Abstract
The quantification of denitrifying bacteria is a component in the further understanding of denitrification processes in the environment. Real-time PCR primers were designed to target two segments of the denitrifier population (cnorBP [Pseudomonas mandelii and closely related strains] and cnorBB [Bosea, Bradyrhizobium, and Ensifer spp.]) in agricultural soils based on functional cnorB (nitric oxide reductase) gene sequences. Total population numbers were measured using 16S rRNA gene real-time PCR. Two soil microcosm experiments were conducted. Experiment 1 examined the response of the indigenous soil microbial population to the addition of 500 mg/kg glucose-C daily over 7 days in soil microcosms. Changes in the total population were correlated (r = 0.83) between 16S rRNA gene copy numbers and microbial biomass carbon estimates. Members of the cnorBP population of denitrifiers showed typical r-strategy by being able to increase their proportion in the total population from starting levels of <0.1% to around 2.4% after a daily addition of 500 mg/kg glucose-C. The cnorBB guild was not able to increase its relative percentage of the total population in response to the addition of glucose-C, instead increasing copy numbers only in proportion with the total population measured by 16S rRNA genes. Experiment 2 measured population dynamics in soil after the addition of various amounts of glucose-C (0 to 500 mg/kg) and incubation under denitrifying conditions. cnorBP populations increased proportionally with the amount of glucose-C added (from 0 to 500 mg/kg). In soil microcosms, denitrification rates, respiration, and cnorBP population densities increased significantly with increasing rates of glucose addition. cnorBB guild densities did not increase significantly under denitrifying conditions in response to increasing C additions.
Nitrogen oxides (nitric oxide [NO] and nitrous oxide [N2O]) are involved in ozone layer depletion and global warming (17). Nitrous oxide has a global warming potential 296 times higher than that of carbon dioxide and is increasing in concentration in the atmosphere due to anthropogenic emissions (17). Most anthropogenic N2O emissions are associated with agricultural production (8). For 2004, the most recent year for which data are available for Canada, about 64% of N2O emissions were derived from agriculture (8). Increased rates of mineral nitrogen fertilizers in crop production systems are an important contributor to N2O emissions (4, 13). Denitrification and nitrification are biological processes that have been implicated in N2O emissions from agricultural soils; however, denitrification has been postulated as the dominant process leading to N2O emissions in agricultural soils (1), particularly under high-soil-moisture conditions (23).
Denitrifying bacteria are widespread in the soil environment and belong to diverse genera (6). Denitrification in bacteria consists of four steps for the reduction of NO3− to N2, catalyzed by nitrate, nitrite, nitric oxide, and nitrous oxide reductases. Denitrification functional genes have been isolated from cultured bacteria, and gene fragments have been amplified from cultured and environmental samples using a range of primers (5, 6, 12, 30). Studies of denitrifying bacteria tend to focus on the steps from nitrite reductase onward, because nitrate reductase is not always linked to complete denitrification but may be related to dissimilatory nitrate reduction to ammonia (27). Broad-range and specific primer sets for real-time quantification of denitrifiers have been developed based on the nitrite reductase genes nirS and nirK (11, 14) and, most recently, the nitrous oxide reductase gene nosZ (15). Nitric oxide reductases are responsible for the conversion of NO to N2O (35). Nitric oxide reductase has two forms in bacteria, cNOR (cytochrome c electron donor) and qNOR (quinol electron donor) (6). cNOR is most commonly associated with denitrifier populations, with qNOR found in some denitrifiers but also in nondenitrifying microorganisms with a detoxification function against NO (6).
Although several researchers have quantified denitrifier populations in soil samples and other environments (11, 14, 18), studies linking analysis of the denitrifying population abundance and actual denitrification rates have yet to be performed. Real-time PCR provides a simple, rapid method for quantifying target genes in complex samples, such as soil and other environmental matrices. The abundance of Pseudomonas stutzeri nirS was quantified using real-time PCR in a range of environmental sample matrices, including groundwater and agricultural soil (11). Broad-range primers to amplify nirK gene fragments from soil samples have been developed previously (14). Henry et al. reported that carbon addition increased the denitrifier population in soil as quantified using the nirK gene; however, the conditions under which this occurred are not well defined and the increased population was not linked to a measure of denitrification activity. Broad-range primers for analysis of the nosZ gene have also been developed (15) and used to quantify these targets and a selection of other denitrification genes using real-time PCR in a range of soil samples.
Literature reviews have identified the need to link the structure, abundance, and function of denitrifying populations to actual denitrification rates to determine the influence of the microbial population in this fundamental process (28, 29). The aims of this study were (i) to develop and validate primers for real-rime PCR quantification based on cnorB genes from cultured, soil-derived denitrifiers and (ii) to use the primers to quantify changes in denitrifier population densities under different carbon addition treatments, chosen to induce different levels of denitrification activity. Denitrification rates were monitored to determine whether population densities were related to actual denitrification in soil microcosms.
MATERIALS AND METHODS
Bacterial cultures and soil samples.
Control denitrifier and nondenitrifier cultures (Table 1) were maintained on appropriate solid media (tryptic soy agar [Sigma-Aldrich, Oakville, Ontario, Canada], nutrient agar [Difco, BD Biosciences, Mississauga, Ontario, Canada], or yeast extract-mannitol agar [YMA medium; DSMZ]) and stored long term in 15% (vol/vol) glycerol at −80°C. Field soil denitrifier isolates (Table 1) (7) were maintained on nutrient agar at 25°C and stored at −80°C as above. Soil for the microcosm experiments described below was sampled between June and July 2006 from a field cropped to spring wheat at the Potato Research Centre, Agriculture and Agri-Food Canada (Fredericton, New Brunswick, Canada) (45°52′N, 66°31′W). Soil was sampled from bulk soil from a depth of 0 to 15 cm. Soil was sieved (<4 mm) and used within 1 to 2 days of sampling in all cases. Soil was stored field moist at 4°C prior to use. The soil texture was determined (hydrometer method) to be 375 g/kg sand, 503 g/kg silt, and 121 g/kg clay. Soil concentrations of organic carbon and total nitrogen (combustion) were 29.3 g/kg and 1.61 g/kg, respectively. The soil pH, determined using a 1:1 water extract, was 7.6.
TABLE 1.
Specificity of PCR amplification of cnorB genes from control denitrifier and field isolatesa
| Strain | Denitrification | cnorBP | cnorBB | Source |
|---|---|---|---|---|
| Azoarcus tolulyticus ATCC 51758 | + | − | − | ATCCb |
| Ochrobactrum anthropi ATCC 49687 | + | − | − | Oxoidc |
| Pseudomonas stutzeri ATCC 17588 | + | − | − | ATCC |
| Pseudomonas sp. strain G-179 | + | − | − | UBCd |
| Achromobacter cycloclastes ATCC 21921 | + | − | − | ATCC |
| Paracoccus denitrificans ATCC 17741 | + | − | − | ATCC |
| Bacillus azotoformans ATCC 29788 | + | − | − | ATCC |
| Paracoccus denitrificans ATCC 19367 | + | − | − | ATCC |
| Paracoccus pantotrophus DSM65 | + | − | − | DSMZe |
| Pseudomonas stutzeri JM300 | + | − | − | UBC |
| Thauera aromatica DSM14793 | + | − | − | DSMZ |
| Pseudomonas stutzeri ATCC 14405 | + | − | − | ATCC |
| Bradyrhizobium japonicum USDA110 | + | − | + | USDA-ARSf |
| Alcaligenes faecalis subsp. faecalis ATCC 8750 | + | − | − | Oxoid |
| Cupriavidus necator DSM428 | + | − | − | DSMZ |
| Escherichia coli ATCC 29425 | − | − | − | ATCC |
| Alcaligenes faecalis ATCC 35655 | − | − | − | Oxoid |
| Enterobacter cloacae ATCC 23355 | − | − | − | Oxoid |
| Ochrobactrum anthropi ATCC 49187 | − | − | − | ATCC |
| Pseudomonas migulae PD1 | + | − | − | PRCg |
| Pseudomonas mandelii PD2 | + | + | − | PRC |
| Pseudomonas sp. strain PD3 | + | − | − | PRC |
| Pseudomonas brassicacearum PD4 | + | − | − | PRC |
| Pseudomonas brassicacearum PD5 | + | − | − | PRC |
| Pseudomonas sp. strain PD6 | + | + | − | PRC |
| Achromobacter sp. strain PD7 | + | − | − | PRC |
| Pseudomonas mandelii PD8 | + | + | − | PRC |
| Pseudomonas grimontii PD9 | + | − | − | PRC |
| Pseudomonas grimontii PD10 | + | − | − | PRC |
| Pseudomonas lini PD11 | + | − | − | PRC |
| Sinorhizobium sp. strain PD12 | + | − | − | PRC |
| Pseudomonas mandelii PD13 | + | + | − | PRC |
| Pseudomonas sp. strain PD14 | + | − | − | PRC |
| Pseudomonas lini PD15 | + | − | − | PRC |
| Pseudomonas sp. strain PD16 | + | − | − | PRC |
| Pseudomonas migulae PD17 | + | + | − | PRC |
| Bosea sp. strain PD18 | + | − | + | PRC |
| Bosea sp. strain PD19 | + | − | + | PRC |
| Achromobacter sp. strain PD20 | + | − | − | PRC |
| Pseudomonas sp. strain PD21 | + | + | − | PRC |
| Pseudomonas sp. strain PD22 | + | + | − | PRC |
| Bosea sp. strain PD23 | + | − | + | PRC |
| Bosea sp. strain PD24 | + | − | + | PRC |
| Achromobacter sp. strain PD25 | + | − | − | PRC |
| Pseudomonas sp. strain PD26 | + | − | − | PRC |
| Achromobacter sp. strain PD27 | + | − | − | PRC |
| Pseudomonas lini PD28 | + | − | − | PRC |
| Ensifer adhaerens PD29 | + | − | + | PRC |
| Pseudomonas mandelii PD30 | + | + | − | PRC |
| Pseudomonas kilonensis PD31 | + | − | − | PRC |
+, visible band of expected size; −, no visible band; ATCC, American Type Culture Collection (Manassas, Virginia); Oxoid, Oxoid Quality Control organisms (Nepean, Ontario, Canada); UBC, University of British Columbia (Vancouver, British Columbia, Canada); DSMZ, Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (Braunschweig, Germany); USDA-ARS, United States Department of Agriculture-Agricultural Research Service (Beltsville, MD); PRC, Potato Research Centre (Fredericton, New Brunswick, Canada).
Design and testing of cnorB-targeted PCR primers.
cnorB gene fragments were amplified and sequenced from a collection of denitrifiers isolated from potato field soils as previously described (7). Isolated denitrifier cnorB gene sequences were aligned using ClustalW, along with closely related database (GenBank) sequences. Two groups were identified for primer design, cnorBP (mostly Pseudomonas mandelii) and cnorBB (Bosea-Bradyrhizobium-Ensifer spp.). Consensus sequences were exported to Primer Express, version 2.0.0 (Applied Biosystems Canada, Streetsville, Ontario, Canada), and primers were selected based on standard conditions for real-time PCR as recommended by Applied Biosystems (Table 2). Primer sequences were compared to those in GenBank using BlastN to confirm specificity compared to database cnorB genes and other sequences. Primer specificity was tested against genomic DNA from a range of cultured denitrifiers and field isolates. Soil DNA was also used as a template for PCRs, and specific PCR products were cloned into PCR cloning vectors (pGEM-T [Promega, Madison, WI] or pCR2.1 TOPO [Invitrogen, Burlington, Ontario, Canada]), sequenced, and compared to denitrifier and database cnorB target sequences. GenBank accession numbers of representative target cnorB genes are DQ420251 (P. mandelii PD30 cnorB) and EF507803 (Bosea sp. PD18 cnorB).
TABLE 2.
Primers/probes and conditions used for real-time PCR amplifications
| Primer/probe | Sequence (5′-3′) | Positiona | Concn (nM) | Reference |
|---|---|---|---|---|
| 1369F | CGG TGA ATA CGT TCY CGG | 1369-1386 | 900 | 33 |
| 1492R | GGW TAC CTT GTT ACG ACT | 1475-1492 | 900 | 33 |
| TM1389Fb | 6-FAM-CTT GTA CAC ACC GCC CGT C-TAMRAc | 1389-1407 | 300 | 33 |
| cnorBPF | CAT GGC GCT GAT AAC GGG | 738-755 | 900 | This study |
| cnorBPR | CTT IAC CAT GCT GAA GGC G | 870-888 | 900 | This study |
| cnorBBF | AIG TGG TCG AGA AGT GGC TCT A | 653-674 | 900 | This study |
| cnorBBR | TCT GIA CGG TGA AGA TCA CC | 810-829 | 900 | This study |
Numbering refers to the Escherichia coli sequence for 16S rRNA gene primers, the Pseudomonas fluorescens cnorB sequence (nucleotides 828 to 2255 of AF197467) for cnorBP primers, and the Bradyrhizobium japonicum sequence (nucleotides 757 to 2115 of AJ132911) for cnorBB primers.
The TaqMan detection system was used for 16S rRNA only.
FAM, 6-carboxyfluorescein; TAMRA, carboxytetramethylrhodamine.
Real-time PCR assay.
Real-time PCR products were amplified on an Applied Biosystems (Streetsville, Ontario, Canada) ABI PRISM 7000 sequence detection system using SYBR green (cnorB primer sets) or TaqMan (16S rRNA gene primer set [33]) probe-based detection. Real-time PCR for cnorB was performed in 25-μl reaction mixtures containing the following components: 12.5 μl 2× SYBR green PCR master mix (containing AmpliTaq Gold DNA polymerase, uracil-N-glycosylase, deoxynucleotide triphosphates with UTP, passive reference dye, and optimized buffer; Applied Biosystems), primers at optimized concentrations (Table 2), and template DNA (5 μl of 100-fold dilutions of soil nucleic acid extracts and 5 μl of 10-fold dilutions of plasmid DNA for standard curves). 16S rRNA gene amplifications were carried out using 2× TaqMan Universal PCR master mix (Applied Biosystems), primers and probe (31) at optimized concentrations, and template DNA as described above. Reaction mixtures using soil DNA as a template also included T4 gene 32 protein to relieve inhibition by soil inhibitory components also obtained in nucleic acid extracts (250 ng per reaction; New England Biolabs, Pickering, Ontario, Canada) (15, 18). T4 gene 32 protein was not included in reaction mixtures used to generate standard curves, as it had no effect on reaction efficiency when purified plasmid DNA was used as a template (data not shown). The TaqMan probe for 16S rRNA gene PCR was obtained from Integrated DNA Technologies (Coralville, IA). Dual-labeled oligonucleotide probe was resuspended in Tris-EDTA buffer, aliquoted, and stored at −20°C in the dark. Primer and probe concentrations were optimized for each reaction individually according to Applied Biosystems guidelines (Table 2). Amplification conditions for cnorB primer sets were as follows: 1 cycle of 95°C for 10 min and then 40 cycles of 95°C for 15 s, 60°C for 30 s, and 80°C for 30 s. Fluorescence data were acquired at 80°C to remove signals that may have been obtained from nonspecific products or primer dimers. Melting temperatures of the specific products were 82 to 83°C (cnorBP) and 84 to 85°C (cnorBB). Amplification conditions for the 16S rRNA gene primer/probe set were as described in reference 31. Soil nucleic acid extracts were tested for the presence of coextracted inhibitory substances by comparing the copy numbers obtained in 10-fold dilutions using primers for 16S rRNA. The template concentrations used from soil nucleic acid extracts were not inhibitory to the real-time PCR (data not shown).
Standard curves for DNA quantitation.
External standard curves were generated for each of the three target sequences, cnorBP, cnorBB, and 16S rRNA genes. cnorB PCR products obtained by amplification with primers cnorB2F and cnorB7R (6) were cloned into pGEM-T according to the manufacturer's instructions (Promega). 16S rRNA genes from a representative denitrifier strain were amplified using primers 27f and 1492r (20) and similarly cloned. Plasmid DNAs were extracted using plasmid Mini or Midi kits (QIAGEN, Inc., Mississauga, Ontario, Canada) and quantified using the fluorescent dye PicoGreen according to the manufacturer's specifications (Invitrogen). Prior to use in standard curves, plasmids were restriction digested with appropriate enzymes (single cut to linearize the vector) and heat treated to inactivate enzymes. The sizes of PCR product inserts used for the generation of standard curves were confirmed by sequencing. Copy numbers of plasmid standards were calculated directly from the concentration and length (base pairs) of the extracted plasmid DNA. Standard curves were generated for each target sequence, showing the relationship between cnorB or 16S rRNA gene copy numbers and threshold cycle values. Standard curves were run on each 96-well plate used for real-time PCR. Where possible, all samples from an experiment were run on a single plate; however, when this was not possible, replicate calibrator samples were run on each plate to adjust values obtained. Copy numbers of target sequences in unknown soil DNA extracts were determined from standard curves. 16S rRNA and cnorB copy numbers were not corrected for DNA extraction efficiency from soil.
Pure culture and soil DNA extraction procedures.
Genomic DNA was extracted from overnight cultures of denitrifiers by using the UltraClean microbial DNA kit according to the manufacturer's instructions (MO BIO Laboratories, Inc., Carlsbad, CA). Soil DNA was extracted using a modified procedure based on those in references 10 and 21. In brief, 0.25-g portions of freeze-dried soils were weighed into 2-ml screw-cap tubes containing 0.1 g each of washed and sterile 0.1-mm-diameter, 0.2- to 0.3-mm-diameter, and 0.7- to 1.2-mm-diameter-glass beads plus one 2.5-mm-diameter glass bead (Sigma-Aldrich Canada Ltd., Oakville, Ontario, Canada). Modified hexadecyltrimethylammonium bromide extraction buffer (0.5 ml; equal volumes of 10% [wt/vol] hexadecyltrimethylammonium bromide σ in 0.7 M NaCl and 240 mM potassium phosphate buffer, pH 8.0, plus 2.5 mg.ml−1 aurintricarboxylic acid σ), aluminum ammonium sulfate (50 μl of 200 mM filter-sterilized solution), and phenol-chloroform-isoamyl alcohol (0.5 ml; 25:24:1) were added to tubes and mixed thoroughly. Tubes were shaken at 20 strokes/s for 10 min (MO BIO 96-well plate shaker with tube adapter set) to lyse cells and then centrifuged (16,000 × g, 10 min, 4°C) before removing the aqueous phase to a new tube. The aqueous phase was reextracted with chloroform-isoamyl alcohol (24:1) and centrifuged as above, and the aqueous phase was removed to a fresh tube. Total nucleic acids were precipitated by the addition of 2 volumes 30% (wt/vol) polyethylene glycol 6000 (Fluka BioChemika)-1.6 M NaCl for 2 h at room temperature and then centrifuged (18,000 × g, 30 min, 4°C). Pelleted nucleic acids were washed in ice-cold 70% (vol/vol) ethanol and air dried prior to resuspension in Tris-EDTA buffer (pH 8.0). Extracted DNA was visualized by agarose (0.8% [wt/vol]) gel electrophoresis. DNA was quantified spectrofluorometrically using the fluorescent dye PicoGreen (Invitrogen).
Detection limits and extraction efficiency.
Denitrifying isolates (Bosea sp. strain PD18 and Pseudomonas sp. strain PD21) were inoculated into sterile and nonsterile soil, and DNA was then extracted to determine the extraction efficiencies and detection limits of pure cultures. Different numbers of cells (approximately 108, 106, 104, and 102 cells/g dry weight soil) were added into the soil samples, and dilution plate counts were used to enumerate the added cell populations. Cell numbers inoculated were compared to copy numbers generated in real-time PCR assays. Gene copy numbers were calculated to be equivalent to cell numbers, as cnorB has only ever been found in single copy in the bacterial genome (35).
Soil microcosm experiments. (i) Experiment 1.
A preliminary experiment was conducted to determine whether real-time PCR primers for denitrifier populations and total bacterial population (16S rRNA genes) were capable of detecting differences in these populations in soil over time and to compare this method with biochemical methods for microbial biomass measurements. The experiment manipulated population densities by the addition of large amounts of readily available carbon to soil (500 mg glucose-C/kg soil per day). Fresh field soil (250 g) was placed in 1-liter glass jars and maintained at a moisture content of 29% (wt/wt) ± 1% for the duration of the experiment by the addition of nutrient or control treatments in distilled water (dH2O). Two nutrient treatments were used, a daily addition of 500 mg/kg glucose-C plus 100 mg/kg nitrate-N and a control with no nutrient addition (dH2O instead of nutrient solutions). Each treatment was manually stirred daily during additions of nutrients/water. Stock solutions of glucose and nitrate (in the form of KNO3) were prepared in dH2O for the addition of nutrients to soil microcosms. Sufficient jars were used to allow four replicates of each treatment to be sacrificed at 0, 1, 2, 3, 5, and 7 days. Soil from each jar was analyzed for denitrifier and total bacterial populations by real-time PCR methods as described above and by microbial biomass carbon as described below.
(ii) Experiment 2.
In contrast to the above experiment, where the primary objective was to evaluate the molecular measurement techniques over time in soil microcosms, this experiment was designed to evaluate the population dynamics of total bacteria and components of the denitrifier population under denitrifying conditions and to compare to actual denitrification rates when differing rates of glucose-C were applied. Soil was sampled as above and stored at 4°C prior to use. Soil was repacked into cores as previously described (9), and soil moisture was adjusted to 70% (wt/wt) water-filled pore space to ensure denitrifying conditions (3). Treatments were additions of 0, 125, 250, and 500 mg/kg of glucose-C. Glucose treatment levels were chosen to reflect amounts of available C equivalent to field application levels (9). Nitrate (KNO3-N) was added to all microcosms at 500 mg/kg to ensure nonlimiting concentrations for the duration of the experiment. Sufficient jars were used to allow four replicates of each treatment to be sacrificed at 0 and 2 days. The soil cores were placed in 1-liter glass jars and sealed, and the atmosphere was supplemented with acetylene to a final concentration of 10% (vol/vol). Acetylene blocks the reduction of N2O to N2. Soil treatments were prepared at 4°C and remained at this temperature overnight to allow for the diffusion of acetylene into the soil cores. Jars were then incubated at 25°C in the dark for up to 2 days. Time zero represents the time at which the treatments were transferred to incubation at 25°C. Samples were analyzed for DNA extraction, real-time PCR, and biochemical measurements as described for experiment 1.
Biochemical and analytical methods.
Microbial biomass carbon was measured using the CHCl3 fumigation-extraction method (34). Fumigated and nonfumigated soil samples (25 g) were extracted using 50 ml 0.5 M K2SO4 and analyzed for extractable organic carbon. Microbial biomass carbon was calculated using a factor of 0.35 (34). Nitrate was determined from K2SO4 extracts of nonfumigated samples. Segmented flow analysis (Technicon Industrial Systems, Tarrytown, MA) was used for the colorimetric determination of extractable organic carbon (Technicon Method 455-76 W/A) and NO3− concentrations (Technicon Method 100-70W). Gas analysis was performed for N2O and CO2 production by using a Varian Star 3800 gas chromatograph (Varian, Walnut Creek, CA) fitted with an electron capture detector and a Combi PAL autosampler (CTC Analytics, Zwingen, Switzerland). The electron capture detector was operated at 300°C with 90% Ar, 10% CH4 carrier gas at 20 ml/min in a HayeSep N 80/100 precolumn (0.32-cm diameter by 50-cm length) and HayeSep D 80/100 mesh analytical columns (0.32-cm diameter by 200-cm length) in a column oven operated at 70°C. The precolumn was used in combination with a four-port valve to remove water from samples.
Data analysis.
Analysis of variance was performed using the general linear model of SAS (version 8; SAS Institute Inc., Cary, NC). All nonnormal data were log transformed. Based on a factorial design, means comparisons were performed using the LSMeans test, although if the interaction of the factors showed no significant difference, treatment means were compared using single-degree-of-freedom contrasts. Treatment means and standard errors presented in the tables and figures are calculated from untransformed data. Significance was accepted at a level of probability (P) of <0.05.
RESULTS
PCR primer design and specificity of amplification.
Real-time PCR primers (Table 2) were designed using nitric oxide reductase (cnorB) sequences obtained from field-isolated denitrifiers (7). Nitric oxide reductase is a central enzyme in the denitrification process, but the genes encoding this enzyme have been the focus of fewer studies about the environment. Database sequences for full-length and environmental clones of nirS, nirK, and nosZ number in the thousands, whereas there are less than 400 full or partial cnorB sequences currently in GenBank. In our set of denitrifying field isolates, cNOR was the more common version of the enzyme and the cnorB sequence data set for these isolates was complete (unlike nirS/nirK, for which not all isolates generated a PCR amplicon with the primers tested) (7). DNA from cultured denitrifying strains was used to test the specificity of the cnorB primers. Specific PCR products of the expected size were obtained with each of the PCR primer sets tested (Table 1). When soil DNA was used as a template, a single band of the correct size and approximate melting temperature (determined from dissociation curve) was obtained for both primer sets. The specificity of amplification from soil DNA was confirmed by the cloning and sequencing of random PCR products obtained from soil (data not shown). In all cases tested, products obtained showed high sequence identity (95 to 98% for cnorBP to cnorB from PD21 and 86 to 90% for cnorBB to cnorB from PD18; GenBank accession numbers EF23568 to EF23585). Alignments of the cnorBP soil PCR products showed very little sequence difference between all clones, due to the strain-specific primer design used in this case, whereas soil clones obtained using cnorBB primers showed more variation. cnorBB primers are able to amplify cnorB DNA from the Bosea, Bradyrhizobium, and Ensifer guild and possibly other closely related genera, which would lead to more variability in the fragments obtained from a mixed soil DNA sample.
Detection limits, extraction efficiency, and standard curves.
Nonsterile soil contained background numbers of cnorBP and cnorBB denitrifiers at 1.7 × 105 and 8.3 × 104 copy numbers/g soil, respectively (Table 3). Freshly grown Pseudomonas or Bosea pure cultures inoculated into this nonsterile soil at 106 and 108 cells/g soil could be detected above background levels (Table 3). Inoculated Pseudomonas cells were extracted and detectable at the levels at which they were inoculated (e.g., 108 gene copies were detected when ∼108 cells were added, or at close to 100% extraction efficiency) (Table 3). Bosea cells were not detected at the levels at which they were added into soil (∼10 to 15% extraction efficiency of inoculated cells), indicating difficulties with cell lysis/DNA extraction from these cells in soil (Table 3). Modifications to the DNA extraction method (including size and amount of glass beads, time of bead beating, volume of soil, and buffers) to improve cell lysis were evaluated, but no improvements could be made to increase the extraction efficiency of these cells. Copy numbers are therefore likely to be underestimated when the cnorBB guild is analyzed in native soils. Autoclaved soil was also tested with inoculated cells, but background levels of amplifiable signal were obtained (data not shown). What the source of amplifiable signal was in those soil DNA extracts is not known. Soils were autoclaved three times over 3 consecutive days with incubation in between to remove viable bacteria, but it is possible that DNA from the background population of denitrifiers was still present in autoclaved soil and was subsequently extracted and amplified. Previous work (26) showed a background copy number of ∼103 for the detection of nahAc in autoclaved soil.
TABLE 3.
Detection of inoculated denitrifier cells in nonsterile soila
| Treatment | Observed copy numbers (real-time PCR) | No. of inoculated cells |
|---|---|---|
| P. mandelii | 1.2 × 106 ± 8.3 × 105 | 1.2 × 106 ± 2.6 × 105 |
| P. mandelii | 2.8 × 108 ± 9.1 × 106 | 1.2 × 108 ± 2.6 × 107 |
| Bosea sp. | 9.2 × 105 ± 1.0 × 105 | 5.3 × 106 ± 6.9 × 105 |
| Bosea sp. | 6.2 × 107 ± 1.2 × 107 | 5.3 × 108 ± 6.9 × 107 |
| Uninoculated (cnorBP) | 1.7 × 105 ± 6.2 × 104 | None |
| Uninoculated (cnorBB) | 8.3 × 104 ± 4.2 × 104 | None |
Values presented are per gram (dry weight) of soil and are means ± standard deviations of three replicates.
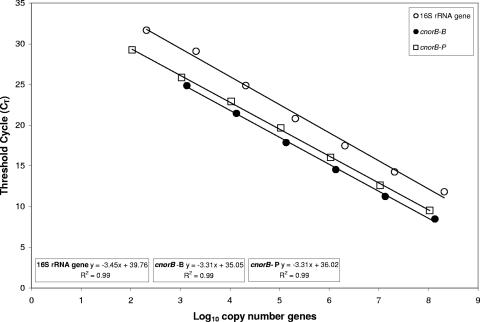
Standard curves for target quantification were prepared from plasmid clones of target cnorB genes. Standard curves were linear over 5 to 6 log dilutions and sensitive down to approximately 100 copies for the two cnorB PCRs and 1,000 copies for 16S rRNA genes (Fig. 1). Although the lowest sensitivity of detection of 16S rRNA genes was higher than that for the cnorB primer sets, other authors have described the difficulty of completely removing all background 16S rRNA gene signal from PCR reagents (31). For 16S rRNA gene quantification in soil, copy numbers were high (108 to 1010) and likely significantly removed from the no-template-control (NTC) signal and, as such, can be adequately quantified. Problems may be encountered if low copy numbers are to be quantified in cases where the signal from the sample may not be adequately removed from the NTC. The detection limit obtained in these experiments was sufficient for the quantification of 16S rRNA gene copy numbers in the soils and conditions tested. NTC reactions generally did not record any signal (threshold cycle of >40) in real-time PCRs conducted with these primer sets, which is equivalent to copy numbers of 2.4 × 103 (16S), 4.7 × 102 (cnorBP), and 5 × 102 (cnorBB) per g dry weight soil. Copy numbers of the denitrification genes were assumed to be equivalent to actual cell numbers based on the premise that the majority of known denitrifiers have a single copy of the denitrification pathway in their genomes (28). It is difficult to relate copy numbers of 16S rRNA genes to actual bacterial cell numbers due to the variable copy numbers (1-15) of this gene in bacterial genomes (19).
FIG. 1.
Standard curves generated from restriction-digested plasmid standards for each of three primer sets, cnorBP, cnorBB, and 16S rRNA genes. Values represent means (n = 3) ± standard errors (error bars are too small to be seen).
Soil microcosm experiments. (i) Experiment 1.
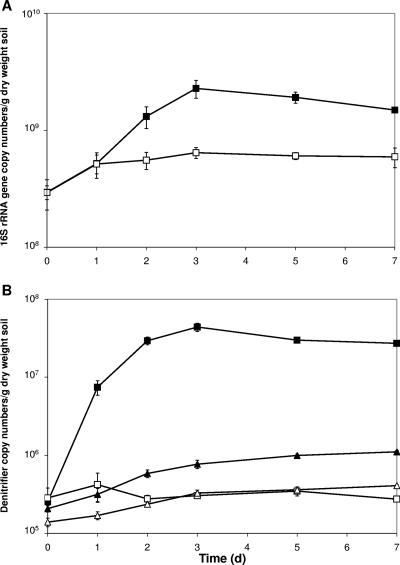
Total bacterial populations, measured using the 16S rRNA gene target population, significantly increased between day 0 and day 1 in both control and carbon-treated microcosms (Fig. 2A). This response reflects more favorable conditions for bacterial growth as a result of the wetting of soil and incubation at a higher temperature (i.e., 25°C during incubation compared with 4°C during storage). The total bacterial population in the control microcosms did not change significantly between day 1 and 7, with an average value of 6 × 108 copy numbers/g soil. In comparison, the total bacterial population in the carbon-amended microcosms increased significantly (P < 0.05) between day 1 and day 3, reaching a maximum value of 2.28 × 109 copy numbers/g soil, and then subsequently decreased over time to a value of 1.49 × 109 copy numbers/g soil at day 7.
FIG. 2.
Copy numbers of (A) total bacterial population (16S rRNA genes) and (B) P. mandelii (cnorBP) (squares) and Bosea-Bradyrhizobium-Ensifer (cnorBB) (triangles) denitrifier populations as measured by real-time PCR after the addition of excess glucose-C to aerobic soil microcosms. Solid symbols indicate the addition of glucose, and open symbols indicate controls (no glucose addition). Values are means (n = 4) ± standard errors (error bars).
The P. mandelii (cnorBP) denitrifier population increased significantly (P < 0.05) in the glucose-C treatment, from a starting population of 2.54 × 105 copy numbers/g soil to a maximum of 4.41 × 107 copy numbers/g soil by day 3 (Fig. 2B). In the control microcosms, cnorBP population did not change over time, with an average value of 3 × 105 copy numbers/g soil at 7 days. The cnorBP population in the carbon-amended microcosms increased its percentage of the total population (measured as a percentage of the total 16S rRNA gene copy numbers obtained) from a starting level of <0.1% to a maximum of 2.4%, whereas in the control microcosms, the cnorBP population remained at <0.1% for the duration of the incubation period.
The growth response of the Bosea-Bradyrhizobium-Ensifer (cnorBB) guild to the addition of glucose-C was different from that of the cnorBP population (Fig. 2B). cnorBB copy numbers in the glucose-treated microcosm increased from 2.1 × 105 copy numbers/g soil at day 0 to a maximum of 1.1 × 106 copy numbers/g soil at 7 days, whereas cnorBB copy numbers in the control microcosms increased from 1.39 × 105 at day 0 to a maximum of 4.1 × 105 copy numbers/g soil at day 7. In contrast to the cnorBP population, the cnorBB guild never increased its relative proportion (expressed as a percentage of total 16S rRNA gene copy numbers) above 0.1% in either the glucose-treated or the control microcosms (data not shown).
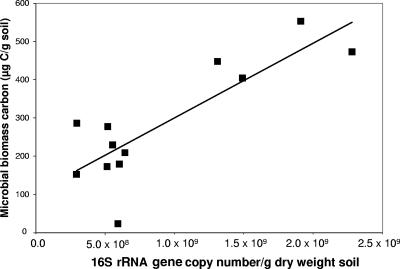
There was a positive correlation (r = +0.83) between the molecular method (16S rRNA gene real-time PCR) for estimating total bacterial population and a biochemical measure (microbial biomass carbon) of microbial biomass in the soil microcosms (Fig. 3). This indicates that the changes in total bacterial population measured using the molecular method are reflected in the biochemical measurement of microbial biomass carbon. It is recognized that the two measures are not equivalent, however, because microbial biomass carbon measures the total microbial biomass in soil, including bacterial, archaeal, and fungal components, whereas the 16S rRNA gene PCR targets only bacterial ribosomal genes. Previous studies examined the relationship between several measures of the microbial population in soil (such as levels of phospholipid fatty acid, substrate-induced respiration, and total DNA extracted [2, 22]), but to the best of our knowledge, this is the first example of this relationship between quantitative measures of 16S rRNA genes and microbial biomass carbon measurements.
FIG. 3.
Relationship between microbial biomass carbon and 16S rRNA gene copy number in soil microcosms. Values are taken from all microcosms at all time points sampled. The line of best fit indicates the linear relationship described by the following equation: y = 1.95 × 10−7x + 105 (r2 = 0.69).
(ii) Experiment 2.
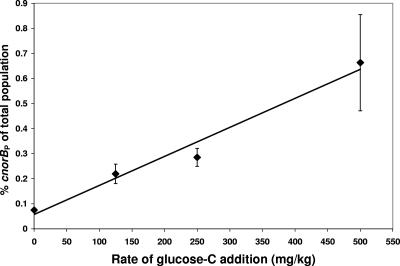
Populations of total bacteria and denitrifiers (i.e., cnorBP and cnorBB) did not change significantly for the 0-mg/kg glucose-C treatment between 0 and 2 days (Table 4). Populations of total bacteria and Bosea-Bradyrhizobium-Ensifer (cnorBB) guild denitrifiers did not change significantly in response to increasing additions of glucose-C. In contrast, the population of P. mandelii (cnorBP) denitrifiers significantly (P < 0.05) increased in size in proportion to the amount of glucose-C added to soil microcosms. Increasing the additions of glucose-C also led to increased denitrification (as measured by total N2O evolved in acetylene-treated microcosms) and increased respiration (as measured by CO2 evolution) at 2 days. There was a positive relationship (r2 = 0.97) between glucose addition and cnorBP population numbers measured at 2 days (Fig. 4). Denitrification (measured by production of N2O) occurred in all treatments with the addition of nitrate and carbon. Nitrate concentrations decreased significantly in carbon-treated samples compared to that in the control soil without added carbon (data not shown), consistent with denitrification.
TABLE 4.
Denitrifier and total bacterial copy numbers and cumulative N2O-N and CO2-C emissions in response to glucose-C additions to soil microcosms
| Treatment | Treatment time (days) | No. of gene copies g−1 of soila
|
Cumulative emissions (mg/kg soil)
|
|||
|---|---|---|---|---|---|---|
| cnorBP | cnorBB | 16S rRNA genes | N2O-N evolved | CO2-C evolved | ||
| Control | 0 | 1.80 × 105 Ab | 1.88 × 105 A | 2.75 × 108 A | 0.1 A | 5.0 A |
| Control | 2 | 1.72 × 105 A | 9.87 × 104 A | 2.29 × 108 A | 3.2 B | 37.7 B |
| Glucose, 125 mg/kg | 2 | 4.95 × 105 B | 1.76 × 105 A | 2.44 × 108 A | 12.6 C | 87.4 C |
| Glucose, 250 mg/kg | 2 | 7.56 × 105 B | 2.56 × 105 A | 2.79 × 108 A | 19.5 D | 146.0 D |
| Glucose, 500 mg/kg | 2 | 1.87 × 106 C | 2.80 × 105 A | 3.21 × 108 A | 32.1 E | 274.2 E |
Standard errors (four replicates for each sample measurement) are 2.76 × 105 for cnorBP, 3.61 × 104 for cnorBB, 4.02 × 107 for 16S rRNA genes, 1.38 for N2O-N, and 4.31 for CO2-C.
The same letter is placed next to values that are not significantly different (P > 0.05).
FIG. 4.
Relationship between rate of glucose-C added (mg/kg) to soil microcosms and the size of the cnorBP population as a percentage of total population measured by 16S gene copy number at time 2 days. The line of best fit indicates the linear relationship described by the following equation: y = 0.00116x + 0.0570. Values are means (n = 4) ± standard errors (error bars).
DISCUSSION
Methods for quantitation of functional guilds in the environment are of interest in our efforts to understand functional diversity and the effects of environmental factors on population densities and activities. The quantitation and analysis of denitrifying bacteria are of particular interest because of the inherent difficulties in analyzing this phylogenetically and genetically diverse functional community (28). Quantitative methods such as real-time PCR allow simple, rapid processing of large numbers of samples, which could not previously be easily processed with culture-based methods (24). Quantitative methods allow the measurement of indigenous soil populations at multiple time points and with multiple treatments, enabling more sophisticated experimental designs to be implemented.
The nitric oxide reductase gene (cnorB) was used as a marker for the denitrification populations targeted in this study. Denitrification gene sequences were previously amplified and sequenced from a culture collection of denitrifiers isolated from potato field soil (7). cnorB was present in a majority of the denitrifiers isolated, and we were able to target specific bacterial denitrifier groups (Pseudomonas mandelii-like strains and the Bosea-Bradyrhizobium-Ensifer guild) using this gene. The primer sets developed were determined to be specific and sensitive enough for the detection of changes in denitrifying bacterial populations in soil microcosms. The sensitivity obtained was similar to that in a previous study (15), where a detection limit of 10 to 100 target molecules per assay was achieved, equivalent to 103 to 104/g dry soil for NTC samples. The use of a third step (80°C) in the PCR cycle was implemented with our cnorB primer sets. The fluorescence data acquired avoided any signal from primer dimers or lower-melting-temperature nonspecific products (15); however, in most cases this was simply a precaution, as few nonspecific products were observed.
The populations analyzed in these experiments were small components of the total bacterial population and small components of the total (as opposed to culturable) denitrifier population, but they are found in a range of soil samples obtained from our potato field sites (7). The response of these populations to denitrifying conditions provides knowledge of denitrifier population dynamics in this process in a natural soil. Depending on the availability and composition of nutrients and habitat in soil, the soil bacterial community may comprise different proportions of r- and K-strategists (32). The P. mandelii (cnorBP) population showed a classical r-strategy response, with rapid growth in response to the addition of glucose, a readily available carbon source. This population increased its relative proportion in the total population, indicating the ability of this population to successfully compete for available resources. In contrast, the Bosea-Bradyrhizobium-Ensifer (cnorBB) guild did not increase in response to glucose addition, indicating a different growth strategy (K-strategist) under these conditions. Glucose is often used as a substrate in soil studies since the majority of soil microorganisms can metabolize it (32). The response to other organic substrates, such as crop residues, may be substantially different for the denitrifying populations identified here.
Although real-time PCR provides a simple rapid method for the quantification of bacterial populations in soil, the values obtained may not be accurate or “absolute” for a number of reasons. The total population measured here was estimated using 16S rRNA gene primers and a TaqMan probe. The use of a probe and the limitations of primer design, while required for PCR specificity, may lead to an underestimation of the total population of bacteria if the probe and primer do not bind to all possible amplified target regions (33). The estimated abundances of denitrifier populations may not equal the true abundance of these groups in soil due to differences and biases in extraction efficiency. We showed that freshly grown Bosea cells inoculated into soil were not extracted efficiently. Whether this problem would also apply to native populations of this bacterium is unknown, but it is likely that extraction bias significantly affects the absolute numbers of targets extracted and subsequently amplified in real-time PCR. Although absolute numbers may not be achievable, gross differences and changes in population size are still detectable. The differences observed between the two denitrifier populations studied are then real differences in the responses of these populations to the conditions tested.
Denitrification was measured and observed to increase in response to increasing glucose additions, along with soil respiration. This increase in denitrification rate may result from the growth and/or revival (25) of denitrifier population (and hence increase in abundance of active denitrifiers), leading to an overall increase in the denitrification rate in soils, or from the growth of the total population, leading to increased respiration, lowered oxygen levels, and subsequent induction of denitrification genes in the denitrifier population. We have shown differences in the growth responses of two groups of the denitrifier population to the addition of an available carbon source. Although we measured overall denitrification activity, the specific contribution of each denitrifier population to denitrification was not measured. mRNA analysis (real-time reverse transcription-PCR) is the next step towards understanding the different factors that influence not only population density but also functional gene activity in response to conditions that influence denitrification and nitrous oxide emissions in soils.
The targeting of specific populations provided knowledge of the responses of subpopulations of denitrifiers to changing conditions and provides insights into factors that contribute to denitrifier population density in the field. An analysis of larger components of the denitrifier population, through the use of broad-range primer pairs for denitrification genes as described in references 14, 15, and 18, would provide a better overall understanding of the relationships of denitrifier communities to denitrifying conditions and carbon amendments. Absolute quantification of total denitrifier populations is still not technically feasible using current techniques due to the diversity of gene sequences for functional genes and to the diversity of phylogenetic genes (i.e., 16S rRNA genes) from which denitrifiers are obtained. Recent work (16) has highlighted the limitations of current broad-range primer sets for nitrite reductase genes (nirK and nirS) in cultured and uncultured denitrifiers. Two primer sets developed for nosZ, while both amplifying a broad range of target genes in soil samples, amplified different sets of sequences, verified when PCR products were sequenced (15). Primers have been developed based on a few conserved full-length genes and are unlikely to cover the entire diversity of denitrifiers in any given environment.
This study demonstrates the feasibility of using a molecular approach to understanding the effects of different treatments on the dynamics of denitrifier populations in incubation experiments. Such approaches should be applicable under field conditions. The application of broad-range primer sets as well as the more specific primer sets developed in this study will allow a comparison of the overall denitrifier population dynamics in response to environmental inputs as well as some of the internal dynamics of denitrifier population turnover in a mixed soil population.
Acknowledgments
We acknowledge Drucie Janes, Jason Dalziel, and Jan Zeng for technical assistance and J. Neufeld for providing denitrifier cultures from the University of British Columbia.
Funding for this project was supplied by the GAPS program of Agriculture and Agri-Food Canada and an NSERC (Canada) team strategic grant. M.N.M. was the recipient of an NSERC postgraduate scholarship.
Footnotes
Published ahead of print on 20 April 2007.
REFERENCES
- 1.Aulakh, M. S., D. A. Rennie, and E. A. Paul. 1984. Gaseous nitrogen losses from soils under zero tillage as compared with conventional tilled system. J. Environ. Qual. 13:130-136. [Google Scholar]
- 2.Bailey, V. L., A. D. Peacock, J. L. Smith, and H. Bolton, Jr. 2002. Relationships between soil microbial biomass determined by chloroform fumigation-extraction, substrate-induced respiration, and phospholipids fatty acid analysis. Soil Biol. Biochem. 34:1385-1389. [Google Scholar]
- 3.Bateman, E. J., and E. M. Baggs. 2005. Contributions of nitrification and denitrification to N2O emissions at different water-filled pore space. Biol. Fertil. Soils 41:379-388. [Google Scholar]
- 4.Bouwman, A. F. 1996. Direct emission of nitrous oxide from agricultural soils. Nutr. Cycl. Agroecosyst. 46:53-70. [Google Scholar]
- 5.Braker, G., A. Fesefeldt, and K.-P. Witzel. 1998. Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl. Environ. Microbiol. 64:3769-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braker, G., and J. M. Tiedje. 2003. Nitric oxide reductase (norB) genes from pure cultures and environmental samples. Appl. Environ. Microbiol. 69:3476-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dandie, C. E., D. L. Burton, B. J. Zebarth, J. T. Trevors, and C. Goyer. 2007. Analysis of denitrification genes and comparison of nosZ, cnorB and 16S rDNA from culturable denitrifying bacteria in potato cropping systems. Syst. Appl. Microbiol. 30:128-138. [DOI] [PubMed] [Google Scholar]
- 8.Environment Canada. 2006. National Inventory Report, 1990-2004—greenhouse gas sources and sinks in Canada. Greenhouse Gas Division, Environment Canada, Gatineau, Quebec, Canada.
- 9.Gillam, K. M. 2006. Factors controlling the amount and partitioning of gaseous nitrogen losses from denitrification in an agricultural soil, p. 27. M.S. thesis. Dalhousie University and Nova Scotia Agricultural College, Halifax, Nova Scotia, Canada.
- 10.Griffiths, R. I., A. S. Whiteley, A. G. O'Donnell, and M. J. Bailey. 2000. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 66:5488-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grüntzig, V., S. C. Nold, J. Zhou, and J. M. Tiedje. 2001. Pseudomonas stutzeri nitrite reductase gene abundance in environmental samples measured by real-time PCR. Appl. Environ. Microbiol. 67:760-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hallin, S., and P.-E. Lindgren. 1999. PCR detection of genes encoding nitrite reductase in denitrifying bacteria. Appl. Environ. Microbiol. 65:1652-1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Helgason, B. L., H. H. Janzen, M. H. Chantigny, C. F. Drury, B. H. Ellert, E. G. Gregorich, R. L. Lemke, E. Pattey, P. Rochette, and C. Wagner-Riddle. 2005. Toward improved coefficients for predicting direct N2O emissions from soil in Canadian agroecosystems. Nutr. Cycl. Agroecosyst. 72:87-99. [Google Scholar]
- 14.Henry, S., E. Baudoin, J. C. Lopez-Gutierrez, F. Martin-Laurent, A. Brauman, and L. Philippot. 2004. Quantification of denitrifying bacteria in soils by nirK gene targeted real-time PCR. J. Microbiol. Methods 59:327-335. [DOI] [PubMed] [Google Scholar]
- 15.Henry, S., D. Bru, B. Stres, S. Hallet, and L. Philippot. 2006. Quantitative detection of the nosZ gene, encoding nitrous oxide reductase, and comparison of the abundances of 16S rRNA, narG, nirK, and nosZ genes in soils. Appl. Environ. Microbiol. 72:5181-5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heylen, K., D. Gevers, B. Vanparys, L. Wittebolle, J. Geets, N. Boon, and P. de Vos. 2006. The incidence of nirS and nirK and their genetic heterogeneity in cultivated denitrifiers. Environ. Microbiol. 8:2012-2021. [DOI] [PubMed] [Google Scholar]
- 17.Houghton, J. T., Y. Ding, D. J. Griggs, M. Noguer, P. J. Van der Linden, X. Dai, K. Maskell, and C. A. Johnson (ed.). 2001. Climate change 2001: the scientific basis. Contribution of Working Group I to the third assessment report of the Intergovernmental Panel on Climate Change (IPCC). Cambridge University Press, Cambridge, United Kingdom.
- 18.Kandeler, E., K. Deiglmayr, D. Tscherko, D. Bru, and L. Philippot. 2006. Abundance of narG, nirK, and nosZ genes of denitrifying bacteria during primary successions of a glacier foreland. Appl. Environ. Microbiol. 72:5957-5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klappenbach, J. A., P. R. Saxman, J. R. Cole, and T. M. Schmidt. 2001. rrndb: the Ribosomal RNA Operon Copy Number Database. Nucleic Acids Res. 29:181-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, New York, NY.
- 21.Lerat, S., L. S. England, M. L. Vincent, K. P. Pauls, C. J. Swanton, J. N. Klironomos, and J. T. Trevors. 2005. Real-time polymerase chain reaction quantification of the transgenes for Roundup Ready corn and Roundup Ready soybean in soil samples. J. Agric. Food Chem. 53:1337-1342. [DOI] [PubMed] [Google Scholar]
- 22.Marstorp, H., X. Guan, and P. Gong. 2000. Relationship between dsDNA, chloroform labile C and ergosterol in soils of different organic matter contents and pH. Soil Biol. Biochem. 32:879-882. [Google Scholar]
- 23.Mathieu, O., C. Hénault, J. Lévêque, E. Baujard, M.-J. Milloux, and F. Andreux. 2006. Quantifying the contribution of nitrification and denitrification to the nitrous oxide flux using 15N tracers. Environ. Pollut. 144:933-940. [DOI] [PubMed] [Google Scholar]
- 24.Okano, Y., K. R. Hristova, C. M. Leutenegger, L. E. Jackson, R. F. Denison, B. Gebreyesus, D. Lebauer, and K. M. Scow. 2004. Application of real-time PCR to study effects of ammonium on population size of ammonia-oxidizing bacteria in soil. Appl. Environ. Microbiol. 70:1008-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oliver, J. D. 2005. The viable but nonculturable state in bacteria. J. Microbiol. 43:93-100. [PubMed] [Google Scholar]
- 26.Park, J.-W., and D. E. Crowley. 2006. Dynamic changes in nahAc gene copy numbers during degradation of naphthalene in PAH-contaminated soils. Appl. Microbiol. Biotechnol. 72:1322-1329. [DOI] [PubMed] [Google Scholar]
- 27.Philippot, L. 2005. Tracking nitrate reducers and denitrifiers in the environment. Biochem. Soc. Trans. 33:200-204. [DOI] [PubMed] [Google Scholar]
- 28.Philippot, L., and S. Hallin. 2005. Finding the missing link between diversity and activity using denitrifying bacteria as a model functional community. Curr. Opin. Microbiol. 8:234-239. [DOI] [PubMed] [Google Scholar]
- 29.Philippot, L. 2006. Use of functional genes to quantify denitrifiers in the environment. Biochem. Soc. Trans. 34:101-103. [DOI] [PubMed] [Google Scholar]
- 30.Scala, D. J., and L. Kerkhof. 1998. Nitrous oxide reductase (nosZ) gene-specific PCR primers for detection of denitrifiers and three nosZ genes from marine sediments. FEMS Microbiol. Lett. 162:61-68. [DOI] [PubMed] [Google Scholar]
- 31.Smith, C. J., D. B. Nedwell, L. F. Dong, and A. M. Osborn. 2006. Evaluation of quantitative polymerase chain reaction-based approaches for determining gene copy and gene transcript numbers in environmental samples. Environ. Microbiol. 8:804-815. [DOI] [PubMed] [Google Scholar]
- 32.Stenström, J., K. Svensson, and M. Johansson. 2001. Reversible transition between active and dormant microbial states in soil. FEMS Microbiol. Ecol. 36:93-104. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki, M. T., L. T. Taylor, and E. F. DeLong. 2000. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 66:4605-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Voroney, R. P., J. P. Winter, and R. P. Beyaert. 1993. Soil microbial biomass C and N, p. 277-286. In M. R. Carter (ed.), Soil sampling and methods of analysis. Canadian Society of Soil Science, Lewis Publishers, Chelsea, MI.
- 35.Zumft, W. G. 2005. Nitric oxide reductases of prokaryotes with emphasis on the respiratory, heme-copper oxidase type. J. Inorg. Biochem. 99:194-215. [DOI] [PubMed] [Google Scholar]