Abstract
A previous survey of Bacteroides isolates suggested that the ermB gene entered Bacteroides spp. recently. Previously, ermB had been found almost exclusively in gram-positive bacteria. In one Bacteroides strain, ermB was located on 100-kb conjugative transposon (CTn) CTnBST. To assess the possible origin of this CTn, we obtained the full DNA sequence of CTnBST and used this information to investigate its possible origins. Over one-half of CTnBST had high sequence identity to a putative CTn found in the genome of Bacteroides fragilis YCH46. This included the ends of the CTn and genes involved in integration, transfer, and excision. However, the region around the ermB gene contained genes that appeared to originate from gram-positive organisms. In particular, a 7-kb segment containing the ermB gene was 100% identical to an ermB region found in the genome of the gram-positive bacterium Arcanobacterium pyogenes. A screen of Bacteroides isolates whose DNA cross-hybridized with a CTnBST probe revealed that several isolates did not carry the 7-kb region, implying that the acquisition of this region may be more recent than the acquisition of the entire CTnBST element by Bacteroides spp. We have also identified other Bacteroides isolates that carry a slightly modified 7-kb region but have no other traces of CTnBST. Thus, it is possible that this 7-kb region could itself be part of a mobile element that has inserted in a Bacteroides CTn. Our results show that CTnBST is a hybrid element which has acquired a portion of its coding region from gram-positive bacteria but which may originally have come from Bacteroides spp. or some related species.
A recent survey of Bacteroides isolates found DNA that hybridized to the erythromycin resistance gene, ermB, only in strains isolated after 1990 (18). Thus, the ermB gene appears to have entered Bacteroides spp. fairly recently. Previously ermB had been found almost exclusively in gram-positive bacteria, the sole exception being an Escherichia coli strain (3). Thus, it was possible that ermB had come into Bacteroides spp. from gram-positive bacteria. A strain of Bacteroides uniformis that carried ermB (WH207) was able to transfer ermB from B. uniformis WH207 to Bacteroides thetaiotaomicron BT4001 by conjugation, raising the possibility that ermB was on a transmissible element (7, 18).
Gupta et al. (7) used pulsed-field gel electrophoresis to show that the B. uniformis element carrying ermB was integrated into the chromosome and was thus probably a conjugative transposon (CTn). The CTn was named CTnBST. CTnBST was estimated to be about 100 kbp in size. A 13-kbp region containing ermB was sequenced and compared to sequences then in the databases. The DNA sequence of the ermB gene was identical to genes found in Clostridium difficile. Outside the ermB gene, however, only a few hundred base pairs downstream of ermB had high sequence identity to DNA from gram-positive bacteria. This did not rule out a gram-positive bacterium origin for CTnBST, however, because at that time, relatively few sequences from gram-positive bacterium transmissible elements were available in the databases.
Bacteroides species are gram-negative bacteria that are normal residents of the human colon. They comprise approximately 25 to 30% of the indigenous microflora (5, 14). The other numerically predominant group of bacteria in the colon consists of gram-positive anaerobes, about which little is known (1, 6, 23). These bacteria comprise at least 65% of the human colonic microflora. Bacteroides strains could have picked up an ermB-containing element from these gram-positive colonic bacteria or from oral gram-positive bacteria that are swallowed on a daily basis. Recently, we succeeded in cloning portions of DNA that covered the entire length of CTnBST and used them to locate and sequence the ends and integrase gene of CTnBST (21). The availability of these clones provided an opportunity to ask whether CTnBST had other regions of sequence similarity to DNA from gram-positive bacteria. In the several years since the study by Gupta et al. (7), many more sequences from gram-positive bacteria have been deposited in the databases, so we considered that expansion and further analysis of the CTnBST sequence could well yield more evidence for the origin of CTnBST. Specifically we looked at the % G+C content of CTnBST in conjunction with BLAST analysis to determine if all, or part of, CTnBST has a gram-positive bacterium origin. In this report we describe an analysis of the entire CTnBST sequence.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Strains of Bacteroides were grown anaerobically at 37°C in prereduced Trypticase-yeast extract-glucose broth (TYG) (8) or plated on TYG agar plates incubated in BBL GasPak jars. Escherichia coli strains were grown aerobically at 37°C in Luria-Bertani (LB) broth or plated in LB agar. Thy− strains of Bacteroides (such as BT4100) were grown in the presence of thymidine (100 μg/ml) and trimethoprim (300 μg/ml).
TABLE 1.
List of strains and plasmids used in this study
| Strain or plasmid | Markera | Description and/or source (reference) |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| DH5(MCR) | RecA− | Gibco BRL |
| S17-1 | RecA− Tpr | Transfer genes from RP4 are integrated into the chromosome (19) |
| Bacteroides spp. | ||
| BT4001 | Rifr | Rifr derivative of B. thetaiotaomicron 5482 |
| BT4100 | Thy− Tpr | Thy− Tpr derivative of B. thetaiotaomicron |
| BT4100ΩxisBST | Emr Tcr | BT4100 wild-type BST with a Cefr insertion in xisBST |
| BT4020 | Emr | BT4001 with CTnBST integrated into the chromosome (7) |
| BF8371 | Emr Tcr | Clinical isolate from WALb |
| Bov7991 | Emr Tcr | Clinical isolate from WAL (18) |
| DH3760 | Emr Tcr | Clinical isolate from VA Hospital, Loyola (18) |
| WAL9063 | Emr Tcr Cmr | Clinical isolate from WAL |
| WH108 | Emr Tcr | Community isolate from Woods Hole |
| WH109 | Tcr | Community isolate from Woods Hole |
| WH202 | Tcr | Community isolate from Woods Hole (18) |
| WH301 | Emr Tcr | Community isolate from Woods Hole |
| WH303 | Emr Tcr | Community isolate from Woods Hole |
| WH714 | Emr Tcr | Community isolate from Woods Hole (18) |
| Plasmids | ||
| pUC19 | Apr | E. coli cloning vector (24) |
| pGEM-T Easy | Apr | Cloning vector (Promega) |
| pLYL001A | Apr (Tcr) | Bacteroides suicide vector (15) |
| pLYLExc | Apr (Tcr) | Used for making disruption in xisBST; this study |
| pLYL05 | Apr (Cefr) | E. coli/Bacteroides shuttle vector (15) |
| pBSTxis1 | Apr (Cefr) | Carries BST002 through BST004; this study |
| pBSTxis2 | Apr (Cefr) | Carries xisBST; this study |
| pBSTxis3 | Apr (Cefr) | Carries xisBST and BST004; this study |
| pBSTxis4 | Apr (Cefr) | Carries BST002 and xisBST; this study |
Bacteroides markers are indicated within the parentheses. Abbreviations: Ap, ampicillin; Cef, cefoxitin; Cm, chloramphenicol; Em, erythromycin; Rf, rifampin; Tc, tetracycline; Thy, thymidine.
WAL, Wadsworth Anaerobe Laboratory.
DNA isolation.
Total DNA was isolated using a modification of the method designed by Saito and Miura (16). Plasmid DNA was isolated from E. coli using the method designed by Ish-Horowitz as described by Sambrook et al. (17) or with a QIAGEN QIAprep Spin Miniprep kit. Restriction digestions, cloning, and ligation reactions were set up as described by the respective manufacturers (New England Biolabs, Promega, and Invitrogen).
Sequencing of CTnBST.
The four fosmid clones containing clones of CTnBST DNA (21) were digested with EcoRI and HindIII, and the resulting fragments (EcoRI/EcoRI, EcoRI/HindIII, and HindIII/HindIII) were subcloned into pUC19 (24) and sequenced from either end of the multiple cloning site using the BigDye Terminator v3.1 cycle sequencing kit from Applied Biosystems. Sequencing was performed by the Core DNA Sequencing Facility of the W. M. Keck Center for Comparative and Functional Genomics at the University of Illinois at Urbana-Champaign. The primer walking method was used to sequence clones that were too large to sequence with a single read from either end of the clone. The sequence fragments were assembled using the program Sequencher 4.5 (Gene Codes Corporation). Tentative open reading frames (ORFs) were identified by using two Web-based ORF finding programs, GeneMark (2) and FGENESB: Bacterial Operon and Gene Prediction (Softberry Inc.). Identified ORFs were assigned putative functions based on BLAST searches.
Localization of the excision region.
A possible excisionase gene was identified on the basis of the properties of its product (small basic protein) and its proximity to the integrase gene. A small internal region of the putative excisionase gene (∼150 bp) of CTnBST was PCR amplified and was cloned with the pGEM-T Easy Vector System I kit from Promega (catalog no. A1360). Unique SacI and SphI restriction sites were engineered into the primers for the PCR. The fragment was subsequently subcloned into the suicide vector pLYL001a (15) between the SacI and SphI restriction sites. The resulting construct was used to transform chemically competent E. coli S17-1 cells (19) to ampicillin resistance. The resulting transformant was used as a donor in a filter mating with a Bacteroides thetaiotaomicron 5482 strain containing CTnBST, BT4021, as the recipient (7). CTnBST can sometimes integrate into multiple locations in the same strain (21). Southern blot analysis had indicated that the strain which was used for the disruption contained only one copy of CTnBST (data not shown). The CTn from the resulting mutant strain, BT4100ΩxisBST, was unable to excise itself from the chromosome, as tested by PCR to detect the joined ends of the CTnBST element.
Portions of the excisionase region from CTnBST were PCR amplified and were cloned with the pGEM-T Easy Vector System I kit from Promega. The primers used for the amplification are shown in Table 2. The gene was then subcloned into the E. coli/Bacteroides shuttle vector pLYL05 (11). The resulting construct, pLYL05xisBST, was used to transform chemically competent E. coli S17-1 cells (19) to ampicillin resistance. The resulting transformant was used as the donor in a filter mating with strain BT4100ΩxisBST as the recipient. Transconjugants were then screened for their ability to excise from the chromosome (by PCR). In addition, pLYL05xisBST was transferred to a strain containing the CTnBST minielement, which cannot excise (21), to see if the cloned DNA causes minielement excision.
TABLE 2.
List of primers used in this study
| Primer | Sequence (5′-3′) |
|---|---|
| CTnBST joined ends | |
| BST6E4FJ | GTTGGTCAGCTCGATAATACG |
| BST10C7RJ1 | GAGTCCACTTTCTGCTGTCT |
| Bacteroides 16S | |
| UnivBactF | GYTACCTTCTTACGACTT |
| UnivBactR | GAGTTTGATYMGGCTC |
| pBSTxis constructs | |
| 6E4-EH3-R.1600 | CACAGGTGTACGCCAAAATCATCAACC |
| 6-E4-EH7-Out-Rev | CATCAAAAAGCCTTGCTTCCGCGG |
| BST-2789(internal)F | GAGAGCTCCGGAAGTATGACACATCG |
| BSTxis-R | TTCCAATCCCATTCCCGTCTTCTGG |
Conjugation assay.
Conjugation was done using a filter mating procedure that has been described previously (20). When matings between Bacteroides donors and Bacteroides recipients were performed, filters were incubated anaerobically. Following incubation, transconjugants were selected for on TYG agar plates that contained the appropriate antibiotics. The antibiotics along with their concentrations used are as follows: for E. coli, ampicillin at 100 μg/ml and streptomycin at 100 μg/ml; for Bacteroides, gentamicin at 200 μg/ml, rifampin at 10 μg/ml, erythromycin at 3 μg/ml, tetracycline at 1 μg/ml, and trimethoprim at 100 to 300 μg/ml.
PCR.
Primers used for PCR in this study are shown in Table 2. For the excision assay, a set of primers was designed for PCR amplifying approximately 800 bp of the joined ends of the excised form of CTnBST. PCRs were done using the specifications provided by the manufacturer of Taq DNA polymerase (Invitrogen). Thermocycler parameters were as follows: 95°C for 5 min; 95°C for 30 seconds, 53°C for 30 seconds, and 72°C for 30 seconds for 30 cycles; and 72°C for 5 min. For PCR products longer than 1 kb the extension time (at 72°C) was increased accordingly (30 seconds per 2 kb).
Nucleotide sequence accession number.
The complete sequence of CTnBST has been submitted to the GenBank nucleotide sequence database under accession number AY345595. Approximately 13 kb of CTnBST (bp 46854 to 59763) was previously published under this accession number (7), which has now been corrected and updated.
RESULTS
CTnBST contains both gram-positive bacterium and Bacteroides sequences.
A total of 100,903 bp was sequenced to obtain approximately onefold coverage of the CTn (GenBank accession no. AY345595). Our strategy was to obtain enough sequence data to allow us to perform database searches for possible homologs, which might provide clues as to the types of genes found on the CTn and the possible origin of the CTn. Since we were primarily interested in obtaining an overview of what was present on the CTn, the level of sequence coverage was sufficient. Sequence analysis revealed a total of 121 possible coding sequences, with an average length of 500 bp. BlastP and BLASTX analyses were able to assign putative functions, such as transfer and integration, to 57 ORFs. In addition, BLAST analysis identified the remnants of a number of ORFs. These coding sequences lacked identifiable start and/or stop codons but showed high levels of homology to gram-positive bacterium genes. Several ORFs contained frameshifts or nonsense mutations; however, this may be a result of sequencing errors. Proteins encoded by 76 ORFs had amino acid sequence identity, ranging between 30 to 95% (with an average of 68% identity), to proteins encoded by ORFs identified in a cryptic CTn found in the genome sequence of Bacteroides fragilis YCH46; however, many of these predicted proteins have no known function (10). The annotators of the YCH46 genome named this putative B. fragilis CTn “CTn3.” Unfortunately, the annotators of the Bacteroides thetaiotaomicron 5482 genome sequence identified a cryptic CTn based on sequence similarities with CTn proteins in the databases that they also designated “CTn3.” The “CTn3” from B. thetaiotaomicron has no sequence similarity to the “CTn3” from B. fragilis. To avoid confusion, we will call the B. fragilis “CTn3” CTn3-Bf.
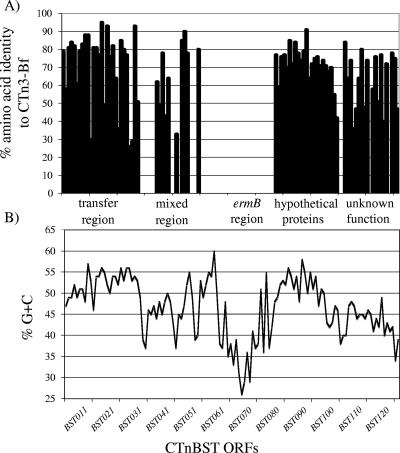
When the deduced amino acid sequences encoded by the 76 ORFs of CTnBST were compared to the corresponding proteins encoded by CTn3-Bf ORFs, an interesting pattern emerged (Fig. 1A). A 17-kb region of CTnBST encoded proteins with no amino acid similarity to any proteins associated with CTn3-Bf. This 17-kb region was localized around the ermB gene. An additional 23-kb region upstream of the 17-kb ermB region had intermittent sequence similarity to CTn3-Bf, but there were several large gaps between the regions of similarity. By contrast, the remainder of CTnBST, comprising the left and right ends of the element, encoded proteins with high levels of amino acid sequence similarity (30 to 95%) to those encoded by the corresponding regions in CTn3-Bf.
FIG. 1.
(A) The percent amino acid identity between the ORFs of CTnBST and CTn3-Bf defines two distinctly gram-positive organism-like regions within CTnBST. The ermB region, as depicted in the graph, is 100% identical on the nucleotide level to the same sequence in Arcanobacterium pyogenes OX-7. The region upstream of the ermB region, labeled as a mixed region, contains a mixture of Bacteroides-like and gram-positive organism-like ORFs. (B) The % G+C content of each ORF in CTnBST is shown. This graph helps to define both functional regions (transfer region) and putative insertions of gram-positive organism-like DNA (in both the mixed region and the ermB region). It should be noted that the average % G+C content of the Bacteroides chromosome is around 42%.
The 40-kb segment of CTnBSt mentioned above (17 kb around the ermB gene and the 23-kb upstream region) contained ORFs that appeared to originate from gram-positive bacteria. In fact, 25 ORFs plus several partial ORFs (BST039 to BST073) showed similarity (products had between 27 and 100% amino acid sequence identity) to sequences from gram-positive organisms such as Clostridium spp., Streptococcus spp., and Enterococcus spp. Interestingly, a 5.5-kb (BST057 to BST070) subsection, included in Table 3, showed 100% DNA sequence identity to an antibiotic resistance element found in the gram-positive bacterium Arcanobacterium pyogenes (9). A. pyogenes is an opportunistic pathogen that is normally found in the intestines of cattle. We obtained A. pyogenes (strain OX-7) DNA from Stephen Billington (Department of Veterinary Sciences and Microbiology, The University of Arizona, Tucson) and have now extended the A. pyogenes region by another 1.5 kb. Results of the analysis of this region revealed that the 100% sequence identity between CTnBST and OX-7 DNA extended beyond the published OX-7 sequence to include DNA through BST073 for a total of approximately 7 kb.
TABLE 3.
List of ORFs from CTnBST that have identical homologs in Arcanobacterium pyogenes OX-7
| Locus taga | CDS (bp)b | No. of amino acids | Database match | % Amino acid identityc | Putative function | Organism | Accession no. |
|---|---|---|---|---|---|---|---|
| BST057 | 55339-55884 | 182 | orf181 | 98 (71/72) | Hypothetical | Campylobacter coli | AAQ94626.1 |
| BST058 | 55981-56346 | 121 | UbiE | 68 (83/121) | N-Methyltransferasee | Clostridium thermocellum | ZP_00510925.1 |
| BST059 | 57368-56346 | 341 | IS4 | 52 (179/341) | IS4d transposase | Chlorobium phaeobacteroides | EAM62306.1 |
| BST060g | 57567-57848 | 94 | UbiE | 67 (63/94) | N-Methyltransferasee | Clostridium thermocellum | ZP_00510925.1 |
| BST061 | 57849-58025 | 59 | TrsK | 100 (55/55) | Conjugation proteinh | Enterococcus faecalis | CAC29188.1 |
| BSTermB | 58061-58798 | 246 | ErmB | 99 (244/245) | Methyltransferase | Streptococcus pyogenes | CAI99373.1 |
| BST063 | 58803-58934 | 44 | ORF3 | 100 (43/43) | Hypothetical | Enterococcus faecalis | AAA27453.1 |
| BST064 | 58968-59213 | 85 | Top beta | 97 (80/82) | Topoisomerase IAh | Enterococcus faecalis | AAC38606.1 |
| BST065 | 59197-59280 | 28 | ORF298 | 88 (24/27) | ATPase involved in chromosome segregationh | Clostridium perfringens | AAC43479.1 |
| BSTPalB | 59282-59301 | PalB | PalB sequence | ||||
| BST066 | 59372-59587 | 72 | Omega | 97 (69/71) | Transcriptional repressor | Streptococcus pyogenes | YP_232757.1 |
| BST067 | 59604-59876 | 91 | Epsilon | 100 (90/90) | Antidote of killing systemf | Streptococcus pyogenes | 63021992 |
| BST068 | 59878-60087 | 70 | Zeta | 91 (64/70) | Toxin of killing systemh | Streptococcus pyogenes | YP_232759.1 |
| BST069 | 60073-60744 | 224 | UbiE | 68 (99/145) | N-Methyltransferaseh | Clostridium thermocellum | ZP_00510925.1 |
| BST070 | 60768-60980 | 71 | RacX | 41 (29/70) | Aspartate racemaseh | Thermosinus carboxydivorans | ZP_01665795.1 |
Name designated by coding sequence (CDS) translated into amino acid sequence.
CDS of the given ORF from the starting base pair to the ending base pair.
Numbers in parentheses indicate the total numbers of identical amino acids/total numbers of amino acids in the region of homology.
This IS is not found in the A. pyogenes OX-7 sequence.
Encoded by a continuous ORF that has been disrupted by an IS (BST059).
CDS has identifiable start and stop codons but, based on homology, does not encode a complete protein.
BST060 has no start codon.
CDS lacks an identifiable stop or start codon or both and represents regions of homology.
% G+C content.
To assess further the possible hybrid nature of CTnBST, the % G+C content of each ORF was calculated (Fig. 1B). The % G+C content fluctuated over the length of the entire element in defined sections (Fig. 1B). The first section defined by the % G+C content contained genes that appeared, from the amino acid sequence similarity of their products to proteins in the databases, to have functions like transfer and mobilization. The next section varied widely in the % G+C content and corresponded to the 40-kb region mentioned above (which includes the 7-kb Arcanobacterium pyogenes sequence) that contained gram-positive bacterium sequences. The remaining two sections identified by this analysis, both approximately 20 kb in length, showed much less variation in the % G+C content.
The average % G+C content of CTnBST in the region where it was most similar to CTn3-bf was 48%, which is higher than the average % G+C content of Bacteroides species (42 to 43% G+C). In addition, as the % G+C content of CTnBST was compared along the entire sequence, a significant change occurred within and around the region of A. pyogenes homology. This region had an average of around 36% G+C, much lower than the rest of CTnBST.
Further analysis of the ermB region.
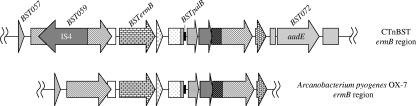
As mentioned above, the ermB region is the region that had 100% DNA sequence identity to a tylosin resistance determinant that carries the ermB gene found in the high-% G+C gram-positive species A. pyogenes (Fig. 2) (9). The identity begins 144 bp into the published A. pyogenes sequence with BST058 in CTnBST (orf181 in the A. pyogenes sequence) and extends all the way through the published A. pyogenes sequence ending with BST070 (Table 3), whose product is most closely related to an aspartate racemase found in Thermosinus carboxydivorans. A detailed description of this intervening region is given by Jost et al. (9).
FIG. 2.
The ermB region of CTnBST and the corresponding region in Arcanobacterium pyogenes OX-7 share 100% identity on the nucleotide level. This identity continues past the published sequence for Arcanobacterium pyogenes OX-7 (shown here) to include the aadE homolog and the flanking partial ORFs (in gray).
The only difference was that CTnBST contains an insertion element (IS) (IS4, BST060) between BST059 and BST061, which most likely represent one continuous ORF. Indeed, these two coding sequences make up one continuous ORF, orfY, in the A. pyogenes sequence. The product of this ORF is closely related to an arginine N-methyltransferase found in Clostridium thermocellum. The A. pyogenes sequence does not contain this IS. Lying downstream of BST069 in CTnBST is a putative streptomycin resistance gene (aadE), which was most closely related to the same gene found in Campylobacter jejuni. This gene was flanked by two high-% G+C partial ORFs, both of which are closely related to two tandem genes found in Clostridium difficile 630. While this sequence is not included in the published A. pyogenes sequence, we have shown that homology extends through BST073 and includes the proposed gene for AadE (BST072).
Comparison with other ermB-containing strains.
Previously, we had screened approximately 300 Bacteroides isolates for ermB by hybridization and, in a subset of cases, PCR. An ermB gene that was identical to that on CTnBST was found in six of these isolates (18). Further hybridization and sequence analysis showed that all six of these isolates carry the same 7-kb ermB region as that found in A. pyogenes (Table 4). However, only the CTnBST-carrying strain (WH207) had an IS between BST059 and BST061. The other five isolates do not contain this IS. Three of the six isolates (WH202, WH714, and DH3760) appear to have had the aadE gene deleted. The three strains that carried the aadE gene (WH207, Bov7991, and BF8371) appeared to be almost 100% identical on the DNA sequence level. A partial sequence analysis of the 16S rRNA genes of these strains revealed that these six isolates represent several Bacteroides species (Table 4).
TABLE 4.
Overview of CTnBST and ermB strainsa
| Strain | 16S identification | Presence of:
|
|||
|---|---|---|---|---|---|
| ermB region (% identity) | CTnBST (% identity)b | IS4 | aadE | ||
| WH207 | B. uniformis | + (100) | + (100) | + | + |
| Bov7991 | B. ovatus | + (99.9) | + (80)c | − | + |
| BF8371 | B. fragilis | + (99.9) | + (98.5) | − | + |
| DH3760 | B. uniformis | + (98.4) | + (80)c | − | − |
| WH202 | B. vulgatus | + (99.0) | − | − | − |
| WH714 | B. vulgatus | + (99.0) | − | − | − |
| WH301 | B. dorei | − | + (97.9) | − | − |
| WAL9063 | B. distasonis | − | + (99.2) | − | − |
+, present; −, absent. The numbers in parentheses represent the percentages of nucleotide identity with respect to the original CTnBST strain, WH207.
Percent identity is based on the percent nucleotide identity outside of the 7-kb ermB region.
The % identity was based on dot blots, which require 80% identity or more to give a positive result.
Strain Bov7991 appeared to carry the ermB region on a copy of a CTnBST-like element. Primers used on any part of CTnBST were able to amplify the corresponding sequences from Bov7991, and these sequences were nearly 100% identical to those on CTnBST. Also, the ermB region was in the same location as it is in CTnBST. However, PCR did not amplify the joined ends of the excised element, nor could this strain transfer erythromycin resistance to a recipient strain via conjugation. Thus, the CTn was unable to excise from the chromosome under the conditions tested.
The remaining isolates (WH202, WH714, DH3760, and BF8371) did not contain any CTnBST sequences outside the ermB region, as detected by PCR and DNA-DNA hybridization. It could be that the ermB region is itself on a transmissible element and has integrated into the chromosomes of Bacteroides spp. several times, only once inserting into an active CTn3-Bf like element.
We also found two other Bacteroides isolates (WH301 and WAL9063) that appeared to contain a close homolog of CTnBST, with more homology to CTnBST than CTn3-Bf and without any trace of the ermB region, as both PCR and DNA-DNA hybridization failed to identify any sequences in this region. Again, this may imply that the ermB region has come from another, presumably gram-positive, bacterial species via several independent transfer events.
Assignment of possible functions to CTnBST genes.
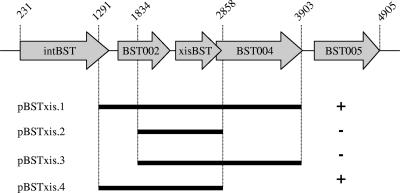
The first ORF in CTnBST, as depicted in Fig. 3, encodes a tyrosine type site-specific recombinase, IntBST, that has been shown to be the CTnBST integrase (21). This ORF, when cloned into a suicide plasmid that also contained the joined ends of CTnBST, was sufficient for integration but not for wild-type levels of excision. Approximately 150 bp downstream of intBST was an ORF (BST002) encoding a hypothetical protein which our sequence analysis shows contains a possible frameshift mutation. The next ORF, 200 bp downstream of BST002, was a small ORF (395 bp) that encoded a basic protein. This gene might encode an excisionase protein for CTnBST. If so, it appeared to be in an operon with a downstream ORF (BST004) which overlapped the putative excisionase by several base pairs.
FIG. 3.
The region that includes the genes necessary for excision of CTnBST. The numbers above the arrows indicate the base pair positions within CTnBST sequence. The “+” and “−” symbols correspond to whether or not complementation of the excisionless phenotype was achieved with the given construct. The results of this experiment lead us to conclude that the promoter for xisBST lies within and/or upstream of BST002.
The next 20 kb contained ORFs that encoded proteins with sequence similarity to transfer proteins from other conjugative elements (tra genes). As stated above, the region between BST039 and BST073 (which includes the A. pyogenes sequence) encoded proteins with a high level of amino acid sequence identity to proteins found in gram-positive bacteria. The next 20 kb showed a high level of similarity to the end of CTn3-Bf and comprised genes encoding hypothetical proteins. The final 20 kb of the element encoded several putative DNA-modifying proteins such as recombinases, single-stranded DNA binding proteins, and transposases.
Identification of the excision region.
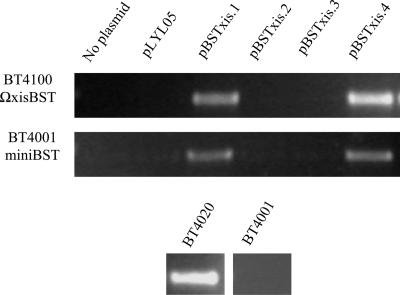
The excision region of the CTn CTnDOT is quite complex, involving three excision genes that work together with the integrase (4). By contrast, the gram-positive CTn Tn916 has only a single excisionase gene (13). We wanted to identify the excisionase gene(s) of CTnBST to determine if its excision mechanism was more like that of CTnDOT or the much simpler xis of Tn916. The putative excisionase gene, tentatively designated xisBST, was disrupted by insertion of a suicide plasmid. A PCR assay was used to determine if the element was able to excise itself from the chromosome, and the result was that CTnBST could no longer excise itself from the chromosome (Fig. 4).
FIG. 4.
The joined ends of the excised circular form of CTnBST were PCR amplified from each of the listed strains carrying each of the listed plasmids shown in Fig. 3 and described in Table 1. BT4020, a strain that carries a wild-type copy of CTnBST, was used as a positive control. BT4001 does not contain CTnBST and was used as a negative control in this experiment. As a positive control for the amplification reaction the gene that encodes sigma 70 in Bacteroides was successfully amplified from each strain but is not shown here.
The putative excisionase region, approximately 5 kbp from the left end of CTnBST, was cloned into a plasmid that replicated in Bacteroides spp. The resulting construct, pBSTxis1, was mated into strain BT4100ΩxisBST, and excision of CTnBST was detected using the PCR excision assay that detects the joined ends of the CTnBST circular form. We were able to show that pBSTxis1 complements the excisionase phenotype of strain BT4100ΩxisBST (Fig. 3 and 4). It is interesting that only one of several xisBST clones was able to complement the excisionless phenotype of strain BT4100ΩxisBST. We started with a construct, pBSTxis1, that contained the xisBST gene plus the upstream (BST002) and downstream (BST004) ORFs. This construct fully complemented strain BT4100ΩxisBST; however, when we tried to further localize the excisionase gene, cloning only xisBST plus a few hundred base pairs of upstream sequence, pBSTxis2, we lost complementation. Complementation was restored only when we included the entire upstream ORF (BST002) with construct pBSTxis4. This gene, BST002, carries a potential frameshift but is still present in BT4100ΩxisBST, implying that the promoter for xisBST may be upstream of the coding sequence of BST002.
In addition, pBSTsxis1 and pBSTxis4 were also able to complement the excision-deficient phenotype of the CTnBST minielement, miniBST (Fig. 4). This minielement contains all of the DNA sequences that are needed for the integration of CTnBST into the chromosome, including the integrase gene intBST plus the joined ends of the excised circular form of CTnBST (21). Together these data imply that the excisionase gene, including the promoter sequence, is the only thing that is required for excision of CTnBST. This is in stark contrast to the complex excision mechanism of CTnDOT, which is tightly regulated and requires multiple proteins (4).
DISCUSSION
CTnBST contains approximately 100 potential protein coding sequences. Several ORFs contained frameshift or nonsense mutations. These could be due to sequencing errors, or they could be real mutations. In either case, the ORFs in which they are located provide some indication of the genetic potential of the CTn because such mutations, if real, could revert to wild type if selection for the gene's function occurred.
Initially, we had hoped to identify the origin of CTnBST by finding the entire CTnBST sequence in genome sequences from other bacteria. It is clear from our results that this hypothesis was too simplistic. Instead, CTnBST appears to be an amalgam of sequences having different origins, some of which may already have been in Bacteroides spp. as a cryptic CTn before acquiring sequences from gram-positive bacteria. It is also possible, however, that cryptic forms of CTnBST such as CTn3-Bf may themselves have had their origin outside of Bacteroides spp., because the % G+C content of this DNA differs from that of the BT4001 chromosome (46 to 50% versus 42 to 43%). In addition to portions of an element like CTn3-Bf, CTnBST seems also to have acquired several regions that could have come from a gram-positive bacterium. Moreover, even this region has itself undergone an IS insertion in one instance and a deletion in the aad gene in three others. Even the region that has sequence similarity to CTn3-Bf has also undergone some alterations. That is, it is not identical in sequence to CTn3-Bf but has experienced a number of changes, some of which are fairly large (Fig. 1A). Our results show how much variation can occur in transmissible elements as they move from one host to another.
The high sequence identity of the ermB region of CTnBST to a similar region in Arcanobacterium pyogenes OX7 strongly suggests a direct genetic conduit between gram-positive species and Bacteroides species. The fact that CTnBST carries an IS in one of the ORFs from this region, whereas similar ermB regions in other Bacteroides isolates do not, implies a direction of transfer. That is, the IS insertion may have occurred after the ermB region arrived in Bacteroides spp. An alternate explanation is that the IS was lost by precise excision in the other Bacteroides spp., but this possibility appears to be less likely than the acquisition of the IS sequence in Bacteroides spp. It is important to note that, even if the argument is accepted that the ermB region moved from a gram-positive bacterium to Bacteroides spp., an intermediate host cannot be ruled out. The fact that this region first appeared in Arcanobacterium pyogenes OX-7 before Bacteroides is also supported by the fact that several isolates of Bacteroides that carry the ermB region have a deletion of the aadE gene. Bacteroides species are naturally resistant to streptomycin, so the fact that this gene was deleted in several isolates is consistent with the idea that there was no selection to maintain the gene in Bacteroides spp.
Approximately 23 kbp of sequence upstream of the ermB region appear to be a mixture of Bacteroides-like sequences and gram-positive bacterium-like sequences. Part of this region is also found in CTn3-Bf, suggesting that the acquisition of some of the gram-positive bacterium-like sequences occurred in a common progenitor of the two CTns. Jost et al. (9) proposed that, in Arcanobacterium pyogenes, a mobile element carrying ermB had inserted into an ORF, designated orfY (homologous to BST059, BST061, and BST070 in CTnBST), causing a duplication of orfY. Subsequent to that event, an internal portion of the trsK gene inserted itself immediately upstream of ermB, forming a putative promoter to drive expression of ermB in Arcanobacterium pyogenes. The evidence they provide is that they have other isolates of Arcanobacterium pyogenes that do not contain ermB but carry an intact orfY and lack any trace of trsK. As noted above, the homology between the A. pyogenes element and CTnBST begins with an ORF immediately upstream of the first orfY. Taken together, these findings suggest that this element came together in Arcanobacterium pyogenes and subsequently, through an as yet unknown mechanism, transferred itself to Bacteroides spp., either directly or indirectly.
Identifying possible CTns in genome sequences has proven to be a challenge because of the very limited number of CTn sequences that are currently available. One way to locate a foreign integrated element has been to calculate the % G+C content, looking for regions that differ from the rest of the chromosome. This is the approach that has been used to identify possible pathogenicity islands (12). Two problems with this approach are illustrated by the CTnBST sequence. First, the % G+C content of most of CTnBST is higher than, but not radically different from, that of the chromosomes of sequenced Bacteroides strains. However, this difference may be significant enough to imply that this part of CTnBST is not native to Bacteroides species. Second, the abrupt drop in % G+C in the middle of CTnBST, which is clearly different in % G+C content, could indicate the presence of an integrated region of some other origin or it could indicate the presence of two CTns, one on either side of the low-% G+C region. It is not clear simply from the % G+C content whether these disparate segments transfer as a unit or as separate units. Either way the drastic change in G+C content (from 48% to 36% G+C) implies that this particular segment of CTnBST is of a different origin than the rest of CTnBST. It is interesting that gram-positive organisms such as Clostridium spp., Enterococcus spp., and Streptococcus spp. have a % G+C content between 28 and 39%. And, as stated above, most of the ORFs in this region show a high level of identity to genes from these organisms. This would imply that this 7-kb ermB region, while it may have entered Bacteroides species via A. pyogenes, did not originate in A. pyogenes, which is a high-G+C gram-positive organism (9). The fact that the same regions from both organisms are 100% identical provides further evidence that CTnBST has picked up an element(s) from a gram-positive organism during its evolution. It also seems likely that the region upstream of the 7-kb ermB region may itself have come in on another gram-positive element. The % G+C content in this area is similar to that of the rest of CTnBST, but many of the ORFs had a high level of identity to gram-positive organism genes. In addition, this area contains a mixture of Bacteroides homologs (a few from CTn3-Bf) and gram-positive organism homologs. Thus, this region may have been present in CTnBST long enough to reshuffle genes and/or to acquire a Bacteroides type codon usage.
A second approach for identifying CTns in a genome sequence is to look for genes known to be associated with known CTns, like transfer (tra) genes. Indeed, this was how CTn3-Bf was identified (10). However, it should be noted that, while the annotators of the YCH46 genome sequence identified CTn3-Bf as a CTn on the basis that some of the sequences encoded by this region had amino acid sequence similarity to known CTn transfer proteins, no evidence was presented to show that this is an active CTn.
CTnBST illustrates a theme that is beginning to emerge among the CTs: their chimeric structure. For example, the region carrying the ermB gene in CTnBST, which extends for at least 7 kb, appears to have come from A. pyogenes or some close relative. Something like this has been seen in CTnDOT, where a 13-kbp region carrying the ermF gene seems to have entered the larger CTn because a very closely related CTn, CTnERL, is virtually identical to CTnDOT except for the addition of the 13-kbp segment in CTnDOT (22). Apparently, the structures of CTns can be very fluid.
CTnBST is only the second type of CTn from the genus Bacteroides that has been sequenced. The other type of sequenced Bacteroides CTn is represented by CTnDOT and CTn341, two CTns with very similar sequences. Other sequenced CTns are Tn916, the enterococcal CTn, and the STX element of Vibrio cholerae, if indeed the STX element is a CTn. As seen from this report, some of the regions of genome sequences tentatively identified as CTns but not shown to be transmissible may prove to be active CTns. If so, the number of CTn sequences will multiply considerably because multiple possible cryptic CTns have been identified in the genome sequences of all three of the sequenced Bacteroides spp. and in the emerging genome sequences of Porphyromonas and Prevotella spp.
It is interesting that the genes on CTnBST that are most like those on CTnDOT are the transfer genes, especially a gene called traG. The fact that the CTnBST sequence is otherwise so different from the sequence of CTnDOT reinforces the use of tra genes to identify a possible CTn, at least in the Bacteroides group of genera. CTnBST turned out to be closely related to a tentatively identified CTn in B. fragilis YCH46, CTn3-Bf. There was no evidence that this putative B. fragilis CTn is transmissible, but the similarity between it and CTnBST, which does excise, suggests that it may be or may have been transmissible. As more CTn sequences become available, it will be easier to identify CTns more precisely in the emerging genome sequences.
Acknowledgments
We thank Stephen J. Billington for providing us with purified chromosomal DNA from Arcanobacterium pyogenes.
This work was supported by a grant from the Ellison Foundation (IDSS-042703) and by a grant from the National Institutes of Health (AI/G M22383).
Footnotes
Published ahead of print on 4 May 2007.
REFERENCES
- 1.Backhed, F., R. E. Ley, J. L. Sonnenburg, D. A. Peterson, and J. I. Gordon. 2005. Host-bacterial mutualism in the human intestine. Science 307:1915-1920. [DOI] [PubMed] [Google Scholar]
- 2.Besemer, J., and M. Borodovsky. 2005. GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 33:W451-W454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brisson-Noel, A., M. Arthur, and P. Courvalin. 1988. Evidence for natural gene transfer from gram-positive cocci to Escherichia coli. J. Bacteriol. 170:1739-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng, Q., B. J. Paszkiet, N. B. Shoemaker, J. F. Gardner, and A. A. Salyers. 2000. Integration and excision of a Bacteroides conjugative transposon, CTnDOT. J. Bacteriol. 182:4035-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finegold, S. M. 1995. Overview of clinically important anaerobes. Clin. Infect. Dis. 20(Suppl. 2):205-207. [DOI] [PubMed] [Google Scholar]
- 6.Gill, S. R., M. Pop, R. T. Deboy, P. B. Eckburg, P. J. Turnbaugh, B. S. Samuel, J. I. Gordon, D. A. Relman, C. M. Fraser-Liggett, and K. E. Nelson. 2006. Metagenomic analysis of the human distal gut microbiome. Science 312:1355-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta, A., H. Vlamakis, N. Shoemaker, and A. A. Salyers. 2003. A new Bacteroides conjugative transposon that carries an ermB gene. Appl. Environ. Microbiol. 69:6455-6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holdeman, L. V., E. P. Cato, et al. 1977. Anaerobe laboratory manual. Anaerobe Laboratory, Blacksburg, VA.
- 9.Jost, B. H., H. T. Trinh, J. G. Songer, and S. J. Billington. 2004. A second tylosin resistance determinant, ErmB, in Arcanobacterium pyogenes. Antimicrob. Agents Chemother. 48:721-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuwahara, T., A. Yamashita, H. Hirakawa, H. Nakayama, H. Toh, N. Okada, S. Kuhara, M. Hattori, T. Hayashi, and Y. Ohnishi. 2004. Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaptation. Proc. Natl. Acad. Sci. USA 101:14919-14924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li, L. Y., N. B. Shoemaker, and A. A. Salyers. 1993. Characterization of the mobilization region of a Bacteroides insertion element (NBU1) that is excised and transferred by Bacteroides conjugative transposons. J. Bacteriol. 175:6588-6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lio, P., and M. Vannucci. 2000. Finding pathogenicity islands and gene transfer events in genome data. Bioinformatics 16:932-940. [DOI] [PubMed] [Google Scholar]
- 13.Marra, D., and J. R. Scott. 1999. Regulation of excision of the conjugative transposon Tn916. Mol. Microbiol. 31:609-621. [DOI] [PubMed] [Google Scholar]
- 14.Moore, W. E., E. P. Cato, and L. V. Holdeman. 1978. Some current concepts in intestinal bacteriology. Am. J. Clin. Nutr. 31:S33-S42. [DOI] [PubMed] [Google Scholar]
- 15.Reeves, A. R., J. N. D'Elia, J. Frias, and A. A. Salyers. 1996. A Bacteroides thetaiotaomicron outer membrane protein that is essential for utilization of maltooligosaccharides and starch. J. Bacteriol. 178:823-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saito, H., and K. I. Miura. 1963. Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochim. Biophys. Acta 72:619-629. [PubMed] [Google Scholar]
- 17.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 18.Shoemaker, N. B., H. Vlamakis, K. Hayes, and A. A. Salyers. 2001. Evidence for extensive resistance gene transfer among Bacteroides spp. and among Bacteroides and other genera in the human colon. Appl. Environ. Microbiol. 67:561-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simon, R., U. Priefer, and A. Piihler. 1982. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 20.Valentine, P. J., N. B. Shoemaker, and A. A. Salyers. 1988. Mobilization of Bacteroides plasmids by Bacteroides conjugal elements. J. Bacteriol. 170:1319-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wesslund, N. A., G. R. Wang, B. Song, N. B. Shoemaker, and A. A. Salyers. 2007. Integration and excision of a newly discovered bacteroides conjugative transposon, CTnBST. J. Bacteriol. 189:1072-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whittle, G., B. D. Hund, N. B. Shoemaker, and A. A. Salyers. 2001. Characterization of the 13-kilobase ermF region of the Bacteroides conjugative transposon CTnDOT. Appl. Environ. Microbiol. 67:3488-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson, K. H., and R. B. Blitchington. 1996. Human colonic biota studied by ribosomal DNA sequence analysis. Appl. Environ. Microbiol. 62:2273-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]