Abstract
The diversity of microorganisms active within sedimentary rocks provides important controls on the geochemistry of many subsurface environments. In particular, biodegradation of organic matter in sedimentary rocks contributes to the biogeochemical cycling of carbon and other elements and strongly impacts the recovery and quality of fossil fuel resources. In this study, archaeal diversity was investigated along a salinity gradient spanning 8 to 3,490 mM Cl− in a subsurface shale rich in CH4 derived from biodegradation of sedimentary hydrocarbons. Shale pore waters collected from wells in the main CH4-producing zone lacked electron acceptors such as O2, NO3−, Fe3+, or SO42−. Acetate was detected only in high-salinity waters, suggesting that acetoclastic methanogenesis is inhibited at Cl− concentrations above ∼1,000 mM. Most-probable-number series revealed differences in methanogen substrate utilization (acetate, trimethylamine, or H2/CO2) associated with chlorinity. The greatest methane production in enrichment cultures was observed for incubations with salinity at or close to the native pore water salinity of the inoculum. Restriction fragment length polymorphism analyses of archaeal 16S rRNA genes from seven wells indicated that there were links between archaeal communities and pore water salinity. Archaeal clone libraries constructed from sequences from 16S rRNA genes isolated from two wells revealed phylotypes similar to a halophilic methylotrophic Methanohalophilus species and a hydrogenotrophic Methanoplanus species at high salinity and a single phylotype closely related to Methanocorpusculum bavaricum at low salinity. These results show that several distinct communities of methanogens persist in this subsurface, CH4-producing environment and that each community is adapted to particular conditions of salinity and preferential substrate use and each community induces distinct geochemical signatures in shale formation waters.
Subsurface sedimentary environments contain an estimated 1/10 to 1/3 of all living biomass on Earth, and the activity of subsurface microorganisms plays an important role in biogeochemical cycling of elements and the global carbon cycle (38, 51, 57). Study of the activity of organisms in deeply buried sedimentary rocks in particular helps inform our understanding of the persistence of isolated microbial communities over geologic time and the cumulative impact that long, sustained activity may have on geochemical signatures trapped in ancient rocks. In certain environments, subsurface biological activity may be directly linked with atmospheric and climate changes. Among these environments are sites where methanogenesis occurs in sedimentary basins, in which activity may be stimulated by a changing surficial hydrologic cycle and release of CH4 may impact the atmospheric composition and the global climate. The Michigan Basin is a ∼600-km-wide, 4-km-deep stratigraphic basin comprising sedimentary rocks ranging from 540 to 145 million years old. Within the rock formations of the basin is the ∼380-million-year-old Antrim Shale, a fine-grained, laminated shale with high concentrations of organic carbon and pyrite. Intense natural gas production has been developed along the northern margin of the basin, where CH4 abundance is greatest; at present, over 10,000 gas- and water-producing wells have been installed in the Antrim Shale along a 50 km-wide north-south band extending across the entire Lower Peninsula of Michigan (Fig. 1).
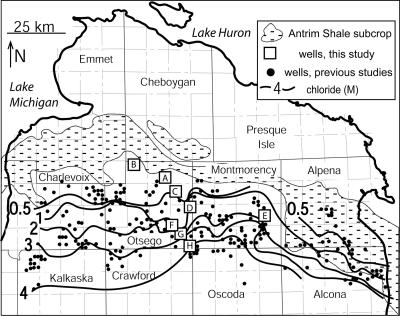
FIG. 1.
Map of study area in northern Michigan, showing the locations of wells used in this study and wells examined in a previous geochemical study, the contours of the Cl− concentration within Antrim Shale formation waters, and the subcrop of Antrim Shale beneath glacial till.
Multiple lines of geochemical evidence suggest that there is or has been substantial biological activity within the Antrim Shale and also indicate that CH4 produced along the northern margin of the basin is primarily biological in origin (29, 30, 31, 34, 35). Pore waters collected from wells along the CH4-rich northern margin of the basin exhibit elevated alkalinity (20 to 60 meq liter−1) which is greater than the alkalinity that can be supported by dissolution of carbonate minerals; this suggests that there is a source of inorganic carbon derived from remineralization of organic matter (29, 30). Gas produced from the Antrim Shale is almost entirely lacking in higher-chain hydrocarbons such as ethane, propane, and butane, which are generated along with CH4 during thermal decomposition of organic matter-rich sedimentary rocks (29, 30, 34, 35). Stable isotope (D/H) ratios of Antrim Shale H2O and CH4 reflect isotopic fractionation induced during methanogenesis (29, 30), as do comparisons of δ13C ratios for coproduced CO2 and CH4 (31).
Antrim Shale pore waters exhibit a sharp salinity gradient ranging from extremely dilute water along the margins of the basin to an NaCl concentration greater than 5 M at the center of the basin (30). The dissolved salts are dominated by chloride and sodium, with lesser contributions from Mg, Ca, and bicarbonate (30). Fluid flow and diffusion are restricted in the Antrim Shale due to very low matrix porosity (34, 35). The salinity gradient in the pore waters appears to correlate with methane production, and there is extremely limited CH4 production from wells with salinity above ∼4 M Cl−. Low salinity in the Antrim Shale results from dilution of deep basin brines with meteoric waters, associated with deep groundwater recharge during Pleistocene glaciation events (34). As Antrim Shale CH4 is isotopically linked with glacial-age waters, methanogenesis in the Antrim Shale may be a geologically recent occurrence (30).
While many methanogens are inhibited by increased NaCl concentrations (5, 26, 36, 47), there are halophilic methanogens that use methanol, methylamine compounds, or dimethyl sulfide to generate methane (43). These substrates yield more energy (−78.7 to −191.1 kJ per mol substrate) than H2/CO2 (−34 kJ/mol substrate) or acetate (−31 kJ mol substrate), which likely allows methylotrophic methanogens to grow at higher salt concentrations by offsetting the energy cost of the increased osmoregulatory burden (44). In subsurface settings, hydrogenotrophic and acetoclastic methanogens are more commonly described (6, 13, 20, 22, 45, 56) but generally are limited to lower-salinity environments than methanogens capable of using methanol and methylamines (5, 40). Methanocalculus halotolerans is the most halotolerant hydrogenotrophic methanogen known and can survive NaCl concentrations in culture of up to ∼2.3 M (40); this organism was isolated from oil field brine with 2,175 mM NaCl. Acetoclastic methanogenesis persists at Cl− concentrations up to ∼1,000 mM, but halotolerance data for the methanogens are scarce (44).
The purpose of this study was to investigate if the diversity of methanogens in the Antrim Shale is associated with salinity. The methane-producing zone in the Antrim Shale covers the entire salinity range known to support methanogenesis, from freshwater to water containing 4,000 mM Cl−. Bacterial rRNA gene sequencing efforts for the Antrim Shale have shown that a variety of fermentative, syntrophic, and homoacetogenic bacteria inhabit the shale (32; M. L. Formolo, A. M. Martini, K. Nüsslein, J. Salacup, and S. T. Petsch, submitted for publication). These organisms are believed to convert bioavailable components of shale organic matter into substrates for methanogenesis, such as H2, CO2, acetate, and methylamines. Because different methanogens require different substrates (acetate, H2, methanol, methylamines), salinity may indirectly control the diversity of organisms responsible for decomposition of shale organic matter and production of substrates necessary for methanogenesis. As the Antrim Shale represents a methane source that may prove to be critical for both ancient climate change and modern resource recovery, salinity constraints on methanogenesis are critical for understanding this and other accumulations of biogenic methane in sedimentary basins.
MATERIALS AND METHODS
Sample collection.
Eight wells that produce gas and water from the Antrim Shale were sampled in June 2005 along a north-south transect in northern Michigan, spanning a gradient from dilute waters in the north to highly saline waters in the south (Fig. 1). In all cases, waters were drawn from within the actively pumping well stream to avoid contact with the atmosphere. Samples of cellular material were collected at each well head by drawing well water into a sterile 60-ml disposable syringe and filtering it through a 0.22-μm (nominal pore size) cellulose acetate filter (Millipore, Billerica, MA) housed in a presterilized 25-mm Swinnex filter holder. Filters used for DNA extraction were frozen on dry ice, transported to the laboratory, and maintained at −80°C until they were processed. Parallel filters used for cell counting were fixed on site by flushing each filter with 5 ml of a solution of 4% paraformaldehyde in phosphate-buffered saline at pH 7.2, incubated at 4°C for 6 h, and then rinsed with 50 ml of a 1:1 ethanol/phosphate-buffered saline solution through the filter holder. The phosphate-buffered saline solution comprised 130 mM NaCl, 15 mM Na2HPO4, and 5 mM NaH2PO4, adjusted to pH 7.2. After rinsing, filter holders were filled with a sterile 1:1 ethanol/phosphate-buffered saline solution, capped, stored on dry ice, transported to the laboratory, and maintained at −30°C until analysis. Enrichment cultures were initiated on site by drawing well water from the actively pumping well stream and slowly dispensing it into prepared serum vials (see “Enrichment cultures” below). Replicate water samples for geochemical analysis were collected from each well head, filter sterilized with 0.22-μm nylon filters on site, and stored in 125-ml high-density polyethylene bottles at 4°C until analysis; samples used for cation analysis were acidified to pH 2 with HNO3 upon collection, while samples used for alkalinity, acetate, and anion analysis were not acidified. The temperature and pH of well waters were measured on site. Additional water samples were collected for most-probable-number (MPN) series analysis by completely filling gas-tight 250-ml polyethylene terephthalate glycol bottles with well water and shipping them at 4°C overnight to the laboratory. These bottles were transferred into an anaerobic chamber, and the contents were dispensed into serum vials capped with butyl rubber stoppers.
Enrichment cultures.
Medium for the enrichment of Antrim Shale microorganisms was developed to mimic the geochemistry of water collected from methane-producing Antrim Shale wells. The media consisted of a degassed basal salts solution containing CaCl2·2H2O (3.6 mM), MgCl2·6H2O (1.6 mM), NH4Cl (1.87 mM), KH2PO4 (0.73 mM), NaBr (0.23 mM), NaHCO3 (25 mM), trace vitamin and mineral solutions (DSMZ media 141 and 318, respectively) (http://www.dsmz.de/), and resazurin (1 mg/liter) and were buffered with 2-morpholinoethanesulfonic acid (10 mM) to pH 6.1. General enrichment media contained the following organic carbon sources: sodium acetate (10 mM), sodium formate (1 mM), trimethylamine (10 mM), methanol (50 mM), yeast extract (0.1%), and tryptic peptone (0.1%). Other than CO2, no external electron acceptors were supplied. Fifteen milliliters of medium was dispensed into 50-ml glass serum bottles, which were capped with butyl rubber stoppers. The medium was degassed by sparging with 80:20 N2/CO2. Fifteen milliliters of medium was dispensed into 50-ml glass serum bottles, which were capped with butyl rubber stoppers and autoclaved, and then the medium was reduced by addition of Na2S·9H2O and cysteine (0.03% each). The headspace was pressurized to 160 kPa with 80:20 H2/CO2 to approximate the measured pore water alkalinity. To determine the methane production and salinity tolerance of different communities along the salinity gradient, a suite of enrichments for fermentative and methanogenic organisms was established by amending the general enrichment medium with NaCl at concentrations of 20, 100, 250, 500, 750, 1,000, 1,500, 2,000, 2,500, and 4,000 mM. Enrichments were inoculated with 1% (vol/vol) Antrim Shale well water at the wellhead. Thus, well water samples were dispensed into medium at each salinity, yielding a matrix of well salinity/enrichment salinity incubations. All incubations occurred at 17°C in the dark and were maintained in triplicate for 6 months.
MPN series.
MPN dilutions were employed to determine the relative numbers of viable, culturable methanogens capable of using different methanogenic substrates present in two wells (well C, 200 mM Cl−; well D, 1,041 mM Cl−). The medium was set up as described above, except that the carbon sources were limited to 0.005% yeast extract and one of the following: sodium acetate (10 mM), trimethylamine (10 mM), or 80:20 H2/CO2 at 160 kPa overpressure in the headspace. Dilutions without H2/CO2 were overpressured with 160 kPa N2/CO2. NaCl concentrations were amended to approximate field conditions, and pore water was serially diluted from 1:10 to 1:10,000. Preparations were kept at room temperature in the dark for 2 months. Growth of methanogens was confirmed by microscopic analysis and by determining the increase in the headspace methane concentration (see “Geochemical analyses” below), and MPN series cell densities were calculated based on dilution to extinction using the software MPN Calculator (version VB6; http://www.i2workout.com/mcuriale/mpn/index.html).
Enumeration of cells.
Fixed filters containing cellular material were removed from Swinnex filter holders, mounted in a glass vacuum filter holder, and wetted with 1 ml phosphate-buffered saline solution. A vacuum was applied to near dryness, and filters were then stained with 500 μl of a 1-μg/ml DAPI (4′,6-diamidino-2-phenylindole) solution in phosphate-buffered saline solution in the dark for 2 min. The filters were washed in 10 ml phosphate-buffered saline solution to remove excess DAPI, dried by applying a gentle vacuum, and examined by epifluorescence microscopy. Cell counts were expressed as the average and standard deviation for 20 field counts per slide, using a 100-μm-diameter field area, a 24-mm-diameter filter area, and 60 ml (total volume) of a filtered water sample. The presence of strongly fluorescing methanogens was confirmed on unstained filters as autofluorescence of cells by excitation at 420 nm; as several groups of methanogens, especially those that utilize acetate and methylated substrate, contain only low levels of the F420 cofactor, these groups are underrepresented by autofluorescence analysis.
DNA extraction and amplification.
DNA was extracted from filters from each well using a QIAamp DNA stool mini kit (QIAGEN, Valencia, CA) and a modified procedure. Briefly, filters were incubated in ASL lysis buffer (QIAGEN, Valencia, CA) at 90°C for 8 min and then vortexed for 20 s. Each filter was removed from the buffer, and for the remaining procedure the protocol provided by QIAGEN was used. DNA was quantified spectrophotometrically using a low-volume system (Nano-Drop, Wilmington, DE) and used at full strength in subsequent PCRs. For a positive control, DNA was extracted from Methanosarcina barkeri DSM 800T (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany) by the same methods. Nucleic acid was successfully extracted from seven of the eight wells. No nucleic acid was recovered from the most saline well sample; extraction of new blank filters also yielded no nucleic acid.
Archaeal 16S rRNA genes from each well and from the M. barkeri control extract were amplified using a nested PCR approach. The first round of PCR used the Archaea-specific primer 21F (5′-TCC GGT TGA TCC YGC C-3′) (8) and the universal primer 1492R (5′-TAC GGY TAC CTT GTT ACG ACT T-3′) (55). The second round used Archaea-specific primers 109F (5′-ACK GCT CAG TAA CAC GT-3′) and 915R (5′-GTG CTC CCC CGC CAA TTC CT-3′) (14). One microliter of sample DNA was used as the template in a 30-μl PCR cocktail containing 2 mM MgCl2 (Sigma, St. Louis, MO), 0.50 μM forward and reverse primers (IDT, Coralville, IA), 0.08 U/μl Taq polymerase, 400 ng/μl bovine serum albumin, and 0.25 mM of each deoxynucleoside triphosphate (all obtained from Promega, Madison, WI). Reactions were performed in a Mastercycler Personal 5332 thermocycler (Eppendorf, Westbury, NY) with the following program: 95°C for 3 min; 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min; and 72°C for 10 min. The resulting PCR product (826 bp) was quantified by comparison to a 100-kb DNA ladder (New England Biolabs, Ipswich, MA) and was gel purified using a MinElute gel extraction kit (QIAGEN, Valencia, CA).
Restriction fragment length polymorphism (RFLP) analysis.
PCR products of 16S rRNA genes obtained from each well and from the M. barkeri control were digested with restriction enzyme HhaI, MspII, or MnlI or a combination of HhaI and MnlI (New England Biolabs, Ipswich, MA) by following the manufacturer's instructions and were electrophoretically separated on an 8% polyacrylamide gel. Gels were stained with 5× SYBR green 1 (Applied Biosystems, Foster City, CA), visualized under UV light, and photographed. Bands were identified on each gel and summarized into a matrix form where rows consisted of well names and columns indicated base sequence length corresponding to individual bands from each restriction digest, determined by comparison with a 100-bp ladder (New England Biolabs, Ipswich, MA). A value of 1 or 0, indicating the presence or absence of a band, respectively, was entered into each cell. Cluster analysis was performed using a Sorensen index distance measure and the nearest-neighbor linkage method based on the software package PC-ORD v. 4.30 (33).
Phylogenetic analysis.
PCR products from well B (12.8 mM Cl−) and well G (2,269 mM Cl−) were chosen for cloning and sequencing. DNA extraction and nested PCR amplification of the 16S rRNA gene were performed as described above, with PCRs using a DNA template from each well performed in triplicate and subsequently pooled. PCR products were cloned into Escherichia coli JM109 competent cells using a pGEM-T Easy vector system (Promega, Madison, WI) according to the manufacturer's instructions. DNA from 150 randomly picked clones was sequenced with primer 21F using a 3730xl DNA analyzer (Applied Biosystems, Inc., Foster City, CA). Sequences were manually edited in Chromas v2.31 (Technelysium Pty. Ltd., Tewantin, Queensland, Australia) and were aligned using the program ClustalX v. 1.83 (52) in the software package BioEdit v.7.0.5 (15). Chimeric sequences were identified using Bellerophon (17), the Chimera_Check program from the Ribosomal Database Project (28), and the software package Mallard (3). Sequences found to be of potential chimeric origin through the analyses described above were excluded from further examination. Operational taxonomic units (OTUs) were assigned using the program DOTUR (48) and a 97% sequence similarity criterion for identical phylotypes. The nearest relative of each OTU was determined by a BLAST search against the NCBI 16S rRNA GenBank database (http://www.ncbi.nlm.nih.gov/BLAST/). Minimum-evolution trees using a Tamura-Nei substitution model were created, and a bootstrap test of phylogeny with 1,000 replicates was performed using the software MEGA v.3.1 (23). SACE and SChao1, which estimate the diversity present in the environment based on sampling, and rarefaction curves were calculated using DOTUR (48).
Geochemical analyses.
Anions in water samples were analyzed by ion chromatography using an AS14 anion column with suppressed conductivity detection (Dionex, Marlton, NJ). Cations were analyzed by inductively coupled plasma-atomic emission spectroscopy (Teledyne-Leeman Labs, Hudson, NH). Concentrations of cations and anions were determined by comparison with gravimetric standards. Alkalinity was determined by endpoint titration, performed within hours of sample collection (12). Concentrations of enrichment culture headspace methane were measured by gas chromatography using a GC-8A (Shimadzu, Columbia, MD) equipped with a thermal conductivity detector and a 100/120 Carbosieve SII column (Supelco, St. Louis, MO) and were quantified by comparison with headspace standards. Total dissolved organic carbon concentrations were determined by catalytic high-temperature combustion using a Shimadzu TOC-VCPH carbon analyzer (Shimadzu USA, Columbia, MD). Acetate concentrations in water samples were determined by solid-phase microextraction/gas chromatography using a method adapted from the method of Ábalos et al. (1). Briefly, volatile fatty acids in an acidified pore water sample were adsorbed to a Carbo-Wax fused-silica fiber and injected into an HP 6890 gas chromatograph equipped with a 30-m NUKOL capillary column and a flame ionization detector; quantification was achieved by comparison with volumetric standards of sodium acetate.
Nucleotide sequence accession numbers.
The phylotypes detected in this study are available from the GenBank nucleotide database (www.ncbi.nlm.nih.gov) under accession numbers DQ830727 to DQ830735.
RESULTS
Geochemistry.
The chlorinity in the eight well water samples ranged from 8 to 3,490 mM Cl−, and the Na+ concentrations ranged from 47 to 1,755 mM (Table 1). The temperature ranged from 14.1 to 15.8°C, and the pH ranged from 5.6 to 8.0. Degassing of CO2 from waters during sampling causes a rise in the pH, especially in waters with elevated alkalinity that are CO2 supersaturated at atmospheric pressure. Thus, pH values greater than ∼7 are likely to be overestimates of the real subsurface conditions. Modeling of the formation water pH using measured water chemistry and Geochemist's Workbench software (Rockware Inc., Golden, CO) predicts a pH range between 5.6 and 7.0. Nonetheless, pH does vary with salinity in this environment and may be responsible for some component of community variability. All wells lacked measurable concentrations of nitrate. The total dissolved Fe concentrations ranged from submicromolar to 2,397 μM, and there was a sharp decrease in the Fe concentration in wells with Cl− concentrations less than 1,000 mM. At circumneutral pH, an appreciable concentration of dissolved free Fe(III) is not supported. A limited amount of total Fe in solution may occur as chelated Fe(III) associated with dissolved organic matter. However, ample pyrite in the shale should buffer the electrochemical potential and preclude accumulation of ferric iron species, and the total dissolved Fe concentrations were greater than the dissolved organic carbon concentrations in all wells with water containing more than 1,000 mM Cl−. The two wells with the lowest salinity exhibited submillimolar SO42− concentrations; the SO42− concentration was below the detection limit, ∼25 μM, in the remaining six wells. With the exception of well B (170 μM SO42−), the characteristic aroma of H2S was absent in all well samples. The alkalinity ranged from 6.3 to 39.5 meq liter− and was negatively correlated with the Cl− concentration (r2 = 0.94). Acetate was detected in four of eight water samples (detection limit, ∼5 μM) at concentrations ranging from 57 to 549 μM. No acetate was detected in wells with water containing less than 1,000 mM Cl−. Thus, while a biological source of acetate in these wells was supported, acetate consumption appeared to be inhibited at higher salinities. The lack of measurable acetate in the well with the highest salinity is consistent with a lack of biological activity at this highly saline site.
TABLE 1.
Sample information, geochemical data, direct cell counts, and nucleic acid extraction
| Well | Depth (m)a | Temp (°C) | pH | Alkalinity (meq liter−1) | [Cl−] (mM) | [Na+] (mM) | [Fe] (mM) | [SO42−] (μM) | [Dissolved organic carbon] (μM) | [Acetate] (μM) | Total counts (103 cells ml−1) | [DNA] (ng ml−1) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 302 | 15.8 | 7.96 | 32.1 | 8.4 | 60.0 | 0.021 | 74 | 169.5 | BDLb | 6.2 | 0.004 |
| B | 280 | 12.3 | 7.49 | 39.5 | 12.8 | 46.5 | 0.0005 | 170 | NDc | BDL | 6.3 | 0.008 |
| C | 296 | 14.4 | 7.59 | 34.1 | 200 | 163 | 0.027 | BDL | 803.4 | BDL | 5.1 | 0.020 |
| D | 442 | 14.5 | 6.71 | 29.8 | 1,041 | 1,617 | 0.337 | BDL | 326.8 | 57 | 8.1 | 0.005 |
| E | 412 | 14.1 | 6.36 | 24.3 | 1,403 | 1,755 | 2.40 | BDL | ND | 199 | 6.1 | 0.009 |
| F | 479 | ND | 6.54 | 25.1 | 1,021 | 970 | 1.24 | BDL | 383.9 | 549 | 4.9 | 0.008 |
| G | 511 | 14.3 | 5.86 | 12.3 | 2,269 | 1,078 | 2.25 | BDL | 307.9 | 306 | 5.3 | 0.011 |
| H | 520 | 14.4 | 5.64 | 6.3 | 3,490 | 1,363 | 1.83 | BDL | 544.3 | Minord | BDL | BDL |
Depth below the land surface.
BDL, below the detection limit.
ND, not determined.
A small acetate peak was detected at the limit of detection (∼5 μM).
Enrichment cultures.
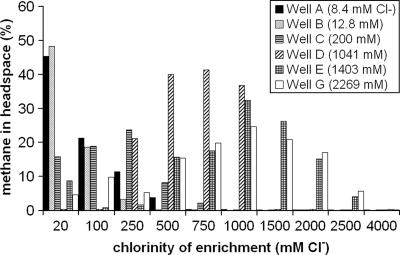
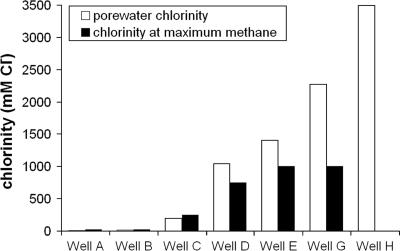
Growth in methanogenic enrichment cultures, assessed as visible turbidity, was observed within 2 to 3 days after initiation. Turbidity was observed in all incubations except those initiated from well H (3,490 mM Cl−). Additionally, no growth was observed in incubations maintained at 2,500 and 4,000 mM Cl− for any of the sampled wells. Incubations derived from wells A through D (8 to 1,041 mM Cl) exhibited turbidity in enrichments with Cl concentrations up to 1,000 mM; turbidity was observed in incubations for well G (2,269 mM Cl−) at up to 2,500 mM Cl−. The headspace gas content of enrichments was measured 6 months after sampling, and the results showed distinct relationships between the headspace methane content, the incubation salinity, and the chloride concentration of the well from which each enrichment series was initiated (Fig. 2). Overall, incubations derived from wells with low salinity generated appreciable methane only at low salinity, while higher-salinity well waters yielded methane over a much greater salinity range, up to 2,500 mM Cl− in the case of incubations derived from well G. When the pore water Cl− concentration of each well was compared to the Cl− concentration in the enrichment derived from that well with the greatest headspace methane concentration, the values were very similar for five of six CH4-producing enrichments (the exception was well G), indicating that the methanogenic community in each well was adapted to or optimized for its native pore water Cl− concentration (Fig. 3). The exception may indicate that there was a shift in the enrichment community composition compared with the inoculum composition in response to high availability of H2 and CO2 in the enrichment or to other enrichment bias. The concentrations of headspace methane in enrichments derived from well H (3,490 mM Cl−) enrichments were below the detection limit (0.1%), demonstrating that methane was not produced in enrichments that exhibited no turbidity.
FIG. 2.
Methane concentrations in enrichments derived from six wells along the Antrim Shale salinity gradient, measured 6 months after initiation. No methane was detected in enrichments derived from highly saline well H (data not shown). The wells and chlorinities of shale pore waters from which enrichments were initiated are indicated.
FIG. 3.
Chloride concentrations of pore waters from seven wells along a salinity gradient compared to the chloride concentrations of the enrichments with the greatest methane production (highest values for seven chloride concentrations in enrichments). No methane was detected in any incubation derived from well H.
Enumeration and nucleic acid extraction.
Microbial cell densities, determined by direct microscopic enumeration, were low but relatively constant along the gradient in seven of the eight wells examined, and the average was approximately 6 × 103 cells ml−1 pore water. Cells were not observed in well H (3,490 mM Cl−), which corresponded to a lack of extractable DNA for this well. The extractable DNA yields from all wells were low, averaging 0.01 ng ml−1.
MPN series.
MPN enrichments for medium-low-salinity well C (200 mM Cl−) and medium-high-salinity well D (1,041 mM Cl−) yielded low counts of culturable cells for all three methanogen substrates examined (Table 2). The numbers of culturable cells are much lower than the direct counts. However, epifluorescence microscopy of fixed water sample filters showed that only 1 to 5% of the total cell population exhibited autofluorescence, indicating that strongly fluorescing methanogens comprised only a small fraction of the total microbial community. Methanogens that grew in the presence of acetate, H2, or trimethylamine could all be cultured from well C, and the greatest number of cells were able to use H2. The relatively high number of cells cultured using trimethylamine may reflect the substrate versatility of many members of the Methanosarcinales (as suggested by cloning results presented below) and may not reflect the fact that trimethylamine is an available substrate in this environment. Note that the low-salinity well used as a sample source for the MPN series (well C) is not the same well that was used for 16S rRNA gene analysis (well B). MPN series showed growth only on trimethylamine for the higher-salinity well D. The limited growth in MPN series dilutions for well D is surprising given the relatively high methane production observed in enrichment cultures derived from this well (Fig. 2). Negative controls which contained only 0.005% yeast extract as a carbon source did not yield any methane.
TABLE 2.
MPN dilution series results, expressed as the estimated range of cell concentrations at the 95% confidence level determined by using MPN Calculator (version VB6)a
| Well | [Cl−] (mM) | Cells ml−1 with the following methanogen substrates:
|
||
|---|---|---|---|---|
| H2/CO2 | Acetate | Trimethylamine | ||
| C | 200 | 60-250 | 2-5 | 15-50 |
| D | 1,041 | 0-2 | 0-2 | 6-25 |
RFLP analysis.
Analysis of restriction fragment lengths revealed strong associations of fragment patterns for wells with similar salinities. With a few exceptions, wells clustered according to pore water salinity; an example is wells F (1,021 mM Cl−) and D (1,041 mM Cl−) (Fig. 4). Wells also appeared to cluster based on salinity instead of geographic proximity, as well G (2,269 mM Cl−) clustered with well E (1,403 mM Cl−) despite a geographic location much nearer well F (1,021 mM Cl−).
FIG. 4.
Distance dendrogram based on RFLP analyses of archaeal 16S rRNA genes from eight sampled wells. The chloride concentration determined for each well is indicated in parentheses. The distance bar indicates the proportion of unique RFLP fragments for two wells.
Cloning and phylogenetic analysis.
Partial 16S rRNA gene sequences from wells B (13 mM Cl−) and G (2,268 mM Cl−) were selected for cloning and sequence analysis. These wells were chosen because they represent the most northern and most southern methane-producing wells along the salinity gradient, have very dissimilar pore water salinities, and exhibited divergent archaeal communities based on the RFLP dendrogram. From the clone library, 89 randomly chosen, nonchimeric sequences were obtained from well B, and 155 sequences were obtained from well G. The individual libraries had no overlapping sequences.
Phylogenetic analysis placed all clones within the Euryarchaeota (Fig. 5). A single archaeal phylotype was observed for well B and grouped within the Methanomicrobiales; the estimated diversity is 1 (SChao1, 100% coverage). This phylotype was closely related to the cultured methanogen Methanocorpusculum bavaricum, with up to 99.5% sequence similarity. Members of the family Methanocorpusculaceae exhibit metabolic versatility and are known to use H2/CO2, formate, and some secondary alcohols to make methane (59). The MPN series for well C indicated use of acetate and trimethylamine substrates, functions which are unknown among the Methanocorpusculaceae. Although wells B and C exhibit similar salinities and pHs, they are separated by 25 km. Thus, we cannot rule out the possibility that these wells contain different communities with different substrate requirements. Sequences related to known acetoclastic methanogens were not found in the well B library.
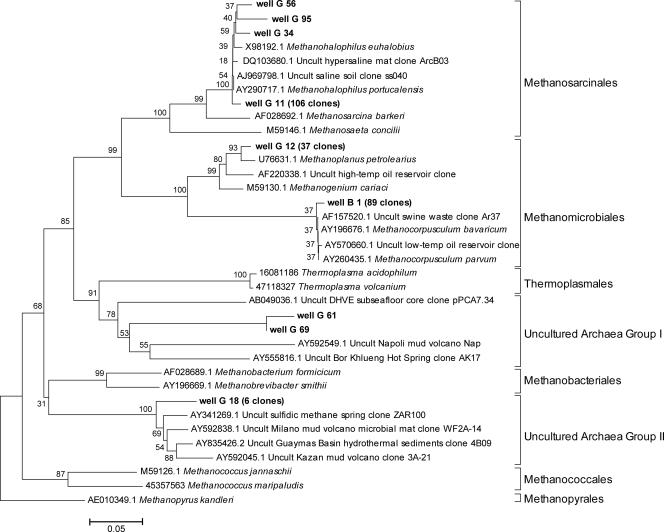
FIG. 5.
Distance tree for archaeal 16S rRNA gene sequences from well B (12.8 mM Cl−) and well G (2,269 mM Cl−) (bold type) compared with closely related sequences and reference strains. The numbers of clones of the same phylotype are indicated in parentheses. The scale bar represents 5% estimated sequence divergence. Bootstrap values (n = 1,000) are indicated at the nodes.
Analysis of well G revealed eight archaeal phylotypes; the estimated phylotype diversity is 11 based on SChao1 (72% coverage) and 18 based on SACE (44% coverage). The great majority of the sequences (109 clones) clustered within the Methanosarcinales (Fig. 5) and were closely related to Methanohalophilus portucalensis, a halophilic methanogen commonly isolated from hypersaline environments which uses methanol and methylamines for growth and methanogenesis (4). Within these sequences, a group of 106 highly similar clones clustered with uncultured Methanosarcinales from other high-salinity sites, such as a hypersaline mat (49), soil surrounding a salt spring (54), and a deep-sea hypersaline anoxic basin in the Mediterranean Sea (58). The second dominant phylotype in the clone library (37 clones) from well G occurred within the Methanomicrobiales and was related to Methanoplanus petrolearius, with up to 97% sequence similarity (Fig. 5). M. petrolearius was isolated from an oil field offshore of Africa and in culture has been shown to tolerate salinities only up to 855 mM Cl− (39). Similar sequences were also found in hypersaline water collected from two oil wells offshore of Monterey, CA (45).
Two distinct groups of well G clones had no cultured relatives. Uncultured Archaea group I consisted of two sequences that were 96% similar to each other but had less than 77% identity with other sequences in the current NCBI database (Fig. 5). The closest related sequences belonged to a clone from a deep-sea hydrothermal vent eukaryotic group sequenced from a subseafloor core (up to 73% similarity) (18) and an environmental sequence from a gas-rich mud volcano in the eastern Mediterranean (up to 76% similarity). Uncultured Archaea group II consisted of a single phylotype of six clones (Fig. 5), whose closest relatives were sequenced from other methane-rich environments, including two different eastern Mediterranean mud volcanoes (up to 91 and 92% sequence similarity) (16), a microbial mat at a hydrothermal vent in the Guaymas Basin (92% similarity) (9), and a sulfidic methane spring in Oklahoma (83% similarity) (10).
Rarefaction curves of the two libraries indicated that clones obtained from well B had been adequately sampled to cover the existing diversity, while well G requires greater sampling to exhaustively describe the community (data not shown). This agrees with the calculated estimates of community richness SChao1 and SACE. The coverage of sampling was estimated as the percentage of identified OTUs relative to the estimated total richness. Due to the lack of diversity in well B, an SACE value for diversity could not be calculated.
DISCUSSION
This study investigated archaeal diversity across a geochemical gradient in the Antrim Shale. The total cell counts, nucleic acid yields, and results of MPN dilutions suggested that microbial populations across the Antrim Shale salinity gradient are small but similar to those found in other subsurface, hydrocarbon-rich sedimentary environments (11, 45). The archaeal diversity found in the pore waters was also low, with eight phylotypes identified in well G and only one phylotype identified in well B. This is consistent with phylogenetic analyses of archaeal communities from other hydrocarbon-rich environments (13, 45) and from other wells that have been sampled in the Antrim Shale (32; Formolo et al., submitted). The archaeal phylotypes represented in the clone libraries were confined to the Euryarcheaota and almost exclusively to the Methanosarcinales and Methanomicrobiales. Within these taxa, however, cultured relatives are known to exhibit functional diversity, with forms capable of utilizing H2/CO2, formate, and alcohols among the Methanomicrobiales, and use of acetate, H2/CO2, alcohols, and methylated amines is known among the Methanosarcinales. Additionally, the nearest relatives of two uncultured archaeal groups are organisms found in sulfidic, methane-rich marine environments, suggesting that there is other functional diversity among these archaea. The lack of external inorganic electron acceptors other than CO2 is uncommon compared with the situation in marine sediments and other sedimentary basins, given that most subsurface hydrocarbon-rich environments exhibit indirect geochemical evidence for common terminal electron-accepting processes, including abundant SO42−/H2S or mineral Fe oxyhydroxides (2, 7, 13, 21, 23, 24, 25, 27, 46, 50). This lack may contribute to the low archaeal diversity in the Antrim Shale compared with the diversity in other subsurface, methanogenic environments (13, 45). The community may be limited by the rate at which substrates for methanogenesis can be generated from shale organic matter solely by fermentation and hydrolysis. The Antrim Shale is a very stable environment and has been hydrologically isolated from surface waters for at least 7,000 years (30, 34), supporting the potential for well-adapted species to dominate. At this point, the metabolic roles which provide the necessary substrates for Antrim Shale methanogens can only be speculated on, although fermentation and/or anaerobic respiration using organic electron acceptors can be considered.
The absence of 16S rRNA gene sequences related to acetoclastic methanogens in well B was surprising since the low salinity of the pore waters coupled with an environment conducive to fermentation should theoretically support acetoclastic growth. Sequences closely related to known acetoclastic methanogens have been recovered from both low-temperature (13) and high-temperature (45) petroleum reservoirs, as well as in subsurface sedimentary environments (56). 16S rRNA gene sequences that are closely related to Methanosaeta concilii were obtained from low-salinity Antrim Shale wells in a previous study (32; Formolo et al., submitted). Due to the low levels of sulfate in well B (∼170 μM), it is possible that sulfate-reducing bacteria are present and outcompete the methanogens for acetate in the pore waters. Acetate is more likely to be a competitive methanogenic substrate at this salinity level, and unlike the situation at high-salinity sites, the noncompetitive substrates methanol, methylated amines, and dimethyl sulfide might not sufficiently support species with a methylotrophic affinity (41). The presence of sulfate in low-salinity Antrim Shale pore waters is a recent occurrence related to industrial-scale dewatering of the Antrim Shale and introduction of water from an underlying sulfate-bearing formation, as sulfate was not detected in this or other high-alkalinity Antrim Shale wells during the previous decade of monitoring of formation water chemistry (30, 32, 34).
The cluster of 16S rRNA gene sequences related to M. petrolearius from well G was also surprising, since the chloride concentration of this well (2,268 mM Cl−) is above the known limit for hydrogenotrophic methanogenesis in culture (2,050 mM Cl−) (40). Although the presence of a 16S rRNA gene sequence does not imply the level of activity or ecological niche, if this phylotype is currently active and utilizing H2 and CO2 to make methane, then this would be the highest known salinity at which hydrogenotrophic methanogenesis can occur. Cultivation and growth range experiments are needed to determine the salinity tolerance and methanogenic substrate use for this organism.
Many factors point to hydrogenotrophic methanogenesis as the main mode of producing methane in the Antrim Shale. Large groups of sequences related to hydrogen-utilizing methanogens were found in clone libraries from both saline and freshwater pore waters. Although enhancement of methanogenesis by a given compound in an enrichment does not mean that the compound is important in this ecosystem, the largest number of methanogens could be cultured with H2 as the electron donor. The δ13C ratios for methane and carbon dioxide and the δD ratios for methane and water also indicate that methanogenesis using H2 is the dominant mode occurring in the basin (31, 32). However, the use of acetate and methyl compounds by methanogens should not be overlooked. In the absence of terminal electron acceptors such as sulfate or ferric iron, fermentation or respiration that employs components of shale organic matter as electron acceptors is likely to be an important mechanism of organic matter degradation in the Antrim Shale; both of these mechanisms may yield acetate. Acetate concentrations were below the detection levels in pore waters with chloride levels below 1,000 mM Cl−, indicating that acetate is consumed in all but the most saline wells. Acetate would build up in these waters unless acetoclastic methanogens (or possibly sulfate reducers in well B) were consuming it. Sequences closely related to methylotrophic methanogens have been identified in Antrim Shale waters; such organisms are likely to be active in pore waters where H2 use by methanogens is inhibited by high salinity or where methyl compounds are generated.
The presence of close relatives of methylotrophic methanogens in the more saline well G suggests a source of the methylamines, methylsulfides, or primary alcohols in this environment. Environmental or geochemical sources of methylamines are not well described, but one possible source is betaine fermentation. In hypersaline environments, betaine is a common osmoregulatory compound synthesized at high concentrations by certain halophilic bacteria (42). Betaine can be fermented to yield acetate and trimethylamine by a variety of fermentative bacteria, including halophilic acetogens and other halophiles (19, 37, 53). Specifically, it has been demonstrated that a methanogen fermenter coculture of Methanohalophilus sp. strain Z-7302 and Acetohalobium arabaticum grown on betaine produces acetate and methane as its only products, presenting a possible trophic link for the hypersaline subsurface pore waters (60).
Regardless of substrate choice or salinity tolerance, the archaeal communities in the Antrim Shale all have the same overall role of generating methane. Yet in detail, each community has adapted to, and exhibits optimum methane generation within, a narrow range of salinities, and the substrates employed in methanogenesis differ in these communities. This observation is critical in guiding exploration of other subsurface environments where methanogenesis may occur. While methanogenesis may develop in environments ranging from freshwater to environments containing more than 3 M Cl−, the substrates that foster methanogenesis at lower salinity are not the same as the substrates used at a higher salinity. The anaerobic degradation of sedimentary organic matter, which may yield acetate, H2, methanol, and/or methylated amines, is a critical component tightly linked with pore water salinity in the subsurface generation of methane in sedimentary basins.
Acknowledgments
This work was supported by NSF Biogeosciences Program grants EAR-0433766 (to S.T.P. and K.N.) and EAR-0433801 (to A.M.M.) from the National Science Foundation and by RPSEA grant R-520 to A.M.M., S.T.P., and K.N.
We acknowledge EnerVest Operating L.L.C. for kindly providing access to wells and especially Jeff Riling for his support in water sampling. We also thank Roger Huang and Matt Stevenson for excellent technical assistance, as well as Jim Holden, Javier Izquierdo, Vicente Gomez-Alvarez, and Lisa Stout for helpful discussions.
Footnotes
Published ahead of print on 27 April 2007.
REFERENCES
- 1.Ábalos, M., J. M. Bayona, and J. Pawliszyn. 2000. Development of a headspace solid-phase microextraction procedure for the determination of free volatile fatty acids in waste waters. J. Chromatogr. 873:107-115. [DOI] [PubMed] [Google Scholar]
- 2.Aitken, C. M., D. M. Jones, and S. R. Larter. 2004. Anaerobic hydrocarbon biodegradation in deep subsurface oil reservoirs. Nature 431:291-294. [DOI] [PubMed] [Google Scholar]
- 3.Ashelford, K. E., N. A. Chuzhanova, J. C. Fry, A. J. Jones, and A. J. Weightman. 2006. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl. Environ. Microbiol. 72:5734-5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boone, D. R., I. M. Mathrani, Y. Liu, J. A. G. F. Menaia, R. A. Mah, and J. E. Boone. 1993. Isolation and characterization of Methanohalophilus portucalensis sp. nov. and DNA reassociation study of the genus Methanohalophilus. Int. J. Syst. Bacteriol. 43:430-437. [Google Scholar]
- 5.Borzenkov, I. A., S. S. Belyaev, Y. M. Miller, I. A. Davydova, and M. V. Ivanov. 1997. Methanogenesis in the highly mineralized stratal waters of the Bondyuzhskoe Oil Field. Mikrobiologiya 66:104-110. [Google Scholar]
- 6.Chapelle, F. H., K. O'Neill, P. M. Bradley, B. A. Methe, S. A. Ciufo, L. L. Knobel, and D. R. Lovley. 2002. A hydrogen-based subsurface microbial community dominated by methanogens. Nature 415:312-315. [DOI] [PubMed] [Google Scholar]
- 7.Coolen, M. J. L., H. Cypionka, A. M. Sass, H. Sass, and J. Overmann. 2002. Ongoing modification of Mediterranean Pleistocene sapropels mediated by prokaryotes. Science 296:2407-2410. [DOI] [PubMed] [Google Scholar]
- 8.DeLong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dhillon, A., M. Lever, K. G. Lloyd, D. B. Albert, M. L. Sogin, and A. Teske. 2005. Methanogen diversity evidenced by molecular characterization of methyl coenzyme M reductase A (mcrA) genes in hydrothermal sediments of the Guaymas Basin. Appl. Environ. Microbiol. 71:4592-4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elshahed, M. S., F. Z. Najar, B. A. Roe, A. Oren, T. A. Dewers, and L. R. Krumholz. 2004. Survey of archaeal diversity reveals an abundance of halophilic Archaea in a low-salt, sulfide- and sulfur-rich spring. Appl. Environ. Microbiol. 70:2230-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fredrickson, J. K., and T. C. Onstott. 2001. Biogeochemical and geological significance of subsurface microbiology, p. 3-38. In J. K. Fredrickson and M. Fletcher (ed.), Subsurface microbiology and biogeochemistry. Wiley-Liss, Inc., New York, NY.
- 12.Gieskes, J. M., and W. C. Rogers. 1973. Alkalinity determination in interstitial waters of marine sediments. J. Sediment. Petrol. 43:272-277. [Google Scholar]
- 13.Grabowski, A., O. Nercessian, F. Fayolle, D. Blanchet, and C. Jeanthon. 2005. Microbial diversity in production waters of a low-temperature biodegraded oil reservoir. FEMS Microbiol. Ecol. 54:427-443. [DOI] [PubMed] [Google Scholar]
- 14.Großkopf, R., P. H. Janssen, and W. Liesack. 1998. Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl. Environ. Microbiol. 64:960-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 16.Heijs, S. K., J. S. Sinninghe-Damste, and L. J. Forney. 2005. Characterization of a deep-sea microbial mat from an active cold seep at the Milano mud volcano in the Eastern Mediterranean Sea. FEMS Microbiol. Ecol. 54:47-56. [DOI] [PubMed] [Google Scholar]
- 17.Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon; a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319. [DOI] [PubMed] [Google Scholar]
- 18.Inagaki, R., K. Takai, T., Komatsu, K. Fujioka, and K. Horikoshi. 2001. Archaeology of Archaea: geomicrobiological record of Pleistocene thermal events concealed in a deep-sea subseafloor environment. Extremophiles 5:385-392. [DOI] [PubMed] [Google Scholar]
- 19.Kevbrin, V. V., T. N. Zhilina, and G. A. Zavarzin. 1995. Physiology of the halophilic homoacetic bacterium Acetohalobium arabaticum. Microbiology 64:134-138. [Google Scholar]
- 20.Kimura, H., M. Sugihara, H. Yamamoto, B. K. C. Patel, K. Kato, and S. Hanada. 2005. Microbial community in a geothermal aquifer associated with the subsurface of the Great Artesian Basin, Australia. Extremophiles 9:407-414. [DOI] [PubMed] [Google Scholar]
- 21.Kleikemper, J., S. A. Pombo, M. H. Schroth, W. V. Sigler, M. Pesaro, and J. Zeyer. 2005. Activity and diversity of methanogens in a petroleum hydrocarbon-contaminated aquifer. Appl. Environ. Microbiol. 71:149-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kotelnikova, S. 2002. Microbial production and oxidation of methane in deep subsurface. Earth Sci. Rev. 58:367-395. [Google Scholar]
- 23.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinformatics 5:150-163. [DOI] [PubMed] [Google Scholar]
- 24.Larter, S., A. Wilhelins, I. Head, M. Koopmans, A. Aplin, R. Di Primio, C. Zwach, M. Erdmann, and N. Telnaes. 2003. The controls on the composition of biodegraded oils in the deep subsurface. Part 1. Biodegradation rates in petroleum reservoirs. Org. Geochem. 34:601-613. [Google Scholar]
- 25.Larter, S., and R. di Primio. 2005. Effects of biodegradation on oil and gas field PVT properties and the origin of oil rimmed gas accumulations. Org. Geochem. 36:299-310. [Google Scholar]
- 26.Liu, Y., and D. R. Boone. 1991. Effects of salinity on methanogenic decomposition. Bioresour. Technol. 35:271-273. [Google Scholar]
- 27.Lovley, D. R., and F. H. Chapelle. 1995. Deep subsurface microbial processes. Rev. Geophys. 33:365-381. [Google Scholar]
- 28.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, P. R. Saxman, R. J. Farris, G. M. Garrity, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martini, A. M., J. M. Budai, L. M. Walter, and M. Schoell. 1996. Microbial generation of economic accumulations of methane within a shallow organic-rich shale. Nature 383:155-158. [Google Scholar]
- 30.Martini, A. M., L. M. Walter, J. M. Budai, T. C. W. Ku, C. J. Kaiser, and M. Schoell. 1998. Genetic and temporal relations between formation waters and biogenic methane: Upper Devonian Antrim Shale, Michigan Basin, USA. Geochim. Cosmochim. Acta 62:1699-1720. [Google Scholar]
- 31.Martini, A. M., L. M. Walter, T. C. W. Ku, J. M. Budai, J. C. McIntosh, and M. Schoell. 2003. Microbial production and modification of gases in sedimentary basins: a geochemical case study from a Devonian shale gas play, Michigan basin. AAPG Bull. 87:1355-1375. [Google Scholar]
- 32.Martini, A. M., K. Nüsslein, and S. T. Petsch. 2005. Enhancing microbial gas from unconventional reservoirs: geochemical and microbiological characterization of methane-rich fractured black shales. GTI-RPSEA GRI-05/0023. Gas Technology Institute, Des Plaines, IL.
- 33.McCune, B., and M. J. Mefford. 1999. PC-ORD. Multivariate analysis of ecological data, version 4. MjM Software Design, Gleneden Beach OR.
- 34.McIntosh, J. C., L. M. Walter, and A. M. Martini. 2002. Pleistocene recharge to midcontinent basins: effects on salinity structure and microbial gas generation. Geochim. Cosmochim. Acta 66:1681-1700. [Google Scholar]
- 35.McIntosh, J. C., L. M. Walter, and A. M. Martini. 2004. Extensive microbial modification of formation water geochemistry: case study from a midcontinent sedimentary basin, United States. Geol. Soc. Am. Bull. 116:743-759. [Google Scholar]
- 36.Mishra, S. R., P. Pattnaik, N. Sethunathan, and T. K. Adhya. 2003. Anion-mediated salinity affection methane production in a flooded alluvial soil. Geomicrobiol. J. 20:579-586. [Google Scholar]
- 37.Moune, S., N. Manac'h, A. Hirschler, P. Caumette, J. C. Willison, and R. Matheron. 1999. Haloanaerobacter salinarius sp. nov., a novel halophilic fermentative bacterium that reduces glycine-betaine to trimethylamine with hydrogen or serine as electron donors; emendation of the genus Haloanaerobacter. Int. J. Syst. Bacteriol. 49:103-112. [DOI] [PubMed] [Google Scholar]
- 38.Nealson, K., and W. A. Ghiorse. 2001. Geobiology: exploring the interface between the biosphere and the geosphere. American Academy of Microbiology, Washington, DC. [PubMed]
- 39.Ollivier, B., J. L. Cayol, B. K. C. Patel, M. Magot, M. L. Fardeau, and J. L. Garcia. 1997. Methanoplanus petrolearius sp nov, a novel methanogenic bacterium from an oil-producing well. FEMS Microbiol. Lett. 147:51-56. [DOI] [PubMed] [Google Scholar]
- 40.Ollivier, B., M. L. Fardeau, J. L. Cayol, M. Magot, B. K. C. Patel, G. Prensier, and J. L. Garcia. 1998. Methanocalculus halotolerans gen. nov., sp. nov., isolated from an oil-producing well. Int. J. Syst. Bacteriol. 48:821-828. [DOI] [PubMed] [Google Scholar]
- 41.Oremland, R. S., and S. Polcin. 1982. Methanogenesis and sulfate reduction: competitive and noncompetitive substrates in estuarine sediments. Appl. Environ. Microbiol. 44:1270-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oren, A. 1990. Formation and breakdown of glycine betaine and trimethylamine in hypersaline environments. Antonie Leeuwenhoek 58:291-298. [DOI] [PubMed] [Google Scholar]
- 43.Oren, A. 1999. Bioenergetic apects of halophilism. Microbiol. Mol. Biol. Rev. 63:334-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oren, A. 2001. The bioenergetic basis for the decrease in metabolic diversity at increasing salt concentrations: implications for the functioning of salt lake ecosystems. Hydrobiologia 466:61-72. [Google Scholar]
- 45.Orphan, V. J., L. T. Taylor, D. Hafenbradl, and E. F. Delong. 2000. Culture-dependent and culture-independent characterization of microbial assemblages associated with high-temperature petroleum reservoirs. Appl. Environ. Microbiol. 66:700-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parkes, R. J., G. Webster, B. A. Cragg, W. J. Weightman, C. J. Newberry, T. G. Ferdelman, J. Kallmeyer, B. B. Jørgensen, I. W. Aiello, and J. C. Fry. 2005. Deep sub-seafloor prokaryotes stimulated at interfaces over geologic time. Nature 436:390-394. [DOI] [PubMed] [Google Scholar]
- 47.Patel, G. B., and L. A. Roth. 1977. Effect of sodium-chloride on growth and methane production of methanogens. Can. J. Microbiol. 23:893-897. [DOI] [PubMed] [Google Scholar]
- 48.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sorensen, K. B., D. E. Canfield, A. P. Teske, and A. Oren. 2005. Community composition of a hypersaline endoevaporitic microbial mat. Appl. Environ. Microbiol. 71:7352-7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Süß, J., B. Engelen, H. Cypionka, and H. Sass. 2004. Quantitative analysis of bacterial communities from Mediterranean sapropels based on cultivation-dependent methods. FEMS Microbiol. Ecol. 51:109-121. [DOI] [PubMed] [Google Scholar]
- 51.Teske, A. P. 2005. The deep subsurface biosphere is alive and well. Trends Microbiol. 13:402-404. [DOI] [PubMed] [Google Scholar]
- 52.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsai, C. R., J. L. Garcia, B. K. C. Patel, J. L. Cayol, L. Baresi, and R. A. Mah. 1995. Haloanaerobium alcaliphilum sp. nov., an anaerobic moderate halophile from the sediments of Great Salt Lake, Utah. Int. J. Syst. Evol. Microbiol. 45:301-307. [DOI] [PubMed] [Google Scholar]
- 54.Walsh, D. A., R. T. Papke, and W. F. Doolittle. 2005. Archaeal diversity along a soil salinity gradient prone to disturbance. Environ. Microbiol. 7:1655-1666. [DOI] [PubMed] [Google Scholar]
- 55.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. A. Lane. 1991. 16S ribosomal RNA for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wellsbury, P., I. Mather, and R. J. Parkes. 2002. Geomicrobiology of deep, low organic carbon sediments in the Woodlark Basin, Pacific Ocean. FEMS Microbiol. Ecol. 42:59-70. [DOI] [PubMed] [Google Scholar]
- 57.Whitman, W. B., D. C. Coleman, and W. J. Wiebe. 1998. Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. USA 95:6578-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yakimov, M. M., L. Giuliano, S. Cappello, R. Denaro, and P. Golyshin. 2007. Microbial community of a hydrothermal mud vent underneath the deep-sea anoxic brine lake Urania (Eastern Mediterranean). Orig. Life Evol. Biosph. 37:177-188. [DOI] [PubMed] [Google Scholar]
- 59.Zellner, G., E. Stackbrandt, P. Messner, B. J. Tindall, E. Conway de Macario, H. Kniefel, U. B. Sleytr, and J. Winter. 1989. Methanocorpuscululaceae fam. nov., represented by Methanocorpusculum parvum, Methanocorpusculum sinense spec. nov. and Methanocorpusculum bavaricum spec. nov. Arch. Microbiol. 151:381-390. [DOI] [PubMed] [Google Scholar]
- 60.Zhilina, T. N., and G. A. Zavarzin. 1990. Extremely halophilic, methylotrophic, anaerobic bacteria. FEMS Microbiol. Rev. 87:315-322. [Google Scholar]