Abstract
Reporter gene fusions are essential tools for the investigation of gene regulation. Such fusions are traditionally generated by transposon mutagenesis and identified by a suitable selection procedure. Alternatively, specific reporter fusions can be generated by cloning of DNA fragments containing promoters or other regulatory elements in reporter plasmids. Here, we describe a novel approach for the rapid generation of reporter gene fusions in single copies at defined positions in bacterial genomes. This technique utilizes the Red recombinase for the homologous recombination of PCR-generated cassettes containing various currently used reporter genes, such as those for β-galactosidase, luciferase, and green fluorescent protein. The approach allows the generation of transcriptional or translational reporter fusions in a single step without the requirement for recombinant DNA constructs and is applicable to various enterobacterial species. Generation of reporter fusions by Red recombination is rapid, overcomes the current limitations of transposon mutagenesis or reporter plasmids, and offers new options for the study of bacterial gene regulation.
For the understanding of biological processes, the analysis of gene regulation is of central importance. Despite the availability of high-throughput approaches to the investigation of global gene regulation, the analysis of individual gene expression is of continuing interest for validating and extending data from microarray analyses or other parallel approaches.
A variety of reporter genes is available to study gene expression, with luciferase and β-galactosidase being the most frequently used enzymes (19) and green fluorescent protein (GFP) being used for multiple applications in living cells (25). Most reporters allow the construction of transcriptional fusions to a regulatory element of interest and the construction of translational fusions, resulting in the synthesis of hybrid proteins. In addition to the study of the regulation of the synthesis of a given protein, translational fusions are also of interest for monitoring the fate of a protein, for example, degradation, subcellular localization, secretion into the extracellular space, or translocation into other cells.
Generation of reporter gene fusions conventionally involves either the cloning and fusion of regulatory elements to a reporter gene or random mutagenesis with transposons harboring the reporter gene followed by identification fusions. The latter approach is undirected but results in the generation of single-copy reporter fusions in the native chromosomal context. A common problem is the introduction of foreign regulatory elements, such as promoters and terminators present in the transposon. In contrast, cloning is directed but usually results in the presence of the regulatory elements in several copies on an episomal element. The directed generation of single-copy chromosomal reporter fusions is also possible. However, such approaches previously involved extensive genetic manipulation, such as the generation of recombinant suicide vectors or bacteriophages, and/or resulted in the integration of the reporter fusion into a different chromosomal context (examples in references 5, 8, 9, 18, and 22).
The development of methods using the Red recombinase for genetic engineering opened a path to rational and rapid deletion of genes, operons, or larger genomic elements (4, 28) in Escherichia coli, Salmonella enterica, and a range of other bacteria. The Red recombinase approach has also been used to epitope tag chromosomal genes (26) and to recombine fusions of bacterial promoter elements with GFP (9) into a defined position on the bacterial chromosome. Another modification allowed Flp recombinase (FLP)-mediated integration of lacZ reporter plasmids into FLP target (FRT) sites that remained on the bacterial chromosome for Red-mediated gene deletions (7). We have also utilized Red-mediated recombination to integrate expression cassettes for heterologous proteins into the chromosome of S. enterica (10).
We previously demonstrated that the Red recombination approach can be used to integrate recombinant expression cassettes into the chromosome of S. enterica serovar Typhimurium (10). We also envisaged the use of this recombination technique for the generation of precise gene fusions with reporter genes. Previous studies showed that Red recombination allowed the precise construction of translational fusions of chromosomal genes to epitope tags (26). Here, we describe a Red recombinase-based approach that allows the rapid and precise construction of transcriptional and translational reporter gene fusions in the bacterial chromosome. The availability of a set of template plasmids will allow the rapid construction of single-copy reporter gene fusions that are precisely targeted to chromosomal locations.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Bacterial strains used in this study are listed in Table 1. Bacterial strains were routinely cultured in Luria-Bertani (LB) medium or on LB agar containing antibiotics (50 μg kanamycin or 50 μg carbenicillin per ml of medium) if required for the selection of plasmids or the chromosomal integration of reporter cassettes.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Strains | ||
| S. enterica serovar Typhimurium | ||
| 12023 | WT | Lab collection |
| EE658 | SL1344 hilA::Tn5 lacZY | 1 |
| E. coli DH5α | General cloning strain | Invitrogen |
| S. flexneri | Virulence plasmid-cured | A. Blocker, Oxford, United Kingdom |
| Plasmids | ||
| p2795 | Core template vector | 10 |
| p3121 | luc template vector | This study |
| p3126 | phoA template vector | This study |
| p3138 | lacZ template vector | This study |
| p3141 | HaloTag template vector | This study |
| p3142 | CAT template vector | This study |
| p3143 | blaM template vector | This study |
| p3174 | GFPmut3 template vector | This study |
| p3176 | DsRed template vector | This study |
| pKD46 | Red-expressing vector | 4 |
| pCP20 | FLP-expressing vector | 4 |
| pFPV25.1 | Constitutive GFPmut3 expression | 3 |
Phosphate-carbon-nitrogen (PCN) minimal media containing 0.4 mM inorganic phosphate (Pi) at pH 5.8 (PCN−P medium) or 25 mM Pi at 7.4 (PCN+P) have been described before (5) and were used to culture S. enterica serovar Typhimurium under conditions inducing the expression of genes of the Salmonella pathogenicity island 2 (SPI2) regulon and under noninducing conditions, respectively. Alternatively, N-minimal media were used with various concentrations of MgCl2 as previously described (5).
Construction of template vectors.
Plasmid p2795 (10) was used for the construction of template vectors. p2795 is derived from pBluescript and harbors the kanamycin resistance gene flanked by FRT sites amplified from pKD4 (4). Various reporter genes were amplified by PCR using primers listed in Table S1 in the supplemental material or were recovered from plasmids by restriction digestion. The resulting products were digested, and fragments were cloned in the multiple cloning site of p2795 as described in detail in the supplemental material. The resulting template plasmids were confirmed by DNA sequencing.
Construction of chromosomal reporter fusions.
A detailed step-by-step protocol for the construction of reporter fusions by Red recombination is available in the supplemental material. Using various plasmids (see Fig. 2) as templates, the targeting constructs were amplified using target gene-specific forward and reverse primers (Table 2). Targeting constructs were generated by PCR (10 min at 95°C; 25 cycles of 45 s at 95°C, 45 s at 70°C, and 2 to 4 [depending on reporter gene size] min at 72°C; 10 min at 72°C). A high-fidelity kit (MBI Fermentas) was used. In order to minimize sequence errors by the PCR procedure, reactions were optimized for maximal amounts of template DNA, low numbers of cycles, and highest annealing temperatures. Extension times of the PCRs were adjusted for the sizes of the reporter genes. PCR products were purified with a QIAGEN PCR purification kit, and residual template plasmid was removed by a DpnI restriction digest. PCR products were analyzed by agarose gel electrophoresis and used for electroporation. Freshly prepared competent cells of S. enterica serovar Typhimurium, E. coli DH5α, and Shigella flexneri harboring pKD46 were electroporated as described before (2, 4). The proper integration of the reporter cassettes was confirmed by colony PCR using a reverse primer specific to the reporter and a forward primer specific to the upstream region of the target gene (see the supplemental material for further details). Confirmed clones were subjected to analyses of gene expression in various assays. For S. enterica serovar Typhimurium, reporter fusions were moved into a fresh strain background or strains harboring other mutations using P22 transduction (14). If appropriate, the aph resistance cassette was deleted by FLP-mediated recombination as described by Datsenko and Wanner (4).
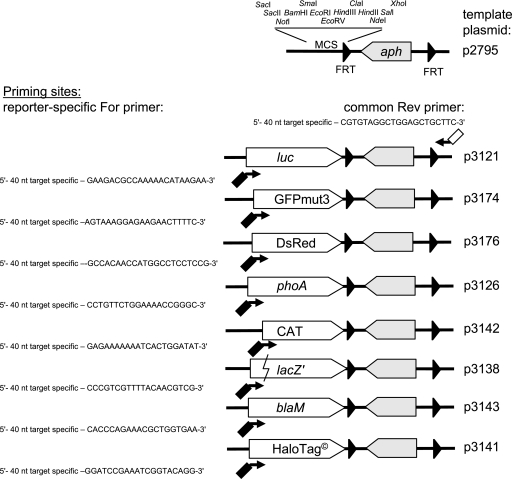
FIG. 2.
Design of template plasmids for various reporter fusions. Core vector p2795 contains the aph resistance cassette flanked by FRT sites. A multiple cloning site (MCS) with a large number of unique sites allows integration of reporter genes. A collection of template vectors with various reporter genes was constructed (plasmids p3121 to p3176). Linear targeting DNA was generated by PCR with primer pairs consisting of forward primers (For) complementary to the reporter and common reverse (Rev) primers. The design of primer pairs for the amplification of targeting DNA is shown. Examples for the generation of translational fusions are shown as in Fig. 1B. Primers typically contain 40-nucleotide (nt) sequences complementary to the target region. The following 20 to 22 nt of the forward primers are specific to one of the reporter genes; the 40-nt sequence was in frame with the ORF of the reporter gene. The reverse primers include a 21-nt sequence complementary to a common priming site of the template plasmids that could be used with each reporter in the series.
TABLE 2.
Oligonucleotides used in this study
| Designation | Sequencea |
|---|---|
| SifA-Red-Luc-For | 5′-ATTATGTAGTCATTTTTACTCCAGTATAAGTGAGATTAATATGGAAGACGCCAAAAACATAA-3′ |
| SifA-Red-LacZ-For | 5′-CAGTATAAGTGAGATTAATATGCCGATTACTATAGGGAATCCCGTCGTTTTACAACGTCG-3′ |
| SifA-Red-Rep-Rev | 5′-ACCCTGAACGTGACGTCTGAGAAAGCGTCGTCTGATTTTAGTGTAGGCTGGAGCTGCTTC-3′ |
| SseJ200-Red-Luc-For | 5′-TTTATCATCTCCACACTTCTTAGGTAAAGAGATGCTTAATGAAGACGCCAAAAACATAA-3′ |
| SseJ-Red-GFPm-For | 5′-TTTGCTAAAGCGTGTTTAATAAAGTAAGGAGGACACTATGAGTAAAGGAGAAGAACTTTTC-3′ |
| SseJ-Red-LacZ-For | 5′-TTAATAAAGTAAGGAGGACACTATGCCATTGAGTGTTGGACCCGTCGTTTTACAACGTCG-3′ |
| SseJ-Red-Rep-Rev | 5′-CGGCACTATGATATTGAGCTGTGTTTTGCTCAAGGCGTACGTGTAGGCTGGAGCTGCTTC-3′ |
| SopE2-Red-Luc-For | 5′-TATAGAAAATATTGCGAATAAGTATCTTCAGAATGCCTCCGAAGACGCCAAAAACATAA-3′ |
| SopE2-Red-Rep-Rev | 5′-GGCAAACCAGCGCCAATGCAGGTGTCGTTACCGTGGCGACCCGTGTAGGCTGGAGCTGCTTC-3′ |
| MgtA-Red-LacZ-For | 5′-TGACTTCGGCGCGGAGGGATTACCTATGCTAAAAATCATTCCCGTCGTTTTACAACGTCG-3′ |
| MgtA-Red-Luc-For | 5′-GAATTTTCTGCGCCTGACTTCGGCGCGGAGGGATTACCTATGGAAGACGCCAAAAACATA-3′ |
| MgtA-Red-Rep-Rev | 5′-TTAAGCACGCTGGCGAATCCCCGACGAAAGTGTTTACTGCGTGTAGGCTGGAGCTGCTTC-3′ |
| EC-MgtA-Red-LacZ-For | 5′-TTCAGACAGTGCGGAGGGACTCCTTATGTTTAAAGAAATTCCCGTCGTTTTACAACGTCG-3′ |
| EC-MgtA-Red-Luc-For | 5′-CATTTCTGTACTGTTTCAGACAGTGCGGAGGGACTCCTTATGGAAGACGCCAAAAACATA-3′ |
| EC-MgtA-Red-Rep-Rev | 5′-GCCGTAACGACGGCTATAGAACCCTTTCACCAACTGGGTCGTGTAGGCTGGAGCTGCTTC-3′ |
| GyrA-Red-SDo-Luc-For | 5′-GGAAAGCGATGACGACGTTGCGGATGACGCTGACGAGTAAAAACTGGAGACTGTCATGGAAGACGCCAAAAACAT-3′ |
| GyrA-Red-Rep-Rev | 5′-AAAGAAAAAGGGCCGGATATCCGGCCCTCGCACAGCAATACGTGTAGGCTGGAGCTGCTTC-3′ |
| Luc-Red-Check-For | 5′-GAGGAGTTGTGTTTGTGGAC-3′ |
| Luc-Red-Check-Rev | 5′-AGTAGTGACAAGTGTTGGCC-3′ |
| GFP-Red-Check-For | 5′-CAGACAACCATTACCTGTCC-3′ |
| GFP-Red-Check-Rev | 5′-AGGTAGTTTTCCAGTAGTGC-3′ |
| LacZ-Red-Check-Rev | 5′-TGTGAGCGAGTAACAACCCG-3′ |
Nucleotides complementary to the template plasmids are set in italics, and the portion containing the RBS is underlined.
Cell culture and infection studies.
The murine macrophage-like cell line RAW264.7 was grown in Dulbecco's modified Eagle medium (DMEM) and used between passages 8 and 20. One day prior to infection, cells were seeded in 24-well plates at about 4 × 105 cells per well and incubated at 37°C in an atmosphere containing 5% CO2. Cells were infected with bacterial strains at a multiplicity of infection of 10 for 30 min. Noninternalized bacteria were removed by washing twice with prewarmed phosphate-buffered saline. DMEM containing 100 μg gentamicin per ml was added for 1 h and replaced by DMEM containing 10 μg gentamicin per ml for the remainder of the incubation time. For cell lysis, the wells were washed twice with prewarmed phosphate-buffered saline, and 100 μl of eukaryotic lysis buffer (Roche) was added and incubated for 15 min with shaking. Samples of the lysate were used to prepare serial dilutions for determination of the number of viable intracellular bacteria by plating onto agar plates. The lysates were transferred to microcentrifuge tubes and centrifuged for 3 min at 13,000 rpm in a microcentrifuge. The supernatants were recovered, and three samples of 25 μl each were used for the luciferase assay as described below. The pellet containing cell debris and bacteria was subjected to the luciferase assay after lysis of cells with bacterial lysis buffer (see below).
Reporter assays.
The luciferase activities in bacterial lysates were quantified as described before (5). Briefly, equal amounts of bacterial cells as adjusted by determination of optical density at 600 nm (OD600) of the cultures were harvested from growth media by centrifugation, and the pellet was resuspended in bacterial lysis buffer (100 mM potassium phosphate buffer [pH 7.8], 2 mM EDTA, 1% [wt/vol] Triton X-100, 5 mg/ml bovine serum albumin, 1 mM dithiothreitol, 5 mg/ml lysozyme). Lysis was performed by a freeze-thaw cycle and incubation for 15 min at room temperature with repeated mixing. Samples (25 μl) of the bacterial lysates or of eukaryotic cell lysates were analyzed by addition of luciferase reagent [20 mM Tricine-HCl (pH 7.8), 1.07 mM (MgCO3)4Mg(OH)2, 100 μM EDTA, 470 μM d(−) luciferin, 33.3 mM dithiothreitol, 270 μM Li3 coenzyme A, 530 μM Mg-ATP). Luciferase activities of eukaryotic or bacterial cell lysates were determined in white microtiter plates (Microfluor; Dynatech) using an Ascent Fluoroscan FL (Labsystems) plate reader or a TopCount instrument (Perkin Elmer) and are expressed as relative light units (RLU). β-Galactosidase activities of bacterial cell lysates were quantified basically as described elsewhere (16) and expressed as Miller units. GFP fluorescence was quantified by flow cytometry on a FACSCalibur (BD) as previously described (11).
RESULTS AND DISCUSSION
Rationale of the Red reporter technique.
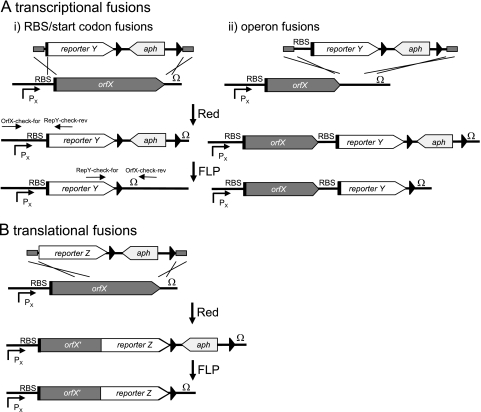
The reporter fusion technique is depicted in Fig. 1. Linear targeting constructs consisting of a promoterless reporter gene and an antibiotic resistance marker flanked by FRT sites were generated by PCR. By the design of the forward primer, it was possible to construct fusions in which the open reading frame (ORF) of the reporter was inserted behind the ribosome binding site (RBS) and start codon of the gene under study (Fig. 1A). This resulted in transcriptional fusions where the expression of the reporter gene was under the control of upstream elements such as the promoter, RBS, etc., of the gene under study. In addition, primers could be designed to generate translational fusions at any position of the ORF of the chromosomal gene (Fig. 1A). A third option was the generation of hybrid operons. For this purpose, targeting constructs were generated with forward primers that contained an RBS and start codon for the reporter gene (Fig. 1B). After integration of the reporter cassettes into the chromosome, P22 transduction was used in S. enterica serovar Typhimurium to move reporter fusions into a fresh strain background or into strains with mutations in other genes. Finally, the aph resistance gene could be deleted by FLP-mediated recombination as described previously (4).
FIG. 1.
Rationale for generation of reporter fusions by Red-mediated recombination. Linear targeting constructs consisting of the reporter, the aph gene flanked by FRT sites, and terminal sequences complementary to the target gene were generated by PCR. The targeting constructs were introduced into various bacterial strains harboring plasmid pKD46 for the expression of Red recombinase. Depending on the design of the targeting construct, Red-mediated recombination results in transcriptional or translational fusions. (A) Transcriptional reporter constructs can be generated by entirely replacing the gene of interest by the reporter gene (i) or by creating hybrid operons where the gene of interest and the reporter gene are under the control of the promoter of interest (Red) (ii). (i) By precise fusion of the reporter at the start codon (black bar) of the gene of interest (orfX), constructs are generated without alterations of regulatory elements such as the RBS or the terminator (Ω). PCR primer pairs for control of integration of reporter cassettes (OrfX-check-for, RepY-check-Rev) and deletion of the aph resistance cassette (OrfX-check-rev, RepY-check-for) are indicated. (ii) Hybrid operons can be generated by integration of reporter cassettes containing an RBS and a start codon for the reporter gene. The expression of orfX and the reporter is under the control of Px, and orfX remains functional. (B) Translational fusions encoding hybrid proteins can be generated at any position of orfX by the design of the forward primer. For all fusions, the aph resistance gene can be removed by FLP-mediated recombination.
The construction of template plasmids for the amplification of targeting constructs is depicted in Fig. 2. Template plasmids are based on the pBluescript derivative p2795, which contains the aph resistance cassette flanked by FRT sites. This basic vector allowed the introduction of various promoterless genes that may function as reporters for gene expression. With this modular approach, various reporter genes were inserted into the multiple cloning site of p2975 to generate a set of template plasmids. Next, primer pairs were designed for the amplification of linear targeting constructs consisting of the aph gene flanked by FRT sites and a reporter gene. While the forward primer is specific for one of the reporter genes and the type and position of the fusion, the reverse primer is specific only for the gene under study and could be used with the entire set of template plasmids.
Applications of the reporter fusion approach.
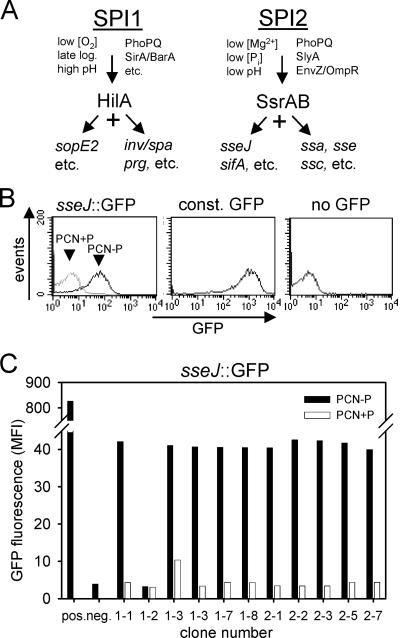
To test the application of Red-mediated reporter fusions to the study of the bacterial gene regulation, we generated fusions to various genes of S. enterica serovar Typhimurium (Fig. 3). S. enterica harbors SPI1 and SPI2, which each encode a type III secretion system (T3SS). In addition to SPI1 and SPI2 genes, genes outside the SPI loci encode effector proteins and are coregulated with SPI1 and SPI2 genes. Both the SPI1 and SPI2 regulons have local and global regulators and are controlled in response to environmental factors (Fig. 3A).
FIG. 3.
Generation of reporter fusions using GFP. (A) Simplified scheme of regulatory cascades of the SPI1 and SPI2 regulons. Various environmental and nutritional factors affect the expression of S. enterica virulence genes. Global regulatory systems, such as PhoPQ, SirA/BarA, or EnvZ/OmpR, modulate expression. The regulatory systems HilA and SsrAB have central roles in controlling expression of genes within SPI1 and SPI2, respectively, or further loci outside of the SPI. (B) A transcriptional fusion (RBS-start codon fusion) of GFPmut3 to the promoter of sseJ was generated by Red-mediated recombination. The resulting strain was grown in PCN−P or PCN+P medium. Growth in media with low concentrations of Pi results in the expression of genes of the SPI2 regulon. S. enterica serovar Typhimurium strains without a GFP reporter or harboring plasmid pFPV25.1 for constitutive expression of GFP were used as negative and positive controls, respectively. Bacteria were analyzed for the GFP fluorescence by flow cytometry. (C) A set of clones harboring an sseJ::GFP fusion was confirmed by PCR for proper insertion of the reporter cassette and analyzed by flow cytometry as described for panel B.
We constructed various reporter fusions of luc, the GFP gene, or lacZ to genes of the SPI1 and SPI2 regulons. Electroporation of linear targeting constructs into competent S. enterica serovar Typhimurium harboring pKD46 routinely resulted in 10 to 100 kanamycin-resistant colonies per 500 ng of PCR product. Linear constructs for generation of lacZ fusions had a size of 5 kb, and targeting integration was also possible, although the frequency of transformation was about 10-fold lower. Screening of a large number of clones by PCR revealed that the vast majority (>90%) of targeting constructs were integrated at the correct position of the chromosome (data not shown).
A translational fusion of GFP to sseJ was analyzed by flow cytometry, and expression of the reporter in response to minimal medium containing small amounts of Pi was observed (Fig. 3B). As generation of targeting constructs by PCR may result in sequence errors during amplification, we initially analyzed the expression characteristics of a larger number of clones for a reporter construct (Fig. 3C). These analyses indicated that the expression levels were very similar for the majority of clones, and less than 10% of the clones showed greatly reduced or undetectable reporter activities. These nonfunctional constructs are likely to result from erroneous oligonucleotide synthesis or from frameshift errors in the reporter ORF, artifacts common to all gene targeting methods that employ PCR amplification. For some of the constructs, the aph resistance cassette was deleted by FLP-mediated recombination (Fig. 1). Comparison of the reporter activities in strains with and without the aph cassette did not indicate any alteration of the expression levels (Fig. 4A) or kinetics (data not shown).
FIG. 4.
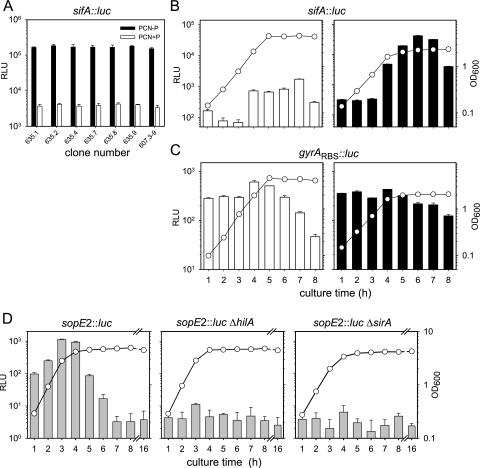
Generation of reporter fusions using luc. Clones harboring a transcriptional fusion of luc to the sifA promoter or the sopE2 promoter were generated by Red-mediated integration. For each fusion, clones were confirmed by PCR for proper insertion of the reporter cassette and a set of confirmed clones were subjected to luciferase activity assays. (A) The expression levels of various clones of the transcriptional sifA::luc fusion (RBS-start codon fusion, strain 635) after FLP-mediated deletion of the aph cassette were compared to those of the parental strain (strain 607.3-9). (B) The expression of the sifA::luc fusion of strain 635 was analyzed during culture in PCN+P (open bars) and PCN−P (filled bars) media. The OD600 of the bacterial culture was determined (circles). (C) A transcriptional fusion (operon fusion) of luc to gyrA was generated, and the resulting strain was analyzed as for panel B. (D) A transcriptional fusion (RBS-start codon fusion) of luc to sopE2, encoding an effector protein of the SPI1 T3SS, was generated. The expression kinetics was analyzed at various time points of culture in LB broth. Expression was analyzed in the wild-type background and in strains deficient in hilA or sirA, encoding regulators of the SPI1 regulon. Luciferase activities are means and standard deviations from three assays. Circles, OD600.
We analyzed the effect of in vitro culture conditions on the expression of S. enterica serovar Typhimurium genes encoding effector proteins of the SPI2 T3SS. As one example, we chose sifA as a member of a regulon under the control of the SsrAB regulatory system. We observed strong induction of the sifA::luc fusion in bacteria entering the stationary growth phase during culture in PCN−P medium or PCN+P medium (Fig. 4B). Only low reporter activities were detected in bacteria cultured in PCN+P medium. These results are in accord with our previous observations on the expression of SsrAB regulon under in vitro conditions (12).
To control the effect of the various growth conditions on the assay system, we generated a fusion to a constitutively expressed gene. Virtually all constitutively expressed genes in the Enterobacteriaceae encode essential housekeeping functions, such as the gyrA gene, which encodes a subunit of the DNA topoisomerase. Since the replacement of such gene by a reporter gene (Fig. 1A, panel i) could affect the viability of the resulting strain, we generated a hybrid operon consisting of gyrA and luc using Red-mediated fusion as outlined in Fig. 1A, panel ii. The resulting strain was not affected in growth rate or viability (data not shown) and was assayed under various growth conditions. An assay for Luc reporter activity indicated the constitutive expression of the gyrA luc hybrid operon during growth in PCN−P and PCN+P media (Fig. 4C). A decrease in Luc activity was observed in the stationary phase, indicating a growth phase-dependent global change in expression levels and protein degradation (see reference 17 for a review).
The analysis of the expression of a luc reporter fusion to the SPI1 effector gene sopE2 indicated that the expression kinetics could be precisely determined (Fig. 4D). Analyses of the expression kinetics of this reporter fusion in strain backgrounds deficient in the SPI1 local regulator hilA or the global regulator sirA revealed greatly reduced levels of expression of the sopE2::luc fusion.
Translational fusions of lacZ behind codon 4 of genes encoding SPI2 effector proteins SifA and SseJ were generated. For comparison, strain EE658 (1), which harbors a Tn5 lacZY transposon insertion in the SPI1 gene hilA, was used, resulting in a transcriptional reporter fusion. The strains were cultured for 4 h or 8 h in medium inducing the expression of genes of the SPI1 regulon (LB broth) or the SPI2 regulon (PCN−P). The expression of lacZ fusion to genes of the SPI1 and SPI2 regulons in response to different environmental conditions was analyzed (Table 3), and the expression pattern was similar to that in previously reported analyses. Red-mediated reporter fusions to various genes under study were generated in an identical fashion and allowed direct comparison of the level of activation. For example, lacZ fusions were generated behind codon 4 of sseJ and sifA, and no alterations to RBSs were introduced. We observed 10-fold-higher expression levels of sseJ than sifA under in vitro conditions inducing the SPI2 regulon. Further reporter strains were constructed using the DsRed gene or the chloramphenicol acetyltransferase gene as the reporter gene. These strains were successfully used to monitor gene expression (data not shown).
TABLE 3.
Generation of reporter fusions using lacZ
| Fusion | β-Galactosidase activity (Miller units) in culturea
|
|||
|---|---|---|---|---|
| LB, 4 h | LB, 8 h | PCN−P, 4 h | PCN−P, 8 h | |
| hilA::Tn5lacZY | 282.3 ± 0.0 | 299.4 ± 10.6 | 9.2 ± 0.7 | 28.2 ± 0.6 |
| sifA::lacZ | 10.0 ± 0.0 | 16.7 ± 0.7 | 31.1 ± 1.6 | 872.7 ± 38.6 |
| sseJ::lacZ | 125.9 ± 3.1 | 479.7 ± 5.4 | 1,226.9 ± 31.7 | 9,795.9 ± 194.8 |
Values are means ± standard deviations from three assays.
The Red reporter technique is applicable to a range of gram-negative bacterial species.
After establishing the Red reporter approach in S. enterica serovar Typhimurium, we tested whether our approach is also useful for the genetic manipulation of other species. We generated targeting constructs for the generation of reporter fusions in a laboratory strain of E. coli as well as the pathogen S. flexneri. For comparison, we analyzed the expression of mgtA. This Mg2+ transport system is conserved in the Enterobacteriaceae, and its regulation has been studied in detail (20). Expression of mgtA is dependent on the concentration of Mg2+ in the growth medium. Transcriptional (RBS start codon) fusions of luc with mgtA were generated in E. coli, S. enterica, and S. flexneri. Growth of the resulting strains in minimal medium with 30 μM Mg2 resulted in strong induction of the reporter, while expression was very low when the reporter strains were grown in medium containing 10 mM Mg2+ (Table 4). Similar results were obtained with lacZ reporter fusions to mgtA in E. coli and S. enterica (Table 4), and additional reporter fusions to environmentally regulated genes could be generated in the three bacterial species (data not shown). The reporter fusion of luc to constitutively expressed gyrA was not affected by the different media.
TABLE 4.
Generation of reporter fusions by Red recombination in E. coli, S. enterica, and S. flexneri
| Activity measured and species | Fusion | Mean ± SD ina:
|
|
|---|---|---|---|
| Low-Mg2+ medium | High-Mg2+ medium | ||
| Luciferase (relative light units) | |||
| S. enterica | gyrA::luc | 1,314 ± 42 | 1,276 ± 62 |
| S. enterica | mgtA::luc | 8,830 ± 70 | 50 ± 9 |
| E. coli | mgtA::luc | 6,821 ± 705 | 30 ± 5 |
| S. flexneri | mgtA::luc | 1,078 ± 59 | 0 ± 0 |
| β-Galactosidase (Miller units) | |||
| S. enterica | mgtA::lacZ | 112.4 ± 24.0 | 2.1 ± 0.0 |
| E. coli | mgtA::lacZ | 82.7 ± 4.0 | 5.5 ± 0.5 |
Strains were grown in N-minimal media containing 30 μM MgCl2 (low-Mg2+ medium) or 10 mM MgCl2 (high-Mg2+ medium).
Use of Red reporter fusions for the generation of luciferase-based translocation assays.
After establishing the construction of translational fusions with various reporter genes, we speculated that these fusions may also be useful for analyzing the fate of proteins synthesized by S. enterica, such as the translocation into eukaryotic cells by a T3SS. The translocation of effector proteins is often quantified by using fusions to the toxin adenylate cyclase (CyaA) from Bordetella pertussis (21). CyaA activity depends on calmodulin, a protein that is present only in the eukaryotic host cell, and only the translocated portion of the CyaA fusion proteins is quantified. Quantification of CyaA activity is based on the determination of cyclic AMP levels in the host cell, but other reporter enzymes may be more convenient to quantify. We decided to construct fusions to luc as a reporter, since firefly luciferase is not present in bacteria or in eukaryotic cells used as host cells. The substrate for Luc, luciferin, cannot penetrate the bacterial cell envelope without permeabilization. This feature allows the translocated fraction of the fusion protein to be quantified, as well as the nontranslocated, bacterium-bound fraction after permeabilization of the bacterial cell. This control is not possible with CyaA, since CyaA activity is dependent on factors present only in the eukaryotic host cell.
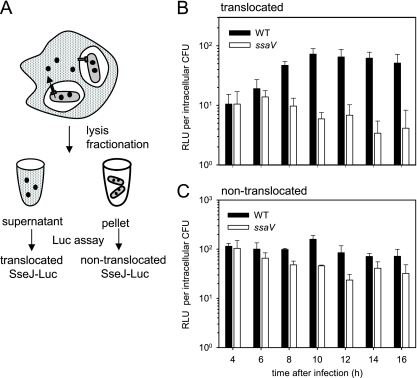
To test the detection of Luc fusions to a translocated protein, we constructed a fusion between the SPI2 effector protein SseJ and Luc. The domain of SseJ required for translocation was determined previously by using the CyaA reporter assay (15). A translational fusion of luc at codon 200 of sseJ was constructed as indicated in Fig. 1B, and the expression pattern of the reporter under in vitro conditions was identical to that of the sseJ::lacZ fusions shown in Table 3. After deletion of the aph resistance marker, an ssaV mutation was transduced into the sseJ::luc reporter strain, resulting in a control strain with a defect of the SPI2-encoded T3SS. Macrophages were infected with the reporter strains and lysed at various time points after infection. For the quantification of translocated SseJ-Luc, the supernatant was used, while the pellet fraction contained the cell debris and intact intracellular bacteria. This fraction was subjected to lysis of bacterial cells followed by quantification of Luc activity (Fig. 5A). The reporter gene was expressed in both wild-type (WT) and ssaV strains (Fig. 5C). Translocation was detectable after 8 h of intracellular life of the S. enterica serovar Typhimurium WT and increased over time (Fig. 5B). In contrast, the Luc activity in the supernatant of lysates of ssaV-infected cells remained at background level, indicating the lack of translocation. We also observed that the nontranslocated portion of the SseJ-Luc fusion in the ssaV strain decreased at longer incubation times, indicating reduced expression levels in the absence of a functional T3SS.
FIG. 5.
Translocation of an SseJ-Luc fusion protein by intracellular S. enterica. (A) Outline of the experimental procedure. RAW264.7 cells were infected with the S. enterica serovar Typhimurium WT or the ssaV strain, deficient in the SPI2-encoded T3SS. Both strains harbored a translation fusion of luc to codon 200 of sseJ, encoding a translocated effector protein of the SPI2-T3SS. At various time points after infection, host cells were lysed and the number of intracellular bacteria was determined. After centrifugation of the lysates, the Luc activities in the supernatant fractions were determined for quantification of translocated SseJ-Luc (B). In addition, the pellet fraction was subjected to lysis of bacterial cells, and Luc assays were performed to quantify cell-associated, nontranslocated SseJ-Luc (C). Luc activities are expressed as RLU per intracellular CFU in order to compensate for intracellular replication over time and different replication rates of WT and ssaV strains. Means and standard deviations for infections with the WT and the ssaV strain are shown for one representative experiment of three.
Our method also allows the generation of translational fusions with other reporters, such phoA, blaM, or HaloTag (Fig. 2), that may be of interest for the investigation of the topology of bacterial membrane proteins, protein secretion, or translocation into host cells. HaloTag is a new reporter enzyme that converts a nonfluorescent substrate into a covalently bound fluorescent product (13).
Concluding remarks.
We have devised a novel approach that allows the generation of reporter gene fusions without the requirement for DNA cloning or random mutagenesis by transposons. Reporter genes can be fused to regulatory elements of genes of interest, and it is also possible to generate translational fusions at any position of a chromosomal gene. The generation of single-copy chromosomal reporter gene fusions in a single step allows the rapid and precise analyses of bacterial gene regulation and eliminates many of the restrictions, intricacies, and uncertainties imposed by previous approaches, such as the presence of the reporter gene fusion in multiple copies, the presence of reporter fusions in a different genetic context, or the lack of upstream regulatory elements. It is also not necessary to define putative promoter regions for cloning. We observed that all currently used reporter genes were efficiently and precisely integrated into the chromosome of Salmonella and other species. Although the PCR-based generation of large targeting constructs might be a limitation, we found that a targeting construct of about 5 kb containing the lacZ gene was integrated efficiently. The Red reporter approach was evaluated in S. enterica, E. coli, and S. flexneri, and it is likely that the technique can be adapted to other bacterial species with minor modifications. Red-mediated recombination has also been described for Yersinia pestis and Yersinia enterocolitica (example in reference 24), pathogenic E. coli strains, such as enteropathogenic E. coli and enterohemorrhagic E. coli, and Citrobacter rodentium (example in reference 6), as well as for the modification of herpesvirus genomes (reviewed in reference 27). We expect that the Red-mediated generation of gene fusions will also be applicable to these organisms. Mammalian genes cloned in bacterial artificial chromosomes have been modified by recombineering (23), and this approach may be extended for the generation of reporter fusions in transgenic animals. We expect that Red-mediated generation of reporter fusions will accelerate the analysis of bacterial gene regulation and enable the analysis of complex regulatory networks.
Supplementary Material
Acknowledgments
This work was supported by grants from the Deutsche Forschungsgemeinschaft (HE 1964) and Elitenetzwerk Bayern Graduate School ‘Lead structures of cell functions’. M.H. was supported by the ‘Fonds der Chemischen Industrie’.
We gratefully acknowledge the technical support from Barbara Bodendorfer and thank Josep Casadesus for comments on the manuscript.
Footnotes
Published ahead of print on 18 May 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bajaj, V., C. Hwang, and C. A. Lee. 1995. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol. Microbiol. 18:715-727. [DOI] [PubMed] [Google Scholar]
- 2.Chakravortty, D., I. Hansen-Wester, and M. Hensel. 2002. Salmonella pathogenicity island 2 mediates protection of intracellular Salmonella from reactive nitrogen intermediates. J. Exp. Med. 195:1155-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 4.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deiwick, J., T. Nikolaus, S. Erdogan, and M. Hensel. 1999. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol. Microbiol. 31:1759-1773. [DOI] [PubMed] [Google Scholar]
- 6.Deng, W., J. L. Puente, S. Gruenheid, Y. Li, B. A. Vallance, A. Vazquez, J. Barba, J. A. Ibarra, P. O'Donnell, P. Metalnikov, K. Ashman, S. Lee, D. Goode, T. Pawson, and B. B. Finlay. 2004. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc. Natl. Acad. Sci. USA 101:3597-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellermeier, C. D., A. Janakiraman, and J. M. Slauch. 2002. Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene 290:153-161. [DOI] [PubMed] [Google Scholar]
- 8.Gunn, J. S., W. P. Loomis, W. J. Belden, and S. I. Miller. 1995. Characterization of the Salmonella typhimurium pagC/pagD chromosomal region. J. Bacteriol. 177:5040-5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hautefort, I., M. J. Proenca, and J. C. Hinton. 2003. Single-copy green fluorescent protein gene fusions allow accurate measurement of Salmonella gene expression in vitro and during infection of mammalian cells. Appl. Environ. Microbiol. 69:7480-7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Husseiny, M. I., and M. Hensel. 2005. Rapid method for the construction of Salmonella enterica serovar Typhimurium vaccine carrier strains. Infect. Immun. 73:1598-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jantsch, J., C. Cheminay, D. Chakravortty, T. Lindig, J. Hein, and M. Hensel. 2003. Intracellular activities of Salmonella enterica in murine dendritic cells. Cell. Microbiol. 5:933-945. [DOI] [PubMed] [Google Scholar]
- 12.Löber, S., D. Jäckel, N. Kaiser, and M. Hensel. 2006. Regulation of Salmonella pathogenicity island 2 genes by independent environmental signals. Int. J. Med. Microbiol. 296:435-447. [DOI] [PubMed] [Google Scholar]
- 13.Los, G. V., and K. Wood. 2007. The HaloTag: a novel technology for cell imaging and protein analysis. Methods Mol. Biol. 356:195-208. [DOI] [PubMed] [Google Scholar]
- 14.Maloy, S. R., V. L. Stewart, and R. K. Taylor. 1996. Genetic analysis of pathogenic bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 15.Miao, E. A., and S. I. Miller. 2000. A conserved amino acid sequence directing intracellular type III secretion by Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 97:7539-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller, J. H. 1992. A short course in bacteria genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 17.Nystrom, T. 2004. Stationary-phase physiology. Annu. Rev. Microbiol. 58:161-181. [DOI] [PubMed] [Google Scholar]
- 18.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 19.Slauch, J. M., and T. J. Silhavy. 1991. Genetic fusions as experimental tools. Methods Enzymol. 204:213-248. [DOI] [PubMed] [Google Scholar]
- 20.Snavely, M. D., S. A. Gravina, T. T. Cheung, C. G. Miller, and M. E. Maguire. 1991. Magnesium transport in Salmonella typhimurium. Regulation of mgtA and mgtB expression. J. Biol. Chem. 266:824-829. [PubMed] [Google Scholar]
- 21.Sory, M. P., and G. R. Cornelis. 1994. Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol. Microbiol. 14:583-594. [DOI] [PubMed] [Google Scholar]
- 22.St. Pierre, R., and T. Linn. 1996. A refined vector system for the in vitro construction of single-copy transcriptional or translational fusions to lacZ. Gene 169:65-68. [DOI] [PubMed] [Google Scholar]
- 23.Testa, G., Y. Zhang, K. Vintersten, V. Benes, W. W. Pijnappel, I. Chambers, A. J. Smith, A. G. Smith, and A. F. Stewart. 2003. Engineering the mouse genome with bacterial artificial chromosomes to create multipurpose alleles. Nat. Biotechnol. 21:443-447. [DOI] [PubMed] [Google Scholar]
- 24.Trülzsch, K., T. Sporleder, E. I. Igwe, H. Rüssmann, and J. Heesemann. 2004. Contribution of the major secreted Yops of Yersinia enterocolitica O:8 to pathogenicity in the mouse infection model. Infect. Immun. 72:5227-5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsien, R. Y. 1998. The green fluorescent protein. Annu. Rev. Biochem. 67:509-544. [DOI] [PubMed] [Google Scholar]
- 26.Uzzau, S., N. Figueroa-Bossi, S. Rubino, and L. Bossi. 2001. Epitope tagging of chromosomal genes in Salmonella. Proc. Natl. Acad. Sci. USA 98:15264-15269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagner, M., Z. Ruzsics, and U. H. Koszinowski. 2002. Herpesvirus genetics has come of age. Trends Microbiol. 10:318-324. [DOI] [PubMed] [Google Scholar]
- 28.Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.