overall clusters are not fuzzy. We suggest an alternative species definition based on gene sequence similarity. In addition, we developed an online identification system and related strain database that will allow the identification of isolates through the Internet.
MATERIALS AND METHODS
Representative strains included in this study are deposited in the BCCM/LMG Bacteria Collection (http://bccm.belspo.be/about/lmg.php) and in the Collection of Aquatic Important Microorganisms (www.ciad.mx/caim), CIAD Mazatlan (Table 1). Isolates originated from a variety of sources, places, and dates, providing a broad representation of the distribution of Vibrio harveyi and V. campbellii (Table 1). Reference and type strains were selected on the basis of previous polyphasic taxonomic analyses using rep-PCR, amplified fragment length polymorphism, and DDH data in order to represent the overall known genomic diversity of Vibrio harveyi and V. campbellii. MLSA was performed essentially as described previously by Thompson et al. (18). Genomic bacterial DNA was prepared using the Promega Wizard DNA purification kit. Approximately 100 ng of DNA was used as a PCR template for amplification of the seven genetic loci: recA, pyrH, gapA, mreB, ftsZ, gyrB, and topA. PCR primers are listed elsewhere (T. Sawabe, K. Kita-Tsukamoto, and F. L. Thompson, submitted for publication). Overall, the primers yielded specific amplicons of 600 to 1,200 bp at 50°C. In a few cases the annealing temperature was adjusted to provide a specific amplification. The PCR products were analyzed on a 1.5% agarose gel with a molecular weight standard for quantification of the PCR yield. The PCR products which produced a single band on agarose gels were purified using Promega gel and PCR purification kits. DNA sequencing was done by Shimadzu Biotech sequencing service (Kyoto, Japan) using a RISA384 sequencer and a Megabase 4000 system (Amersham Biosciences).
TABLE 1.
Strain list
| Strain(s) (synonym[s]) | Isolation
|
Host/origin | |
|---|---|---|---|
| Place | Date | ||
| V. harveyi | |||
| Australia | |||
| 1977 (CRC D53), 1978 (CRC D52) | Tasmania | 1994 | Atlantic salmon (Salmo salar) |
| 1979 (99/0805-1-1), 1847 (99/0425-4) | Tasmania | 1999 | Abalone (Haliotis sp.) |
| 1000 (02/1034) | South Australia | 2002 | Abalone (Haliotis sp.) |
| 0436 (93/0793-1) | Tasmania | 1993 | Abalone (Haliotis sp.) |
| 0802 (00/0163-T5b) | Tasmania | 2000 | Abalone (Haliotis sp.) |
| Brazil | |||
| R-623 | Praia Portinho, São Sebastião | 2006 | Mucus of coral (Palithoa caribaeorum) |
| R-626 | Praia Preta, São Sebastião | 2006 | Mucus of coral (Mussismilia hispida) |
| Ecuador | |||
| R259, R264 | NDa | ND | Shrimp (Penaeus sp.) |
| Japan | |||
| LMG 19643, LMG 19714 | Shizuoka | 1995 | Diseased horse mackerel (Trachurus japonicus) |
| AS11, AS59, AS71, AS93, AS131, S20, S30, S35, S92, 913SDZ20, 818ODDZ2, 823DZ6 | Kumaishi | 2002 | Internal organs of dead abalone (Haliotis discus hannai) |
| 99WT11 | Kumaishi | 2003 | Seawater |
| 913BZ62 | Kumaishi | 2004 | Seawater tank of abalone (Haliotis discus hannai) |
| 720WT44 | Kumaishi | 2005 | Seawater before outbreak |
| 89SF200-3 | Kumaishi | 2005 | Sand-filtrated seawater before outbreak |
| 823BAZ5 | Kumaishi | 2005 | Seawater tank of abalone (Haliotis discus hannai) |
| 823WBZ7 | Kumaishi | 2005 | Seawater |
| 818ODEZ12 | Okushiri | 2005 | Gill of abalone (Haliotis discus hannai) |
| 823TEZ2, 823TEZ1, 823TDZ13 | Taisei | 2005 | Gill of moribund abalone (Haliotis discus hannai) |
| 913SDZ12, 913SDZ14, 913SDZ16, 913SDZ18 | Kumaishi | 2004 | Internal organs of dead abalone (Haliotis discus hannai) |
| 818ODDZ6, 818ODDZ8, 818ODDZ10, 818ODDZ14 | Okushiri | 2005 | Internal organs of abalone (Haliotis discus hannai) |
| 823DZ1, 823DZ2, 823DZ3, 823DZ5 | Kumaishi | 2005 | Internal organs of abalone (Haliotis discus hannai) |
| Mexico | |||
| CAIM 1 | Mazatlan, Sinaloa | 1998 | Seawater from shrimp (Litopenaeus stylirostris) broodstock tank |
| CAIM 2 | Mazatlan, Sinaloa | 1998 | Diseased shrimp nauplii (Litopenaeus stylirostris) |
| CAIM 79 | Gulf of Santa Clara, Sonora | 1999 | Shrimp (Litopenaeus stylirostris) hatching system |
| CAIM 107 | Rosario, Sinaloa | 1999 | Diseased shrimp (Penaeus sp.) hemolymph |
| CAIM 973 | Mazatlan, Sinaloa | 2004 | Snapper (Lutjanus guttatus) kidney |
| CAIM 1075 | Huatabampo, Sonora | 2003 | Oyster (Crassostrea gigas) |
| CAIM 1266 | Mazatlan, Sinaloa | 2004 | Snapper (Lutjanus guttatus) liver |
| CAIM 1159, CAIM 1173, CAIM 1333 | Mazatlan, Sinaloa | 2004 | Snapper (Lutjanus guttatus) spleen |
| CAIM 1508, CAIM 1511 | Mazatlan, Sinaloa | 2005 | Diseased puffer fish (Spheroides annulatus), external lesion |
| CAIM 1510 | Mazatlan, Sinaloa | 2005 | Diseased puffer fish (Spheroides annulatus), anus |
| CAIM 1614 | San Quintin Bay, Baja California | 2004 | Oyster (Crassostrea gigas) |
| CAIM 1761 | Mazatlan, Sinaloa | 2005 | Puffer fish (Spheroides annulatus), kidney |
| CAIM 1766 | Mazatlan Aquarium, Sinaloa | 2005 | Sea horse (Hipocampus ingens), liver |
| New Zealand | |||
| 0772 (99/1052), 1976 (99/0736-4) | ND | 1999 | Rock lobster (Jasus verreauxi) |
| Spain | |||
| R825, R826, R827, R828, R829, R830, R831 | Mediterranean coast | 2005 | Internal organs or ulcers of cultured gilthead sea bream (Sparus aurata) and European sea bass (Dicentrarchis labrax) |
| United States | |||
| LMG 4044T (CAIM 513T) | Massachusetts | 1936 | Dead plankton (Talorchestia sp.); type strain of species |
| LMG 7890 (CAIM 517) | Baltimore | 1982 | Dead brown shark (Carcharhiunus plumbeus) |
| V. campbellii | |||
| Brazil | |||
| R-603 | Praia Grande, São Sebastião | 2006 | Mucus of coral (Mussismilia hispida) |
| R-644 | Praia Portinho, São Sebastião | 2006 | Mucus of coral (Mussismilia hispida) |
| China | |||
| CAIM 392 (STD3-1002) | Feng Cheng | ND | Diseased protozoea stage 3 shrimp (Penaeus chinensis) larvae |
| Mexico | |||
| CAIM 4 | Bay of Santa Barbara, Sonora | 1998 | Seawater |
| CAIM 109 | Matatipac shrimp farm, Nayarit | 1999 | Shrimp (Litopenaeus sp.) hemolymph |
| CAIM 115 | Santa Rosalia shrimp farm | 1999 | Shrimp (Litopenaeus sp.) hemolymph |
| CAIM 134 | La Brecha shrimp farm, Guasave, Sinaloa | 2000 | Diseased shrimp (Litopenaeus sp.) hemolymph |
| CAIM 149, CAIM 150, CAIM 155 | Costa Azul shrimp farm | 1995 | Diseased shrimp (Litopenaeus sp.) hemolymph |
| CAIM 198 | Clementina shrimp farm, Sinaloa | 1999 | Shrimp (Litopenaeus sp.) hepatopancreas |
| CAIM 249 | Costa Azul shrimp farm | 1995 | Diseased shrimp (Litopenaeus sp.) hepatopancreas |
| CAIM 757 | Oyster culture, La Cruz, Sinaloa | 2003 | Oyster (Crassostrea gigas) |
| CAIM 1283, CAIM 1500 | Mazatlan, Sinaloa | 2004 | Snapper (Lutjanus guttatus) liver |
| CAIM 1558 | Oyster culture, La Cruz, Sinaloa | 2004 | Oyster (Crassostrea gigas) |
| CAIM 1074 | Oyster culture, Huatabampo, Sonora | 2003 | Oyster (Crassostrea gigas) |
| Philippines | |||
| CAIM 9 | Negros Island | ND | Juvenile shrimp (Penaeus monodon) hepatopancreas |
| CAIM 372 | Negros Island | ND | Lymphoid organ of a diseased shrimp (Penaeus monodon) juvenile |
| CAIM 401 | Iloilo | ND | Seawater |
| Spain | |||
| R300, R376 | Oyster culture, Bay of Alfacs, Catalonia | 2005 | Oyster (Crassostrea gigas) |
| R633 | Oyster culture, Bay of Fangar, Catalonia | 2005 | Oyster (Crassostrea gigas) |
| United States | |||
| LMG 11216T (CAIM 519T) | Hawaii | 1971 | Seawater; type strain of species |
| Thailand | |||
| CAIM 3 | ND | ND | Diseased shrimp (Penaeus monodon) |
| V. rotiferianus | |||
| Belgium | |||
| LMG 21460T (CAIM 577T) | Artemia Reference Center | 1999 | Rotifer (Brachionus plicantilis) flowthrough system; type strain of species |
| Brazil | |||
| R-601, R-646 | Praia Portinho, São Sebastião | 2006 | Mucus of coral (Mussismilia hispida) |
| Mexico | |||
| CAIM 994* | Mazatlan, Sinaloa | 2004 | Snapper (Lutjanus guttatus) kidney |
| 1975 (99/0736-1a) | ND | 1999 | Snapper (Pagrus auratus) |
ND, not determined.
The sequences were aligned using ClustalX, and phylogenetic trees were constructed by the neighbor-joining (16) and maximum-parsimony methods (MEGA version 3.0; close neighbor interchange). The robustness of each topology was checked by 500 bootstrap replications. Trees were drawn by using MEGA version 3.0 (11).
Recombination in each gene was analyzed by SplitsTree version 4 (9); the phi statistical test (3); and the suite of tests including MaxChi2, SIscan, Bootscan, and Sawyers that are implemented in Recombination Detection Program version 2.0 (13). The GC content, index of association (Ia), and ratio of mean synonymous substitutions per synonymous site/mean nonsynonymous substitutions per nonsynonymous site (ds/dn) were calculated using the software package START, which was obtained from http://pubmlst.org/software/analysis/start/ (10). The Ia was also evaluated by the online tool Lian3.5 (http://pubmlst.org/perl/mlstanalyse/mlstanalyse.pl?site=pubmlst&page=lian&referer=pubmlst.org).
Nucleotide sequence accession numbers.
The gene sequences determined in this study are deposited in the GenBank under the accession no. EF596024 to EF596735. The sequence data are also available at our website, http://www.taxvibrio.lncc.br/.
RESULTS
The genetic loci chosen in this study are typical housekeeping genes, as is evident from the very low dn/ds values (Table 2). Little gene sequence variation was observed in ftsZ, gapA, and pyrH. The topA, pyrH, ftsZ, and mreB trees provided clear-cut discrimination of the species V. harveyi and V. campbellii, with strains falling into quite tight clusters (see Fig. S1 in the supplementary material). These clusters shared mutual gene sequence similarities of 94 to 96% (except for topA, which was 90%), while strains within each species cluster had less than 1.5% gene sequence variation, with most of the substitutions located in the third position of each codon. The topA gene has, thus, the highest resolution, followed by mreB, ftsZ, and pyrH, for both species and strain differentiation. For instance, V. harveyi LMG 19643, LMG 19714, and 823WBZ7 were identical in all loci except in topA, where 823WBZ7 accumulated three silent point mutations.
TABLE 2.
Summary of gene featuresa
| Gene | Length (nt) | dn/ds | % GC | Nucleotide diversity/site (π) | No. of alleles | Recombination (P) |
|---|---|---|---|---|---|---|
| ftsZ | 445 | 0.010 | 47.7 ± 0.6 | 0.027 | 22 | 0.03 |
| topA | 583 | 0.023 | 48.7 ± 0.1 | 0.058 | 49 | 0.009 |
| pyrH | 360 | 0.000 | 48.5 ± 0.3 | 0.027 | 22 | 0.04 |
| mreB | 507 | 0.000 | 49.1 ± 0.4 | 0.033 | 44 | 0.6 |
| recA | 498 | 0.002 | 46.2 ± 0.3 | 0.028 | 56 | 0.08 |
| gyrB | 596 | 0.000 | 46.7 ± 0.6 | 0.04 | 41 | 0.001 |
| gapA | 607 | 0.045 | 46.4 ± 0.2 | 0.01 | 20 | 0.12 |
Phi recombination statistics were used (3).
On the other hand, the recA, gyrB, and gapA trees revealed a somewhat more complex grouping. Vibrio campbellii formed a cluster nested within V. harveyi on the basis of recA gene sequences, with strains sharing at least 94% similarity, whereas V. harveyi appeared in a tight nested cluster (99.5% sequence similarity) within V. campbellii by gapA. V. campbellii was more heterogeneous in the gapA gene, with strains grouping as low as 97.7% gene sequence similarity. These two species seem to be indistinguishable by gyrB sequences though. Among these three genes, recA has the highest resolution for differentiating isolates at the strain and species levels. Overall, V. harveyi strains seemed more homogeneous in all seven loci examined than V. campbellii strains, indicating some sort of unknown environmental selection more effectively acting on V. harveyi. For instance, R-623 and R-626, both isolated from cnidarians in Brazil, were nearly identical to the Japan strain 823DZ5 and to the Ecuador strain R264, respectively. On the other hand, every V. campbellii strain produced a new sequence type, defined by the number of alleles found in each locus (our unpublished data). The gene with the lowest discriminatory power was gyrB, for which V. campbellii CAIM 1173, CAIM 757, R300, CAIM 249, CAIM 134, CAIM 1073, CAIM 401, CAIM 372, CAIM 109, CAIM 115, and R633 had up to 99.5% sequence similarity with the type strain of V. harveyi and several reference strains. Indeed Vibrio harveyi and V. campbellii isolates were rather mixed on the basis of gyrB.
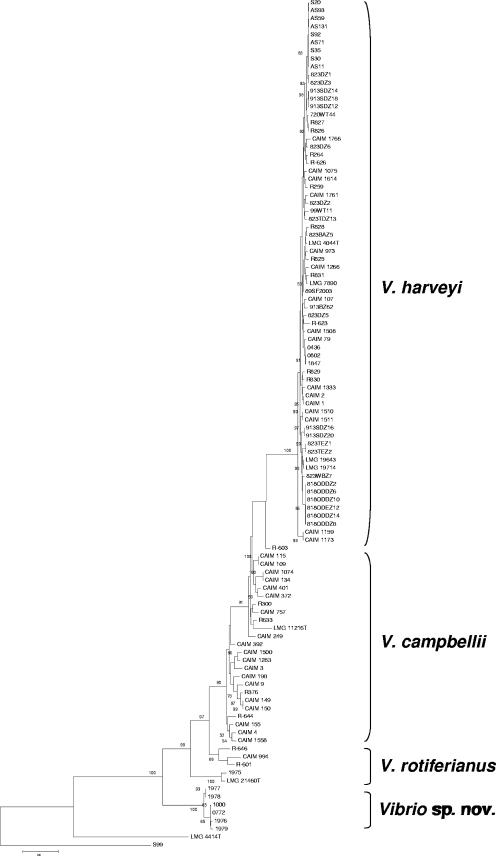
V. harveyi and V. campbellii formed separated phylogenetic groups when a tree was constructed using concatenated sequences (Fig. 1). This consensus tree also suggests that V. harveyi and V. campbellii form monophyletic groups, possibly with V. harveyi evolving from V. campbellii. This finding supports the notion that vibrios form species and that species are not fuzzy in nature. The analysis of concatenated sequences also revealed some interesting groups of Vibrio harveyi. For instance, the strains 823DZ1, 823DZ3, 913SDZ12, AS11, AS59, AS71, AS93, AS131, S20, S30, and S92 represent a persistent clone, observed from 2002 until 2005, associated with mass mortalities of abalones in Japan. We also disclosed a widespread clone present in Spain and Japan, comprising 720WT44, 913SDZ12, 913SDZ14, 913SDZ18, R826, and R827. Another interesting aspect of the genetic diversity of vibrios relates to the fact that often in environmental surveys some strains appear in intermediate (taxonomic) positions and thus may be hard to identify on the basis of single genes. For instance, strain CAIM 994 had different species type strains as its closest neighbor; it showed 100% ftsZ and 98.4% gapA sequence similarity to V. harveyi LMG 4044T; 96.5% mreB and 97% pyrH sequence similarity to V. campbellii LMG 11216T; and 97% recA, 95.5% gyrB, and 91% topA sequence similarity to V. rotiferianus LMG 21460T. Overall, the groups obtained by gene sequence data revealed a complete agreement with the groups obtained in previous studies using rep-PCR and DDH (7, 8) (see Fig. S2 in the supplemental material; http://www.taxvibrio.lncc.br/).
FIG. 1.
Consensus maximum-parsimony phylogenetic tree based on the concatenated gene sequences of the seven loci (3,596 nt). Values of bootstrap after 500 repetitions are shown at the nodes. Tree length, 2,762. Scale bar, 100 substitutions. Vibrio fischeri LMG 4414T and Photobacterium profundum SS9 were used as out-groups.
DISCUSSION
DDH has a number of limitations, but it is clear that this tool has been instrumental to prokaryotic taxonomy so far, in that it established cohesive, stable taxonomic groups, based upon their overall genomic similarity. DDH data have underpinned also the concepts for interpreting environmental microbiological sequence data such as those for the 16S rRNA gene. However prokaryotic taxonomy is in flux in the last years due to the new avenues of research opened up by whole-genome sequencing and the development of multilocus sequence typing (6). Here we found complete agreement between the DDH values and grouping obtained in previous studies (7, 8) and the gene sequence similarity values and grouping observed in the V. harveyi species group (Fig. 1). Strains 1000, 0772, and 1976 to 1979 form a clearly separated phylogenetic clade. These strains belong to a new species related to V. harveyi (J. Carson et al., unpublished data). These strains form a homogeneous group on the basis of DDH, having around 50% DDH similarity to V. harveyi, V. campbellii, and V. rotiferianus. It is particularly informative and reassuring to confirm that groups based on DDH correlate well with gene sequence data shown here. It is evidence of the power of MLSA as an alternative to DDH for defining the boundaries of species. V. harveyi and V. campbellii species can be defined as strains sharing around 95% gene sequence similarity in their concatenated loci. This threshold should not be taken strictly though, because it may vary as new isolates are analyzed. Eventually the significant phylogenetic clades, obtained by different tree-building methods, will equate species and will lead to an alternative bacterial species definition based on evolutionary relationships.
Much has been argued in the literature for the role of recombination in species diversification (4). We analyzed several features of our gene sequences in order to evaluate possible cases of horizontal gene transfer (HGT) that would lead to hybrid, intermediate strains, e.g., CAIM 994. The GC content of the genes is within the range observed in vibrios, suggesting no signs of HGT. The GC content alone is not a good predictor of gene transfer, as genes may undergo the process of amelioration (12). For this reason, we also analyzed the linkage disequilibrium within and between species on the basis of allelic profiles. The observed values of variance (2.00 for V. harveyi and 1.10 for V. campbellii) were always higher than the expected variance (0.96 for V. harveyi and 0.52 for V. campbellii) if the strains were experiencing free recombination. The phi recombination test (3) did point to recombination though. Recombination might have occurred in several loci, including gyrB, ftsZ, topA, and pyrH, with a high statistical significance level according to this test (Table 2), but we should look at these analyses with the greatest caution. The phi recombination test takes polymorphic sites in an alignment with opposing incongruent phylogenetic signals as breakpoints of recombination, but this assumption may lead to false positives, possibly explaining the results found here. The nucleotide substitution patterns observed in this study suggest that recombination is not the main evolutionary force leading to speciation in V. harveyi and V. campbellii. Although we cannot rule out the effect of recombination in the reassortment of genomic sequences in the V. harveyi group, the fuzziness in the gyrB, gapA, and recA trees, our data show, is due to slower molecular clocks, not to recombination. Possibly in these loci there occurred a smaller number of accumulated nucleotide substitutions, leading to a close relationship between species.
The use of MLSA has far-reaching beneficial consequences in environmental microbiology. First, researchers will be able to readily identify their isolates through the Internet using a common tool that is highly reliable taxonomically. Second, this type of portable data will allow for testing new concepts and standards for culture-dependent and -independent studies, particularly concerning the definition and diversification of species. We developed a new online identification scheme that will allow the end users of the taxonomy of vibrios to promptly identify their isolates (http://www.taxvibrio.lncc.br/). V. harveyi and V. campbellii are very intriguing animal pathogens. Apparently they are both able to cause disease in a wide range of aquatic organisms by complex mechanisms involving quorum sensing, toxin (hemolysin) production, and biofilm formation. The present study shows the usefulness of a simple electronic tool for comparing vibrio strains on the molecular level which will certainly be useful for pinpointing the widespread successful clones associated with animal health worldwide.
Supplementary Material
Acknowledgments
F.L.T. acknowledges young researcher grant FAPESP (no. 2004/00814-9). This work was supported by the Japan Society for the Promotion of Science, Institute of Fermentation of Osaka, Goho Life Science Foundation (Japan), and the FOSEMARNAT-2004-01-33 project (Mexico).
We thank Jeremy Carlson and Maria de Jesus Pujalte for providing some strains. We also thank Renato Camara da Silva, Vicente de Araujo Calfo, Yasuhiro Tsuruya, and Youhei Fukui for their excellent technical assistance. We thank Jean Swings for fruitful discussions. We appreciate the comments of the two referees.
Footnotes
Published ahead of print on 4 May 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alsina, M., and A. R. Blanch. 1994. A set of keys for biochemical identification of environmental Vibrio species. J. Appl. Bacteriol. 76:79-85. [DOI] [PubMed] [Google Scholar]
- 2.Austin, B., and D. A. Austin. 1999. Bacterial fish pathogens: disease of farmed and wild fish, 3rd ed. Springer, Berlin, Germany.
- 3.Bruen, T. C., H. Philippe, and D. Bryant. 2006. A simple and robust statistical test for detecting the presence of recombination. Genetics 172:2665-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doolittle, W. F., and R. T. Papke. 2006. Genomics and the bacterial species problem. Genome Biol. 29:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fraser, C., W. P. Hanage, and B. G. Spratt. 2007. Recombination and the nature of bacterial speciation. Science 315:476-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gevers, D., F. M. Cohan, J. G. Lawrence, B. G. Spratt, T. Coenye, E. J. Feil, E. Stackebrandt, Y. Van de Peer, P. Vandamme, F. L. Thompson, and J. Swings. 2005. Opinion: re-evaluating prokaryotic species. Nat. Rev. Microbiol. 3:733-739. [DOI] [PubMed] [Google Scholar]
- 7.Gomez-Gil, B., F. L. Thompson, C. C. Thompson, and J. Swings. 2003. Vibrio rotiferianus sp. nov., isolated from cultures of the rotifer Brachionus plicatilis. Int. J. Syst. Evol. Microbiol. 53:239-245. [DOI] [PubMed] [Google Scholar]
- 8.Gomez-Gil, B., S. Soto-Rodríguez, A. García-Gasca, A. Roque, R. Vazquez-Juarez, F. L. Thompson, and J. Swings. 2004. Molecular identification of Vibrio harveyi-related isolates associated with diseased aquatic organisms. Microbiology 150:1769-1777. [DOI] [PubMed] [Google Scholar]
- 9.Huson, D. H. 1998. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics 14:68-73. [DOI] [PubMed] [Google Scholar]
- 10.Jolley, K. A., E. J. Feil, M. S. Chan, and M. C. Maiden. 2001. Sequence type analysis and recombinational tests (START). Bioinformatics 17:1230-1231. [DOI] [PubMed] [Google Scholar]
- 11.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 12.Lawrence, J. G., and H. Ochman. 1997. Amelioration of bacterial genomes: rates of change and exchange. J. Mol. Evol. 44:383-397. [DOI] [PubMed] [Google Scholar]
- 13.Martin, D. P., C. Williamson, and D. Posada. 2005. RDP2: recombination detection and analysis from sequence alignments. Bioinformatics 21:260-262. [DOI] [PubMed] [Google Scholar]
- 14.Owens, L., and M. Busico-Salcedo. 2006. Vibrio harveyi: pretty problems in paradise, p. 266-280. In F. L. Thompson, B. Austin, and J. Swings (ed.), The biology of vibrios. ASM Press, Washington, DC.
- 15.Rosenberg, E., O. Koren, L. Reshef, R. Efrony, and I. Zilber-Rosenberg. 2007. The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 5:355-362. [DOI] [PubMed] [Google Scholar]
- 16.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 17.Thompson, F. L., and J. Swings. 2006. Taxonomy of vibrios, p. 29-43. In F. L. Thompson, B. Austin, and J. Swings (ed.), The biology of vibrios. ASM Press, Washington, DC.
- 18.Thompson, F. L., D. Gevers, C. C. Thompson, P. Dawyndt, S. Naser, B. Hoste, C. B. Munn, and J. Swings. 2005. Phylogeny and molecular identification of vibrios on the basis of multilocus sequence analysis. Appl. Environ. Microbiol. 71:5107-5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vidgen, M., J. Carson, M. Higgins, and L. Owens. 2006. Changes to the phenotypic profile of Vibrio harveyi when infected with the Vibrio harveyi myovirus-like (VHML) bacteriophage. J. Appl. Microbiol. 100:481-487. [DOI] [PubMed] [Google Scholar]
- 20.Weil, E., G. Smith, and D. L. Gil-Agudelo. 2006. Status and progress in coral reef disease research. Dis. Aquat. Organ. 69:1-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.