Abstract
Thermophilic cyanobacteria of the genus Synechococcus are major contributors to photosynthetic carbon fixation in the photic zone of microbial mats in Octopus Spring, Yellowstone National Park. Synechococcus OS-B′ was characterized with regard to the ability to acclimate to a range of different light irradiances; it grows well at 25 to 200 μmol photons m−2 s−1 but dies when the irradiance is increased to 400 μmol photons m−2 s−1. At 200 μmol photons m−2 s−1 (high light [HL]), we noted several responses that had previously been associated with HL acclimation of cyanobacteria, including cell bleaching, reduced levels of phycobilisomes and chlorophyll, and elevated levels of a specific carotenoid. Synechococcus OS-B′ synthesizes the carotenoids zeaxanthin and β,β-carotene and a novel myxol-anhydrohexoside. Interestingly, 77-K fluorescence emission spectra suggest that Synechococcus OS-B′ accumulates very small amounts of photosystem II relative to that of photosystem I. This ratio further decreased at higher growth irradiances, which may reflect potential photodamage following exposure to HL. We also noted that HL caused reduced levels of transcripts encoding phycobilisome components, particularly that for CpcH, a 20.5-kDa rod linker polypeptide. There was enhanced transcript abundance of genes encoding terminal oxidases, superoxide dismutase, tocopherol cyclase, and phytoene desaturase. Genes encoding the photosystem II D1:1 and D1:2 isoforms (psbAI and psbAII/psbAIII, respectively) were also regulated according to the light regimen. The results are discussed in the context of how Synechococcus OS-B′ may cope with high light irradiances in the high-temperature environment of the microbial mat.
Laminated microbial mats are present in the channels emanating from Octopus Spring, an alkaline, siliceous hot spring in Yellowstone National Park (44). Several studies have focused on the community structure (15, 32, 35) and the physiological/ecological features (34, 44) of the microbial biota of the mat and have helped to elucidate fundamental principles of community ecology and microbial diversity in this environment (2, 31, 43, 44). Recent metagenomic analyses of these consortia are also beginning to reveal ways in which individuals of this community contribute to overall population metabolism (3a). Microbial hot spring communities represent excellent model systems for exploring how environmental parameters, such as light levels, temperature, nutrient availability, and oxic/anoxic conditions, shape the structural and functional aspects of the community. The photic zone of the microbial mat present in Octopus Spring comprises the uppermost 1 to 2 mm. In this region, cyanobacteria, represented by the genus Synechococcus, evolve O2 and fix inorganic carbon (2). During the day, O2 levels may reach >600% air saturation in the photic zone as a consequence of high rates of O2 evolution and diffusion barriers associated with the polysaccharide matrix of the mat (34). Conversely, as light levels decline in the evening, the O2 concentration rapidly diminishes and the mat becomes anoxic (34).
Recent observations based on denaturing gradient gel electrophoresis and 16S rRNA analyses indicated that the Synechococcus organisms of Octopus Spring are represented by different ecotypes present in different physical zones of the mat (14, 15). These habitats are associated with different temperatures (horizontal distribution of organisms in the mat), light conditions (vertical distribution of organisms in the mat), and perhaps nutrient availability (13, 32). In the temperature range of 53 to 63°C, Synechococcus OS-B′ appears to be the most abundant ecotype (15, 43) and is present at all vertical depths within the cyanobacterial layer (∼1.5 mm) of the mat (32), even the top 0.2 mm of the mat, where light irradiances can be >1,000 μmol photons m−2 s−1. At higher temperatures (58 to 75°C), other ecotypes, such as Synechococcus OS-A, are more abundant (15, 43). Recent work has shown that the optimum temperature for photosynthesis in Synechococcus OS-A is slightly higher than that for Synechococcus OS-B′ (1). Several different Synechococcus ecotypes were recently isolated from mats by dilution enrichment methods and have been characterized with respect to growth and photosynthetic function (1).
Ward and colleagues reported light- and temperature-dependent gross and net rates of oxygenic photosynthesis of various Synechococcus sp. isolates in cultures enriched for different cyanobacterial ecotypes from hot spring mats (1). They found that Synechococcus OS-B′ exhibited a growth optimum of between 50 and 55°C, while Synechococcus OS-A showed optimal growth at 55 to 60°C, which is consistent with the temperatures of the mats from which these organisms were originally isolated, suggesting a possible temperature adaptation of these ecotypes (1).
We recently obtained full genome sequences for Synechococcus OS-A and Synechococcus OS-B′, and from the genes associated with each genome we have inferred various physiological functions, including the potential for both of these organisms to fix nitrogen (3a, 40). To learn more about the metabolic potential of the Synechococcus ecotypes, we generated an axenic culture of Synechococcus OS-B′ and have begun to explore how this organism responds to changes in the light environment with respect to growth, pigment composition, the composition of the photosynthetic apparatus, and the expression of genes encoding polypeptides associated with the photosynthetic apparatus and light acclimation.
MATERIALS AND METHODS
Cultivation conditions.
Synechococcus OS-B′ enriched cultures were obtained by a filter cultivation approach (1) and were grown in DH10 medium (medium D [8] supplemented with 10 mM HEPES, pH 8.2). Enriched cultures (designated CIW-5) were maintained at 52.5°C, exposed to fluorescent light at 75 μmol photons m−2 s−1 (plant and aquarium light; Philips Lighting Company, Somerset, NJ), and bubbled with air enriched with 3% CO2. Growth at different light irradiances was achieved by positioning the culture flasks at different distances from the light source.
To generate axenic cultures of Synechococcus OS-B′ (designated CIW-10), we repeatedly streaked the enriched cultures (CIW-5 with Synechococcus OS-B′ as the single cyanobacterium contaminated by heterotrophic microbes) on DH10 medium solidified with 0.4% ultrapure agarose (Invitrogen, Carlsbad, CA). Synechococcus OS-B′ cells are phototactic, and we were able to exploit their movement toward a directional light source to separate them from heterotrophs; this procedure was repeated several times to ensure that the strain was axenic. The growth of the axenic cyanobacterial isolates was slow (e.g., >1-week doubling time) but increased considerably if the DH10 medium was supplemented with 0.05% (wt/vol) Bacto tryptone (BT; BD Diagnostic Systems, Sparks, MD). After 2 weeks of growth, the cultures became chlorotic, but the bleaching was reversed by the addition of fresh BT, suggesting that the medium was being depleted of an essential nutrient that could be supplied by BT. Axenic cultures were examined periodically by light microscopy and plated on DH10-BT agarose solid medium to ensure that the cultures were not contaminated with heterotrophic bacteria.
Growth measurements of Synechococcus OS-B′.
Axenic cultures were grown in DH10-BT medium in continuous light of different irradiances (25, 50, 75, 130, 200, and 400 μmol photons m−2 s−1) and bubbled with 3% CO2 in air. Aliquots of the culture were collected every day at the same time (2:00 p.m.), and cell densities were estimated by using an Ultraplane Neubauer counting chamber (Hausser Scientific, Horsham, PA) and by monitoring the A750 of the culture. Analysis of these parameters revealed a relatively constant relationship between cell number and the optical density at 750 nm (OD750) (∼1.2 × 107 cells/OD750 unit); therefore, cell densities were estimated by measurement of A750. Chlorophyll (Chl) content was quantified by measuring the absorption of acetone-extracted pigments (90% acetone, 10% water [vol/vol]) at 620, 680, and 750 nm; the calculation used for quantification was as follows: mg Chl a = 0.01497(OD680 − OD750) − 0.000615(OD620 − OD750) (11). The relative phycobilisome (PBS) content was estimated by analysis of whole-cell absorbance spectra normalized to the OD750, essentially as described by Collier and Grossman (12). The peak absorbance at 620 nm (peak absorbance of phycocyanin) was compared to a line drawn between the spectral valleys at 550 and 650 nm. The maximum PBS content was found in cells grown at the lowest light irradiance (25 μmol photons m−2 s−1); this value was set at 100%.
Light experiments.
For isolation of RNAs from cells maintained under constant conditions, cultures were grown at various light irradiances (low light [LL] = 25 μmol photons m−2 s−1, medium light [ML] = 75 μmol photons m−2 s−1, and high light [HL] = 200 μmol photons m−2 s−1) to mid-logarithmic phase (OD750, ∼0.5). One hundred fifty milliliters of culture was decanted into a precooled Erlenmeyer flask and rapidly cooled to ∼4°C (the flask was immersed in liquid N2), and the cells were collected by centrifugation (3,000 × g, 4°C, 10 min), frozen in liquid nitrogen, and stored at −80°C until use. For light shift experiments, mid-logarithmic-phase cultures of Synechococcus OS-B′ were shifted to different light levels (as indicated in the figures). Aliquots of the cultures were collected for RNA isolation immediately before the light shift and 5, 10, 15, and 60 min after the light shift. Dark-adapted cells were maintained in the dark (but bubbled with CO2-enriched air) for 16 h after growth to mid-logarithmic phase at an irradiance of 75 μmol photons m−2 s−1. For carotenoid analysis, cells were treated as described above, except that cultures analyzed over a time course of more than 1 day were diluted once per day to maintain logarithmic cell growth and to avoid dramatic changes in light exposure caused by self-shading among individuals within cultures.
Isolation of RNA, reverse transcription, and q-PCR.
Whole-cell RNAs were purified as previously described (40). Prior to use for quantitative reverse transcriptase PCR (q-PCR), the RNAs were subjected to DNase digestion (Turbo DNase; Ambion, Austin, TX), purified by phenol-chloroform extraction followed by ethanol precipitation, and then tested for residual DNA contamination by PCR. For q-PCR, Superscript III RT (Invitrogen, Carlsbad, CA) was used to reverse transcribe 4 μg of DNA-free RNA, using 200 ng random primer hexamers (MBI, Burlington, Canada) in a reaction volume of 20 μl. The reaction was performed as recommended by the manufacturer (Invitrogen, Carlsbad, CA) in a PTC thermocycler (MJ Research Inc., St. Paul, MN) according to the following temperature regimen (after denaturation of the RNA and addition of the reaction mixture): 5 min at room temperature, 5 min at 37°C, 10 min at 45°C, 40 min at 50°C, 20 min at 55°C, and 15 min at 75°C for inactivation of the RT. The single-stranded cDNA product of this reaction was diluted 1:10 in nuclease-free water (final volume, 200 μl), and 2.5 μl of the diluted sample was used in a 20-μl qPCR mix, as previously described (40). The specificity of the qPCR was evaluated by using the melting curve for the product (e.g., single or multiple products with different melting temperatures [Tm]). The relative levels of RNA were quantified by comparing the cycle threshold (CT) values determined for the reactions with the CT value differences obtained for a 10-fold dilution series of a specific target DNA template (40). The genes encoding the transcripts analyzed by qPCR and the primers used for the analyses are given in Table 1.
TABLE 1.
Gene names, locus tags, and primers used for qPCR analysis of Synechococcus OS-B′ transcript levels
| Gene | Locus taga | Encoded protein | Primer sequenceb
|
|
|---|---|---|---|---|
| Forward | Reverse | |||
| apcA | CYB_1438 | αAP | ACGAAATCGATTGTGAACGCGG | AGTTGGTCGGCAGCTTGTTTGA |
| apcB | CYB_1439 | βAP | ||
| apcC | CYB_1440 | LC9.7 | ATGTTCAAGATCACGGCCTGTGTG | AGTCCTACGTCCATCCCAGGCTTA |
| apcD | CYB_0186 | β19.5 | CGCTTGCAAATCGCTCAGGTGTTA | TAATGCCATAGGTCACCAGACGCA |
| apcE | CYB_0431 | LCM101 | TAGCGATTTGCGCATGTTGGCA | ATTAAGCGCTTGGTTCGCCAGA |
| apcF | CYB_2211 | αAPB | TTCAAGCTGGGTACTGCCTTCT | TAGCGATTTGCGCATGTTGGCA |
| cpcA1 | CYB_0940 | αPC1 | ATTGCTGCCGCTGACAATCAAG | TTGAACACTTCCTGCACCGCTT |
| cpcB1 | CYB_0939 | βPC1 | ||
| cpcA2 | CYB_2738 | αPC2 | ||
| cpcB2 | CYB_2740 | βPC2 | GTTGTTGCCCAAGCTGACTCCAAA | GTTCAAATTGGGCTCCTCGGCAAA |
| cpcC | CYB_2737 | LR31.6 | TGCTGGGCAACGACCATTTGTT | TTGAAATTGAGCTCGATGGCCC |
| cpcD | CYB_0941 | LR13.7 | CGATGTTTGGTCAAAGCACCACCA | ATCTCGGCAGACAACCGACTGTA |
| cpcE1 | CYB_0942 | Phycocyanobilin lyase α subunit | ||
| cpcE2 | CYB_2736 | Phycocyanobilin lyase α subunit | ||
| cpcF | CYB_0943 | Phycocyanobilin lyase β subunit | ||
| cpcG | CYB_0944 | LR27.9 | ATCTGGTCATGGTTCCCAACGACA | AGGCGGCATTGTACTCTTCGCTAT |
| cpcH | CYB_0568 | LR20.5 | ATTAAGCGCTTGGTTCGCCAGA | AAAGGCATTGACCATGGCCTGCAA |
| coxA | CYB_2698 | Cytochrome c oxidase, subunit I | GAACTTCCTGCTGAGCCTTG | GTAAAGGCACCGTTGTACCG |
| crtO | CYB_0017 | Putative β,β-carotene ketolase | TAGTTTGCTGCGACCGGAGATCAT | AGGAAGGTTGCTTGGCATAGACCT |
| crtP | CYB_1694 | Phytoene desaturase | TTCAACGCCATTGTCGGCATCTTG | AAATATCGGTGTTGACCCGCTCGT |
| CYB_0605 | CYB_0605 | Tocopherol cyclase | TACCAATACACCGCCCACCTCAAT | TGAATGCGGTAAAGCCAGCGAATG |
| cydA | CYB_1208 | Cytochrome b,d-quinol oxidase, subunit I | TGTGGCCGGTCTTAACAACAGGAA | GTGCCCAGTTCATGCCAAACTGAA |
| sodB | CYB_2514 | Superoxide dismutase | GATGGCACGCTGAAGGTTACCAAA | AGTGCTCCAGGTAGGCATTGATGA |
| psbAI | CYB_0216 | D1:1 protein | CAGCTCTGGAGTTGGAATGGCTTT | GCCGCCGAGATGAGATTATTCC |
| psbAII | CYB_0371 | D1:2 protein | AGTTTGGGCAAGAGGAAGAGACCT | ATTCCGCTCGTGCATCACTTCCAT |
| psbAIII | CYB_0433 | D1:2 protein | AGTTTGGGCAAGAGGAAGAGACCT | ATTCCGCTCGTGCATCACTTCCAT |
The accession number for the Synechococcus OS-B′ genome is CP000240.
Primer sequences are shown for the transcripts analyzed in this work, which encode proteins associated with PBS, photosynthesis, respiration, oxygen and light stress, and carotenoid biosynthesis.
Carotenoid analysis.
Mid-logarithmic-phase cells grown at different light irradiances were collected by centrifugation (10 min, 3,000 × g, 4°C) and stored at −80°C under nitrogen gas until subjected to further analysis. Pigments were extracted by vigorously vortexing the cells in 100% methanol, followed by a 10-min incubation in the dark. The supernatant was recovered by a 10-min centrifugation step (Eppendorf microcentrifuge; maximal speed), transferred into 2-ml amber high-pressure liquid chromatography (HPLC) vials (National Scientific, Rockwood, TN), bubbled with molecular nitrogen, sealed, and immediately used for HPLC analysis. All extracted pigments were maintained at 4°C in the dark.
HPLC analysis.
Pigments were analyzed on an Agilent 1100 series HPLC system equipped with a Nucleosil 300-5 C18 column (Macherey-Nagel, Easton, PA). Methanol (100%)-extracted pigments were adsorbed to the column in acetonitrile-water-triethylamine (9:1:0.01 [vol/vol/vol]) at a flow rate of 0.5 ml/min and then eluted with a gradient of 0 to 100% ethyl acetate over a period of 30 min. To refresh the column, the gradient was reversed for 2 min and then reequilibrated with acetonitrile-water-triethylamine for at least 3 min. Eluted pigments were detected by a diode array detector measuring the A440; absorbance spectra of the eluate were collected from 400 to 800 nm (every 0.4 s) in 1-nm steps. The relative quantity of each pigment resolved by HPLC (see Fig. 3A and B) was determined by integration of the area under the 440-nm peak and by applying absorption coefficients as described previously (25); the absorption coefficient of the pigment designated P1 was estimated to be 986.6 g/cm by (i) assuming a similar molar absorption to that of β,β-carotene at its peak absorption, (ii) recalculating the relative absorbance at 440 nm, and (iii) applying the molar weight of P1 identified in this study. Eluted pigments were collected, vacuum dried, and stored at −80°C under a nitrogen atmosphere until they were used for further analysis.
FIG. 3.
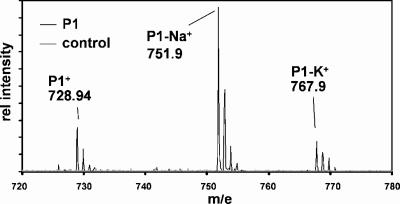
MALDI-TOF MS analysis of purified P1. Putative ion species are indicated. Secondary peaks correspond to different natural isotopes, as discussed in the text.
MALDI-TOF MS analysis.
Matrix-assisted laser desorption ionization-time-of-flight mass spectroscopy (MALDI-TOF MS) was performed using a Voyager DE Pro biospectrometry workstation (Applied Biosystems, Inc.). HPLC-purified pigments were resuspended in 75% acetone and serially diluted (1:1) in 75% acetone. Eight microliters of each of the samples was mixed with 2 μl matrix solution (10 mg/ml 2,5-dihydroxybenzoic acid [Fluka, Buchs SG, Switzerland] in 75% acetone). The sample-matrix mixture (1.5 μl) was spotted onto the target SS plate (Applied Biosystems, Foster City, CA), vacuum dried, and subjected to MALDI-TOF MS. Known control pigments (zeaxanthin and Chl a) were similarly collected and treated. The Voyager DE Pro workstation was operated in the positive reflection mode at 11,500 V with no molecular size gate but with a cutoff at ≤400 Da and a delay time of 93 ns. Grid and guide wire voltages were 76% and 0.005%, respectively. The laser intensity was varied until major peaks were observed, and 200 spectra were collected and averaged per sample. Control spots containing known pigments or only the matrix were treated identically.
77-kelvin fluorescence emission spectra.
Fluorescence emission spectra were obtained in a single-beam fluorometer (Photon Technology International, New Brunswick, NJ). Cells in the logarithmic growth phase were collected by centrifugation (2,500 × g, 5 min) at room temperature, concentrated ∼30-fold, and placed back into their respective growth light irradiances for at least 10 min. The cell suspensions were then incubated for 10 min in the dark or for 10 min in the dark followed by a 10-min exposure to light at 25 μmol photons m−2 s−1, 75 μmol photons m−2 s−1, or 200 μmol photons m−2 s−1, transferred to a measuring cuvette, and immediately frozen in liquid nitrogen. Fluorescence emission spectra from 670 to 750 nm were determined at an excitation wavelength of 435 or 620 nm.
RESULTS
Effect of light irradiance on growth rate and pigmentation.
Growth rates and pigmentation of Synechococcus OS-B′ were monitored in continuous light of different irradiances (25, 50, 75, 130, 200, and 400 μmol photons m−2 s−1, where 25 μmol photons m−2 s−1 is LL, 75 μmol photons m−2 s−1 is ML, and 200 μmol photons m−2 s−1 is HL) over a period of 9 days. At irradiances of 75 and 130 μmol photons m−2 s−1, the cells had the highest growth rates, with doubling times of ∼17 h (Fig. 1B, inset). Cells exposed to the highest irradiances (400 μmol photons m−2 s−1) grew slowly within the first 3 days and then stopped growing; after ∼5 days, the cells appeared to be dead, based on microscopic examinations and a decrease in the OD750.
FIG. 1.
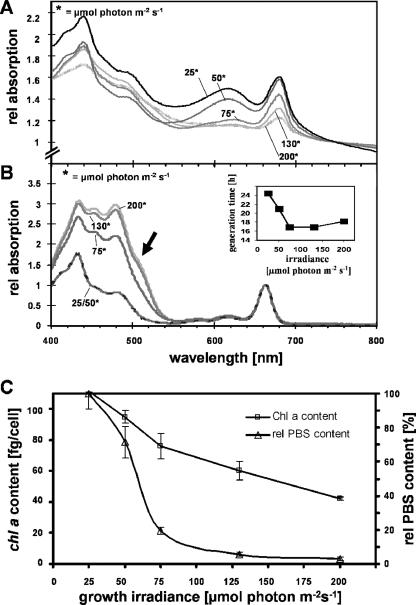
Absorption spectra and growth rates of Synechococcus OS-B′. (A) Whole-cell absorption spectra at different light irradiances. (B) Absorption spectra of acetone-extracted pigments for cultures grown at different light irradiances. (Inset) Doubling times at the different irradiances. (C) Chl and PBS contents of cultures grown at different light irradiances. Spectra from whole cells and acetone-extracted pigments were normalized to the OD750 and OD663, respectively. The absorption shoulder at ∼507 nm in the spectrum of acetone-extracted pigments from HL-grown cells is marked with an arrow in panel B. Error bars on the graphs indicate the means ± standard deviations.
Relative PBS, Chl, and carotenoid contents were estimated from absorption spectra of whole cells and acetone extracts of cell cultures. Cells growing at the different light irradiances exhibited dramatic differences in their pigment contents and compositions. Cultures grown at relatively low light irradiances (25 and 50 μmol photons m−2 s−1) were blue-green, while cultures grown at relatively high light irradiances (75, 130, and 200 μmol photons m−2 s−1) became chlorotic. This difference in pigmentation was reflected by the whole-cell absorption spectra (Fig. 1A) as well as the spectra of acetone-extracted pigments (Fig. 1B). Cells grown in HL had small amounts of Chl a and PBS compared to LL-grown cells (Fig. 1A and C). The decline in PBS levels was more pronounced than that of Chl a levels as the light irradiance for growth was increased (Fig. 1C). Chl a levels were 111 and 45 fg/cell for cultures grown in LL and HL, respectively (Fig. 1C), while the PBS content was ∼5-fold higher for cells grown in LL than for those grown in ML; only very low levels of PBS remained in HL-grown cells (Fig. 1A and C). Cultures shifted from LL to HL exhibited a progressive decline in Chl a and PBS contents; Chl a levels became nearly identical to those of cultures grown in constant HL after 3 days, at which point the PBS levels were almost completely absent (data not shown). Spectroscopic examination of acetone-extracted pigments showed an accumulation of carotenoids relative to Chl a, with the highest relative carotenoid levels in cells grown in HL, as presented in Fig. 1B, which was reflected by a relative change in the ratio of the carotenoid peak at ∼490 nm to the Chl a peak at 430 nm. Furthermore, the spectrum of acetone-extracted pigments demonstrated the emergence of an absorbance shoulder at 507 nm (arrow in Fig. 1B), possibly reflecting the synthesis of an additional carotenoid that is present only in cells grown at higher light irradiances (see below).
HPLC analysis of pigments.
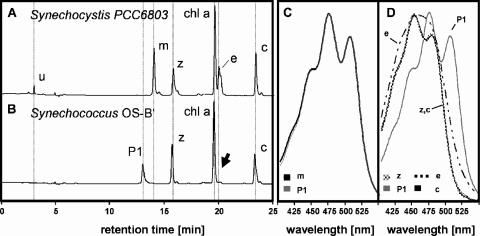
Synechococcus OS-B′ was grown in LL, ML, and HL, and the pigments were extracted in methanol and analyzed by HPLC. Retention times and spectra of individually eluted pigments were compared to those of previously identified pigments of Synechocystis sp. strain PCC6803 grown at 200 μmol photons m−2 s−1 (Fig. 2A and B). Synechocystis sp. strain PCC6803 and Synechococcus OS-B′ both contain Chl a, zeaxanthin (peak z), and β,β-carotene (peak c). However, Synechocystis sp. strain PCC6803 also contains the pigments myxoxanthophyll (peak m) and echinenone (peak e), which are absent from Synechococcus OS-B′. Synechococcus OS-B′ accumulates a pigment of unknown structure with an absorption spectrum that is virtually identical to the spectrum of myxoxanthophyll from Synechocystis sp. strain PCC6803, suggesting that the carotenoid moiety might be identical or highly related (Fig. 2C).
FIG. 2.
HPLC analysis of pigments extracted from Synechocystis sp. strain PCC6803 and Synechococcus OS-B′. Elution profiles for Synechocystis sp. strain PCC6803 (A) and Synechococcus OS-B′ (B) are shown. Peaks corresponding to an unknown pigment (u) in Synechocystis sp. strain PCC6803, myxoxanthophyll (m), zeaxanthin (z), Chl a (chl a), echinenone (e), β,β-carotene (c), and the pigment P1 are indicated. The arrow indicates that no echinenone was detected in the profile of eluted pigments from Synechococcus OS-B′. (C) Overlay of absorption spectra of myxoxanthophyll and pigment P1. (D) Overlay of absorption spectra of echinenone, zeaxanthin, β,β-carotene, and pigment P1.
We subjected the P1 pigment of Synechococcus OS-B′ to MALDI-TOF MS analysis and identified three major signals, at 728.9, 751.9, and 767.9 m/e, corresponding to the pigments P1+, P1-Na+, and P1-K+, as shown in Fig. 3. Na and K adducts are frequently observed by MALDI-TOF MS with the matrix system employed in this study. In addition, we identified minor peaks at m/e 583.4, 567.3, 582.3, and 566.3, corresponding to fragments of P1 (data not shown). The average molecular weight of myxol (C40H56O3) is 584.42 (583.4 if the hydrogen atom of the glycosidic bond is subtracted), and therefore these fragments might correspond to C40H55O3+, C40H55O2+, C40H54O3+, and C40H54O2+, respectively, depending on whether or not the oxygen of the glycosidic bond was retained at the myxol backbone (m/e 583.4 versus 567.3) and whether or not another hydrogen atom was dissociated (m/e 582.3 versus 566.3). These results indicate that myxol is associated with the carotenoid moiety of the pigment and that the attached sugar has a molecular mass of ∼162 g/mol. This molecular mass is congruent with C6H10O5 as the chemical formula for the sugar, suggesting that the attached sugar moiety is an anhydrohexoside. The formula for the entire myxolglycoside would be C46H64O7. Calculations of the isotopic m/e distribution of the putative myxolglycoside with this chemical formula (www.chemcalc.org) revealed a pattern that was virtually identical to the results obtained for P1 MALDI-TOF MS analysis.
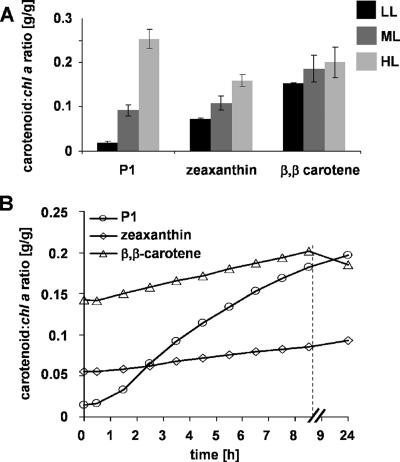
The absorption spectra of the other pigments from Synechococcus OS-B′ are shown in Fig. 2D; note that β,β-carotene and zeaxanthin also have nearly identical absorption spectra in the organic solvent used for extraction. We also investigated the relative carotenoid content compared to the Chl a content of Synechococcus OS-B′ cultures grown in LL, ML, and HL. Interestingly, while there were only minor differences in the levels of β,β-carotene in cells grown under the three different light conditions, zeaxanthin levels were highest in cells grown in HL (with about 50% of the HL level in LL-grown cells), while the myxol-anhydrohexoside P1 was greatly elevated in cultures exposed to HL (Fig. 4A). In order to monitor the kinetics of change in carotenoid pools following a shift in light conditions, we transferred LL-grown Synechococcus OS-B′ cultures to HL and analyzed the relative pigment content over a period of 4 days. The P1 content per g Chl a exhibited a rapid increase (observed within 2 h) that reached ∼14 times the initial level by 9 h following the transfer, as shown in Fig. 4B; the relative level of the myxol carotenoid became similar to that of β,β-carotene. The β,β-carotene and zeaxanthin contents per g Chl a also increased to levels similar to those for cells grown in HL within the first 24 h (Fig. 4B). However, the absolute levels of β,β-carotene and zeaxanthin per cell stayed almost unchanged within the first 24 h and reached the levels for cells grown in HL within 3 days (data not shown). These results demonstrate that only the myxol-anhydrohexoside content of the cells changed rapidly in response to high irradiances, suggesting an important role in the light acclimation response.
FIG. 4.
Carotenoid contents of Synechococcus OS-B′ cells grown at different light irradiances and following a shift in the light conditions. (A) Carotenoid/Chl a ratios (g carotenoid/g Chl a) for P1, zeaxanthin, and β,β-carotene in Synechococcus OS-B′ cultures grown in LL, ML, and HL. Growth light irradiances are indicated in the graph. (B) Kinetics of change of carotenoid/Chl a ratios in cultures grown in LL and then transferred to HL. Note that the time axis is discontinuous. Error bars are given as described in the legend to Fig. 1.
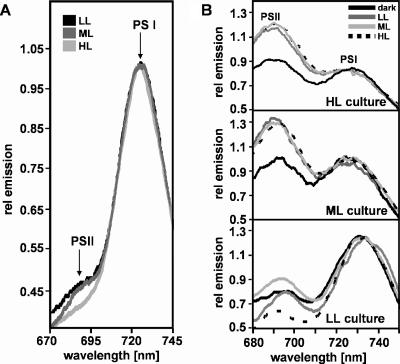
Ratio of PS I to PS II.
We measured 77-K fluorescence emission spectra of Synechococcus OS-B′ cultures grown in LL, ML, or HL, using an excitation wavelength of 435 nm, as shown in Fig. 5A. This excitation wavelength is specific for Chl a and is not absorbed by PBS. The relative Chl fluorescence emissions at ∼693 nm and ∼727 nm provide rough estimates of the relative amounts of photosystem II (PS II) and PS I in cells grown at different light irradiances. The relative amount of PS II in Synechococcus OS-B′ appeared to be low for cultures grown in LL and was further reduced in cells grown in ML, while HL-grown cells exhibited extremely low PS II-specific emission, as shown in Fig. 5A. We also purified the photosystems by sucrose gradient ultracentrifugation and found that PS I-associated Chl a was present in large excess relative to PS II-associated Chl a (≫20-fold) (data not shown). These results suggest that Synechococcus OS-B′ has unusually low levels of PS II relative to PS I and that the PS II/PS I ratio further declines as the light irradiance is increased.
FIG. 5.
77-K fluorescence emission spectra of Synechococcus OS-B′ cells. (A) Spectra of cells grown at different light irradiances, using an excitation wavelength of 435 nm. (B) Spectra of identical cultures measured following excitation at 620 nm. The cultures were grown in LL, ML, and HL (indicated for each set of spectra) and then either pretreated with different light irradiances or kept in the dark (as indicated in the figure) prior to analysis, as described in Materials and Methods. Peak emissions from PS I and PS II are labeled in panel A and in the top panel of panel B.
State transitions.
We analyzed cultures grown at different light irradiances for the ability to perform state transitions. Concentrated cell suspensions were incubated in the dark for 10 min and then subjected either directly or after treatment with light of different irradiances to 77-K fluorescence emission analysis utilizing an excitation wavelength of 620 nm (Fig. 5B, top two panels), which primarily excites PBS but also excites Chl a to some extent. Cells grown in ML or HL and then incubated in the dark for 10 min prior to analysis of the emission characteristics showed similar levels of fluorescence emission from PS II and PS I. However, the cells showed increased PS II emission relative to PS I emission following exposure to LL, ML, or HL after the dark treatment, indicating that cells maintained in the dark were in state II and that light caused a transition to state I (i.e., more PBS was associated with PS II). In contrast, we could not observe a clear state transition in cultures that were grown in LL, as shown in Fig. 5B (bottom panel).
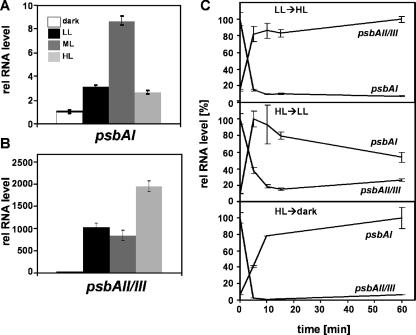
Differential expression of psbAI, psbAII, and psbAIII transcripts.
Cyanobacterial genomes usually encode multiple isoforms of the PS II D1 proteins, and the genes encoding these isoforms are differentially regulated with respect to light conditions (9, 10, 37). We investigated the steady-state levels of transcripts from the three psbA genes of Synechococcus OS-B′, psbAI, psbAII, and psbAIII, in cultures maintained in the dark (bubbled with air) for 16 h or grown in LL, ML, or HL. The psbAII and psbAIII genes (encoding identical D1:2 polypeptides) are 99.8% identical to each other at the nucleotide level and only 69% identical to psbAI (encoding the D1:1 polypeptide). As shown in Fig. 6A, the level of psbAI mRNA in cells grown in ML was ∼9-fold higher than that in dark-adapted cells, while the levels of mRNA in cells grown in LL and HL were only ∼3-fold higher. In contrast, Fig. 6B shows that the levels of the psbAII and psbAIII transcripts were elevated ∼1,000-fold in cells grown in LL and ML and ∼2,000-fold in cells grown in HL relative to those in cells maintained in the dark.
FIG. 6.
Quantification of relative psbAI and psbAII/III RNA levels in Synechococcus OS-B′ cultures. Steady-state transcript levels of (A) psbAI and (B) psbAII/III in cells grown in LL, ML, and HL or kept in the dark for 16 h are shown. (C) Kinetics of change (over a period of 1 h) in the levels of psbAI and psbAII/III transcripts following transfer of cultures from their growth light irradiance to LL, HL, or the dark, as indicated. Values were normalized to the largest amount of each transcript (100%) that was measured over the course of the experiment. Error bars are given as described in the legend to Fig. 1.
We also monitored time-dependent changes in psbA mRNA levels in cultures shifted from LL to HL and from HL to either LL or the dark, as presented in Fig. 6C. The psbAI transcript level increased ∼6- and ∼16-fold in cells transferred from HL to LL and from HL to the dark, respectively, for 1 h (Fig. 6C, middle and bottom panels). During the same shifts, the psbAII/III transcript levels dropped to 1/5 (HL to LL) or <1/20 (HL to dark) of the initial value. Cultures shifted from LL to HL showed a dramatic decrease in the psbAI transcript level, whereas the psbAII/III transcripts increased ∼7-fold (Fig. 6C, top panel). These results demonstrate a strong modulation of the psbA transcript levels by light irradiance and show that the effect of light on psbAI transcript accumulation (most probably representing gene activity) is opposite to that on psbAII/psbAIII.
Transcripts encoding PBS polypeptides.
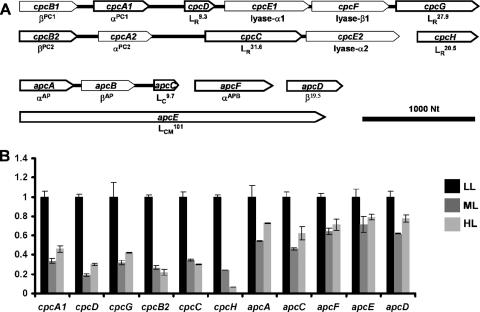
PBS, a light-harvesting complex regulated by many different environmental conditions, is composed of allophycocyanin (AP), phycocyanin (PC), and a number of specific nonpigmented linker polypeptides (18, 45). The genes and operons encoding the different polypeptides of the PBS are presented in Fig. 7A. Based on sequence similarities, Synechococcus OS-B′ has two sets of genes encoding βPC and αPC, designated cpcB1A1 and cpcB2A2. Rod linker peptides (LR20.5, LR27.9, LR31.6, and LR9.3, encoded by cpcH, cpcG, cpcC, and cpcD, respectively, in Synechococcus OS-B′) have been associated with the assembly of the rod substructure (16, 17). In Synechococcus OS-B′, cpcG and cpcD appear to be in the same operon as cpcB1A1, while cpcC appears to be in the same operon as cpcB2A2. Interestingly, the cpcH gene product is truncated at both its N terminus and C terminus relative to the CpcH linker polypeptides in other cyanobacteria. The cpcE and cpcF gene products are required for the attachment of the bilin chromophores to the apophycobiliprotein subunits (18).
FIG. 7.
Organization of genes encoding PBS of Synechococcus OS-B′ and their relative transcript levels in cultures grown in LL, ML, and HL. (A) Organization of genes encoding polypeptides of the PBS core and rods. The direction of transcription is indicated by the direction of the arrow representing each of the genes, and the size of the arrow has been scaled to the length of the gene. The names of the gene products are indicated underneath the respective gene representations, and those genes whose steady-state RNA levels were analyzed are outlined with a thick black line. Contiguous genes are shown connected. (B) Quantification of steady-state levels of apc and cpc transcripts. Transcript levels were normalized to the levels present in LL-grown cells. Growth light irradiances are indicated, and error bars are given as described in the legend to Fig. 1.
The PBS core harbors AP subunits, encoded by apcA and apcB, as well as proteins with AP characteristics, such as αAPB and β19.5, encoded by apcF and apcD, respectively. These proteins may serve as terminal energy acceptors in the complex (18, 45). The core linker peptides, LC9.7 and LCM101, are encoded by apcC and apcE, respectively (42). Among the genes encoding core substructures, only apcA, apcB, and apcC (encoding αAP, βAP, and LC9.7, respectively) are arranged in a putative operon, while genes encoding the other PBS core components (apcE, apcD, and apcF) appear to be transcribed as monocistronic mRNAs.
As shown in Fig. 7B, we analyzed the steady-state transcript levels for genes encoding several of the PBS polypeptides. The transcripts from all of these genes showed significant reductions when the cells were grown in ML or HL relative to those in cells grown in LL. Strikingly, transcripts encoding AP core components (apcA, apcC, apcD, apcE, and apcF) were less reduced in ML and HL than were transcripts encoding components of the rod substructure (cpcA1, cpcB2, cpcC, cpcD, cpcG, and cpcH). The level of the cpcH transcript was particularly sensitive to HL, dropping to almost undetectable levels in cultures grown in HL.
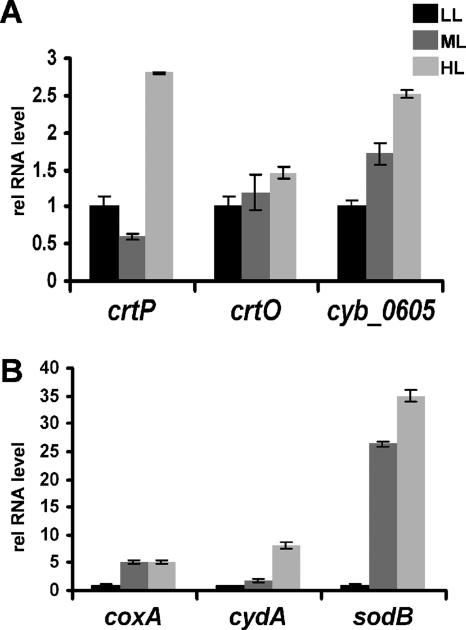
Transcripts related to carotenoid and tocopherol synthesis, respiration, and oxidative stress.
We analyzed cultures grown in LL, ML, and HL for the abundance of transcripts involved in carotenoid and tocopherol synthesis. The steady-state levels of mRNAs for phytoene dehydrogenase (CrtP), a putative β-carotene ketolase (CrtO), and tocopherol cyclase (CYB_0605) were quantified in cultures grown in LL, ML, and HL (Fig. 8A). The levels of the crtP transcript were similar in cells grown in LL and ML but increased ∼2.7-fold in cultures grown in HL. The levels of crtO mRNA were nearly identical in cells grown at all three light irradiances, while the transcript encoding tocopherol cyclase was ∼1.7-fold and ∼2.5-fold more abundant in cultures grown in ML and HL, respectively, than in cultures grown in LL.
FIG. 8.
Relative steady-state transcript levels of genes encoding proteins associated with (A) antioxidant function or (B) oxidative stress. Growth light irradiances are indicated, and error bars are given as described in the legend to Fig. 1.
We also quantified transcripts from the coxA (for subunit I of the cytochrome c oxidase), cydA (for subunit I of the cytochrome b,d-quinol oxidase), and sodB (for a superoxide dismutase) genes, encoding proteins involved in respiration and oxidative stress. CoxA is part of the terminal respiratory oxidase, while CydA is a subunit of an alternative oxidase that transfers electrons from the plastoquinol (PQ) pool to oxygen (19). SodB catalyzes the reduction of superoxide anions to hydrogen peroxide, which may be either thermally degraded or metabolized to water plus oxygen by catalase, preventing the accumulation of reactive oxygen species (ROS). The levels of coxA and sodB transcripts were ∼5-fold and 25- to 35-fold higher, respectively, in cultures grown in ML and HL than in cultures grown in LL (Fig. 8B). The levels of the cydA transcript were similar in cells grown in LL and ML but increased eightfold when the cells were maintained in HL (Fig. 8B).
DISCUSSION
To define how thermophilic cyanobacteria in microbial mats respond to fluctuating environmental parameters, we used an isolate of Synechococcus OS-B′, one of the dominant ecotypes in the 53°C-to-63°C region of Octopus Spring microbial mats. The ability to monitor the growth of this isolate under environmentally relevant temperature conditions, coupled with the availability of the complete genome sequence, provided some of the necessary tools for this investigation, although there are many caveats associated with studies of microbes in axenic cultures. Since much is known about how cyanobacterial cells respond to HL conditions at the biochemical and gene regulation levels, we were able to monitor several key indicators of the physiological state of the cells (7, 21). To our surprise, Synechococcus OS-B′ did not appear to be able to cope with continuous HL irradiances (400 μmol photons m−2 s−1 or higher, although the surfaces of microbial mats are subject to very high irradiance during the day, with large and rapid fluctuations in light intensity associated with cloud cover. We also observed notable differences in photosynthetic function and capacity between Synechococcus OS-B′ cells and well-studied mesophilic cyanobacteria, which are discussed in the context of cyanobacterial physiology and acclimation responses below.
Axenic cultures of Synechococcus OS-B′ grew optimally at light fluence rates of between 75 and 130 μmol photons m−2 s−1 and had a doubling time of ∼17 h. At 200 μmol photons m−2 s−1, the cells exhibited signs of HL acclimation responses, and cells grown in continuous light at an irradiance of 400 μmol photons m−2 s−1 stopped growing after 3 days and then died, suggesting that Synechococcus OS-B′ is unable to tolerate long periods of HL under laboratory conditions. In contrast, mesophilic cyanobacteria, such as Synechocystis sp. strain PCC6803 and Synechococcus sp. strain PCC6301, grow well at light fluence rates of ∼500 μmol photons m−2 s−1 (20). In situ, cells at the surfaces of microbial mats are potentially exposed to light irradiances that are considerably higher than 400 μmol photons m−2 s−1, and thus it may have been expected that Synechococcus OS-B′ cells would survive relatively high light irradiances. It has been speculated that cyanobacterial cells may be able to actively position themselves within the vertical light gradient of the mat to optimize light conditions for photosynthesis (33). It has been demonstrated that in the mat there are distinct cyanobacterial populations that exhibit positive and negative phototaxis (movement toward and away from the light, respectively), suggesting that different cell populations may actively position themselves in different light environments of the mat (33). Furthermore, it has been shown that light is rapidly attenuated as it passes through the top microbial layers of the mat, particularly in the blue and red regions of the spectrum; cells just below the surface may be shielded from high-irradiance surface light (13, 43). The presence of other members of the microbial community, particularly the abundant filamentous bacterium Roseiflexus sp., and the dense polysaccharide matrix of the interstices of the mat are likely to profoundly affect the in situ light environment. In contrast, the cultures grown in the laboratory were dispersed, aerated, and analyzed in mid-log phase (OD750, <0.5), so there was reduced self-shading. Finally, Synechococcus cells growing as axenic cultures under controlled laboratory conditions (continuous fluorescent light, bubbled with CO2-enriched air) in the absence of other members of the microbial community may exhibit different growth responses than cells living in the stratified layers of the mat, where metabolite exchange may play an important role in photosynthetic function.
Photosynthetic organisms can acclimate to the potentially damaging consequences of the absorption of excess light energy in a number of ways, which include a marked decline in light-harvesting pigments, changes in the level and composition of photosynthetic reaction centers, the development of sinks to efficiently remove electrons from the electron transport chain, the establishment of mechanisms to eliminate ROS that might accumulate, and the ability to repair damaged cellular components. We observed many of these acclimation responses in the laboratory-based experiments conducted in this study.
We observed a striking difference in pigmentation among cells grown at different light irradiances. At light irradiances above 50 μmol photons m−2 s−1, the cells had reduced PBS and Chl a contents, while the cell size remained approximately constant, at ∼10 μm by 2 μm. Bleaching was more pronounced at 200 μmol photons m−2 s−1, and cells at 400 μmol photons m−2 s−1 were bleached severely and then died. The loss of PBS reflects the reduced requirement for light-harvesting capacity as environmental light levels increase. Quantification of the abundance of transcripts encoding the polypeptides that make up the PBS was consistent with this observation. Transcripts encoding the individual PC and AP subunits, which are components of PBS rods and cores, respectively, were more abundant in cells grown in LL than in cells grown in ML and HL, but the levels of transcripts encoding Cpc polypeptides declined significantly more than did those encoding Apc polypeptides. These results suggest that at higher light irradiances, there is a reduction in the absorbance cross section of the light-harvesting antenna, reflected by a pronounced decrease in the amount of PC hexamers, while the core polypeptides may be much less reduced; the number of PBS units may not change as dramatically as the absorption cross section of the complex. Furthermore, cpcH mRNA, which encodes an LR20.5 linker polypeptide, was barely detected in cultures grown at a high irradiance. These results suggest that LR20.5 is the most distal rod linker polypeptide and the first to be lost as the rod length decreases at increasing light irradiances.
Analyses of 77-K fluorescence emission spectra following excitation of the cells at 435 nm suggested a pronounced excess of PS I relative to PS II in cells grown under LL, ML, or HL; the ratio of PS II to PS I appeared highest in LL-grown cells. These results contrast with results from studies with Synechocystis sp. strain PCC6714 and Synechococcus sp. strain PCC7942, in which there was an increase in the PS II/PS I ratio with increasing growth light irradiances (28, 36). The number of Chl a molecules bound to individual PS I and PS II reaction centers in Synechococcus sp. strain PCC7942 (former name, Anacystis nidulans) was reported to be 118 and 52, respectively, regardless of growth light and CO2 conditions (24, 29). A decrease in the PS II/PS I ratio in conjunction with a decline in Chl a levels in HL-grown Synechococcus OS-B′ indicates that cells acclimated to HL conditions are likely to have reduced levels of both PS I and PS II. An increased loss of PS II in HL-grown cells may be beneficial because of the potential for PS II photochemistry to generate increased oxidative damage, especially at elevated temperatures. Finally, the fluorescence emission data following excitation of cells with 620-nm light after growth at various light irradiances suggest that the cells (at least those grown in ML and HL) are capable of performing state transitions; we were not able to determine whether or not state transitions were occurring in LL-grown cells.
Carotenoids are an important group of lipid-soluble antioxidants in photosynthetic membranes (30). Synechococcus OS-B′ synthesizes zeaxanthin, β,β-carotene, and a myxolglycoside in which the dimethyl fucoside moiety of myxoxanthophyll (41) is likely replaced by an anhydrohexose. Myxoxanthophyll is critical for stabilization of thylakoid membranes and the formation of the S layer in Synechocystis sp. strain PCC6803 (27). It also increases 1.5- and 2.5-fold after 20 and 45 h, respectively, of exposure of Synechocystis sp. strain PCC6803 to HL (23). In Synechococcus OS-B′, HL triggers a >14-fold increase in the myxolglycoside/Chl a ratio within 9 h, but the response is apparent within the first 2 h of HL, suggesting an important photoprotective function. Furthermore, myxolglycoside accumulation is likely to be a consequence of de novo synthesis, since the levels of β,β-carotene and zeaxanthin remained nearly constant or increased only slightly during the 9-h HL treatment. An increase in carotenoid synthesis during this period is also reflected by an increase in the level of the crtP mRNA (encoding phytoene desaturase).
The development of photoinhibition due to hyperexcitation of the photosynthetic apparatus at high irradiances has been associated with a rapid turnover of the PS II reaction center polypeptide D1 (10). Some cyanobacteria, including Synechococcus sp. strain PCC7942, acclimate to UV radiation by replacing the D1:1 protein isoform with the D1:2 isoform (5, 6, 39), while others, such as Synechocystis sp. strain PCC6803, express only the D1:2 isoform under laboratory conditions (26, 38). In Synechococcus OS-B′, the levels of the transcripts for the three psbA genes (psbAI, psbAII, and psbAIII) were regulated in a light-dependent manner, similar to what has been reported for Synechococcus sp. strain PCC7942 (5). In Synechococcus OS-B′, steady-state psbAI transcript levels were highest in ML (threefold more abundant than those in LL and HL). One explanation for this finding might be the existence of several acclimation processes. More PS II is present in LL-acclimated cells, and with increasing growth light irradiances, elevated turnover of the D1 protein likely occurs, which causes an increase in expression of the psbA1 gene. However, as the light irradiances are increased further, there is more demand for the phototolerant D1:2 isoform. Thus, psbAI transcript levels decline while psbAII/III transcript levels increase. The levels of transcripts from psbAI and psbAII/III appear to be controlled in a reciprocal manner with respect to light irradiance. Interestingly, light shift experiments revealed that psbAI and psbAII/III transcript levels measured 1 h following a shift of the cultures to HL were different from those observed in cultures maintained in HL or LL, suggesting that acclimation was not complete within this time period. Previous work with Synechococcus sp. strain PCC7942 demonstrated rapid transient changes in psbA transcript levels, which slowly stabilized (22). The shift from LL to HL conditions may cause a relatively quick turnover of D1:1, and the consequences of this turnover (e.g., loss of PS II activity) may trigger transcription of the psbAII/III genes. However, the levels of the psbAI and psbAII/II transcripts may begin to stabilize only as the D1:2 protein integrates into the PS II reaction center; other, longer-term processes, including the adjustment of the ratio of the photosystems (see above) and the expression of proteins that can ameliorate the effects of oxidative stress, are also likely to influence the final levels of the psbA transcripts.
The dense polysaccharide matrix of the mat is a diffusion barrier allowing the O2 concentration in the mat to rise well above air saturation during the day (30). An elevated level of O2 in the mat would likely increase photorespiration and cause a dramatic change in the cellular redox state, which in turn might stimulate the generation of ROS, resulting in photoinhibition and damage to many cellular components (2, 26). ROS accumulation and its damaging consequences may be exacerbated by the high temperatures of the hot spring environment (4). Cyanobacteria have evolved many strategies to cope with the potential damaging effects of excess excitation energy. Tocopherol and carotenoids (see below) are antioxidants that help to ameliorate the consequences of ROS accumulation during periods of excess excitation (23). The level of transcript from the CYB_0605 gene, encoding tocopherol cyclase in Synechococcus OS-B′, rises with increasing growth light irradiances. Although we did not directly measure tocopherol concentrations in the cell, these results suggest that tocopherol may accumulate in Synechococcus OS-B′ in response to elevated irradiance. Furthermore, harmful superoxide radicals can be deactivated by superoxide dismutase (32), and indeed, we observed an increased accumulation of sodB mRNA in Synechococcus OS-B′ cells grown in ML and HL. Photodamage might also be reduced by decreasing the redox poise of the cellular quinone pool. This can be achieved by promoting cyclic over linear photosynthetic electron transport and/or by using electrons generated by the splitting of water to reduce alternative electron acceptors (acceptors other than NADP), such as O2 (to regenerate water in a water-to-water cycle). As discussed above, HL-grown cells accumulate much more PS I (involved in cyclic electron flow) than PS II. Furthermore, transcripts for at least two oxidases, namely, the cytochrome b,d-quinol oxidase encoded by cydA, which transfers electrons from the PQ pool to O2 (3, 19), and the respiratory cytochrome c oxidase encoded by coxA, increase following exposure of the cyanobacterial cells to HL. These results suggest that Synechococcus OS-B′ can adjust to HL conditions by both elevating cyclic relative to linear photosynthetic electron transport and diverting electrons from the reduced PQ pool to alternate electron sinks, such as O2. This would be especially important for mat cyanobacteria, since high O2 and/or low CO2 concentrations in the polysaccharide matrix of the mat may limit photosynthetic CO2 fixation. Promoting the flow of electrons from the PQ pool to O2 would help to reduce electron pressure in HL, which in turn would generate a greater proportion of open PS II traps, thereby reducing photoinhibition.
Concluding remarks.
Little is known about how thermophilic cyanobacteria in microbial mats respond to changes in light irradiance, O2 levels, nutrient conditions, and gradients of metabolites synthesized by other bacteria of the mat consortium. Full genome sequences and detailed analyses of the physiological capacity of individual members of the mat community, along with studies to elucidate ways in which the different organisms respond to changing environmental conditions, will greatly contribute to our understanding of the structure and dynamics of microbial consortia and the underlying regulatory processes.
Acknowledgments
We thank Seth DeBolt, Stefan Bauer, Ted Raab, and Staffan Persson (Carnegie Institution of Washington) for helpful discussions and for assistance with the pigment analysis. We thank Dave Ward and Mary Bateson for helpful discussions and for providing the initial Synechococcus OS-B′ enrichment cultures.
This research was funded by the Frontiers in Integrative Biology Program of the National Science Foundation (grants EF-0328698 and OCE-0450874).
Footnotes
Published ahead of print on 4 May 2007.
REFERENCES
- 1.Allewalt, J. P., M. M. Bateson, N. P. Revsbech, K. Slack, and D. M. Ward. 2006. Effect of temperature and light on growth of and photosynthesis by Synechococcus isolates typical of those predominating in the Octopus Spring microbial mat community of Yellowstone National Park. Appl. Environ. Microbiol. 72:544-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bateson, M. M., and D. M. Ward. 1988. Photoexcretion and fate of glycolate in a hot spring cyanobacterial mat. Appl. Environ. Microbiol. 54:1738-1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berry, S., D. Schneider, W. F. J. Vermaas, and M. Rogner. 2002. Electron transport routes in whole cells of Synechocystis sp. strain PCC 6803: the role of the cytochrome bd-type oxidase. Biochemistry 41:3422-3429. [DOI] [PubMed] [Google Scholar]
- 3a.Bhaya, D., A. R. Grossman, A.-S. Stenou, N. Khuri, F. M. Cohan, N. Hamamura, M. C. Melendrez, M. M. Bateson, D. M. Ward, and J. F. Heidelberg. Population level functional diversity in a microbial community revealed by comparative genomic and metagenomic analyses. Int. Soc. Microb. Ecol. J., in press. [DOI] [PubMed]
- 4.Bruskov, V. I., L. V. Malakhova, Z. K. Masalimov, and A. V. Chernikov. 2002. Heat-induced formation of reactive oxygen species and 8-oxoguanine, a biomarker of damage to DNA. Nucleic Acids Res. 30:1354-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bustos, S. A., M. R. Schaefer, and S. S. Golden. 1990. Different and rapid responses of four cyanobacterial psbA transcripts to changes in light intensity. J. Bacteriol. 172:1998-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell, D., M. J. Eriksson, G. Oquist, P. Gustafsson, and A. K. Clarke. 1998. The cyanobacterium Synechococcus resists UV-B by exchanging photosystem II reaction-center D1 proteins. Proc. Natl. Acad. Sci. USA 95:364-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell, D., V. Hurry, A. K. Clarke, P. Gustafsson, and G. Oquist. 1998. Chlorophyll fluorescence analysis of cyanobacterial photosynthesis and acclimation. Microbiol. Mol. Biol. Rev. 62:667-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castenholz, R. W. 1969. Thermophilic blue-green algae and the thermal environment. Bacteriol. Rev. 33:476-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clarke, A. K., V. M. Hurry, P. Gustafsson, and G. Oquist. 1993. Two functionally distinct forms of the photosystem II reaction-center protein D1 in the cyanobacterium Synechococcus sp. PCC 7942. Proc. Natl. Acad. Sci. USA 90:11985-11989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clarke, A. K., A. Soitamo, P. Gustafsson, and G. Oquist. 1993. Rapid interchange between two distinct forms of cyanobacterial photosystem II reaction-center protein D1 in response to photoinhibition. Proc. Natl. Acad. Sci. USA 90:9973-9977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collier, J. L., and A. R. Grossman. 1992. Chlorosis induced by nutrient deprivation in Synechococcus sp. strain PCC 7942: not all bleaching is the same. J. Bacteriol. 174:4718-4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collier, J. L., and A. R. Grossman. 1994. A small polypeptide triggers complete degradation of light-harvesting phycobiliproteins in nutrient-deprived cyanobacteria. EMBO J. 13:1039-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferris, M. J., M. Kuhl, A. Wieland, and D. M. Ward. 2003. Cyanobacterial ecotypes in different optical microenvironments of a 68°C hot spring mat community revealed by 16S-23S rRNA internal transcribed spacer region variation. Appl. Environ. Microbiol. 69:2893-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferris, M. J., G. Muyzer, and D. M. Ward. 1996. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl. Environ. Microbiol. 62:340-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferris, M. J., and D. M. Ward. 1997. Seasonal distributions of dominant 16S rRNA-defined populations in a hot spring microbial mat examined by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 63:1375-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glazer, A. N., D. J. Lundell, G. Yamanaka, and R. C. Williams. 1983. The structure of a “simple” phycobilisome. Ann. Microbiol. (Paris) 134B:159-180. [DOI] [PubMed] [Google Scholar]
- 17.Grossman, A. R., D. Bhaya, K. E. Apt, and D. M. Kehoe. 1995. Light-harvesting complexes in oxygenic photosynthesis: diversity, control, and evolution. Annu. Rev. Genet. 29:231-288. [DOI] [PubMed] [Google Scholar]
- 18.Grossman, A. R., M. R. Schaefer, G. G. Chiang, and J. L. Collier. 1993. The phycobilisome, a light-harvesting complex responsive to environmental conditions. Microbiol. Rev. 57:725-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hart, S. E., B. G. Schlarb-Ridley, D. S. Bendall, and C. J. Howe. 2005. Terminal oxidases of cyanobacteria. Biochem. Soc. Trans. 33:832-835. [DOI] [PubMed] [Google Scholar]
- 20.He, Q., N. Dolganov, O. Bjorkman, and A. R. Grossman. 2001. The high light-inducible polypeptides in Synechocystis PCC6803. Expression and function in high light. J. Biol. Chem. 276:306-314. [DOI] [PubMed] [Google Scholar]
- 21.Hsiao, H. Y., Q. He, L. G. Van Waasbergen, and A. R. Grossman. 2004. Control of photosynthetic and high-light-responsive genes by the histidine kinase DspA: negative and positive regulation and interactions between signal transduction pathways. J. Bacteriol. 186:3882-3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kulkarni, R. D., and S. S. Golden. 1994. Adaptation to high light intensity in Synechococcus sp. strain PCC 7942: regulation of three psbA genes and two forms of the D1 protein. J. Bacteriol. 176:959-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maeda, H., Y. Sakuragi, D. A. Bryant, and D. Dellapenna. 2005. Tocopherols protect Synechocystis sp. strain PCC 6803 from lipid peroxidation. Plant Physiol. 138:1422-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manodori, A., and A. Melis. 1984. Photochemical apparatus organization in Anacystis nidulans (Cyanophyceae): effect of CO2 concentration during cell growth. Plant Physiol. 74:67-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mantoura, R. F. C., and C. A. Llewellyn. 1983. The rapid determination of algal chlorophyll and carotenoid pigments and their breakdown products in natural waters by reverse-phase high-performance liquid chromatography. Anal. Chim. Acta 151:297-314. [Google Scholar]
- 26.Mate, Z., L. Sass, M. Szekeres, I. Vass, and F. Nagy. 1998. UV-B-induced differential transcription of psbA genes encoding the D1 protein of photosystem II in the cyanobacterium Synechocystis 6803. J. Biol. Chem. 273:17439-17444. [DOI] [PubMed] [Google Scholar]
- 27.Mohamed, H. E., A. M. van de Meene, R. W. Roberson, and W. F. Vermaas. 2005. Myxoxanthophyll is required for normal cell wall structure and thylakoid organization in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 187:6883-6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murakami, A., and Y. Fujita. 1991. Regulation of photosystem stoichiometry in the photosynthetic system of the cyanophyte Synechocystis PCC 6714 in response to light-intensity. Plant Cell Physiol. 32:223-230. [DOI] [PubMed] [Google Scholar]
- 29.Myers, J., J. R. Graham, and R. T. Wang. 1980. Light harvesting in Anacystis nidulans studied in pigment mutants. Plant Physiol. 66:1144-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niyogi, K. K. 1999. Photoprotection revisited: genetic and molecular approaches. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50:333-359. [DOI] [PubMed] [Google Scholar]
- 31.Nold, S. C., E. D. Kopczynski, and D. M. Ward. 1996. Cultivation of aerobic chemoorganotrophic proteobacteria and gram-positive bacteria from a hot spring microbial mat. Appl. Environ. Microbiol. 62:3917-3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramsing, N. B., M. J. Ferris, and D. M. Ward. 2000. Highly ordered vertical structure of Synechococcus populations within the one-millimeter-thick photic zone of a hot spring cyanobacterial mat. Appl. Environ. Microbiol. 66:1038-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramsing, N. B., M. J. Ferris, and D. M. Ward. 1997. Light-induced motility of thermophilic Synechococcus isolates from Octopus Spring, Yellowstone National Park. Appl. Environ. Microbiol. 63:2347-2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Revsbech, N. P., and D. M. Ward. 1984. Microelectrode studies of interstitial water chemistry and photosynthetic activity in a hot spring microbial mat. Appl. Environ. Microbiol. 48:270-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruff-Roberts, A. L., J. G. Kuenen, and D. M. Ward. 1994. Distribution of cultivated and uncultivated cyanobacteria and Chloroflexus-like bacteria in hot spring microbial mats. Appl. Environ. Microbiol. 60:697-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Samson, G., S. K. Herbert, D. C. Fork, and D. E. Laudenbach. 1994. Acclimation of the photosynthetic apparatus to growth irradiance in a mutant strain of Synechococcus lacking iron superoxide dismutase. Plant Physiol. 105:287-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schaefer, M. R., and S. S. Golden. 1989. Differential expression of members of a cyanobacterial psbA gene family in response to light. J. Bacteriol. 171:3973-3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sicora, C. I., S. E. Appleton, C. M. Brown, J. Chung, J. Chandler, A. M. Cockshutt, I. Vass, and D. A. Campbell. 2006. Cyanobacterial psbA families in Anabaena and Synechocystis encode trace, constitutive and UVB-induced D1 isoforms. Biochim. Biophys. Acta 1757:47-56. [DOI] [PubMed] [Google Scholar]
- 39.Sippola, K., and E. M. Aro. 2000. Expression of psbA genes is regulated at multiple levels in the cyanobacterium Synechococcus sp. PCC 7942. Photochem. Photobiol. 71:706-714. [DOI] [PubMed] [Google Scholar]
- 40.Steunou, A. S., D. Bhaya, M. M. Bateson, M. C. Melendrez, D. M. Ward, E. Brecht, J. W. Peters, M. Kuhl, and A. R. Grossman. 2006. In situ analysis of nitrogen fixation and metabolic switching in unicellular thermophilic cyanobacteria inhabiting hot spring microbial mats. Proc. Natl. Acad. Sci. USA 103:2398-2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takaichi, S., T. Maoka, and K. Masamoto. 2001. Myxoxanthophyll in Synechocystis sp. PCC 6803 is myxol 2′-dimethyl-fucoside, (3R,2′S)-myxol 2′-(2,4-di-O-methyl-alpha-l-fucoside), not rhamnoside. Plant Cell Physiol. 42:756-762. [DOI] [PubMed] [Google Scholar]
- 42.Wanner, G., and G. Kost. 1980. Investigations on the arrangement and fine structure of Porphyridium cruentum phycobilisomes. Mol. Microbiol. 102:97-109. [Google Scholar]
- 43.Ward, D. M., M. M. Bateson, M. J. Ferris, M. Kuhl, A. Wieland, A. Koeppel, and F. M. Cohan. 2006. Cyanobacterial ecotypes in the microbial mat community of Mushroom Spring (Yellowstone National Park, Wyoming) as species-like units linking microbial community composition, structure and function. Philos. Trans. R. Soc. Lond. B 361:1997-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ward, D. M., M. J. Ferris, S. C. Nold, and M. M. Bateson. 1998. A natural view of microbial biodiversity within hot spring cyanobacterial mat communities. Microbiol. Mol. Biol. Rev. 62:1353-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu, M. H., and A. N. Glazer. 1982. Cyanobacterial phycobilisomes. Role of the linker polypeptides in the assembly of phycocyanin. J. Biol. Chem. 257:3429-3433. [PubMed] [Google Scholar]